Abstract
Adaptation to fluctuations in nutrient availability is a fact of life for single-celled organisms in the ‘wild’. A decade ago our understanding of how bacteria adjust cell cycle parameters to accommodate changes in nutrient availability stemmed almost entirely from elegant physiological studies completed in the 1960s. In this Opinion article we summarize recent groundbreaking work in this area and discuss potential mechanisms by which nutrient availability and metabolic status are coordinated with cell growth, chromosome replication and cell division.
The life of a bacterial cell is feast or famine. To survive the bacterium must rapidly adapt to changing environmental conditions. Colonization of the mammalian gut provides an enteric organism with an abundant source of carbohydrates, whereas a flash flood instantly depletes the nutrient supply for a soil bacterium. Nutrient-rich conditions lead to a decrease in mass doubling time and an increase in cell size, whereas nutrient-poor conditions curtail growth and reduce cell size1,2. Changes in growth rate must be accompanied by changes in the cell cycle to ensure that cell division stays coordinated with mass doubling, chromosome replication and chromosome segregation.
How organisms adjust their cell cycle dynamics to compensate for changes in nutritional conditions is an important outstanding question in bacterial physiology. Recent work, reviewed here, suggests that multiple signalling pathways transmit nutritional and growth rate information directly to the cell cycle machinery. Multiple signalling pathways permit cells to constantly sample their environments and fine-tune cell cycle processes, a substantial advantage under challenging conditions.
The bacterial cell cycle
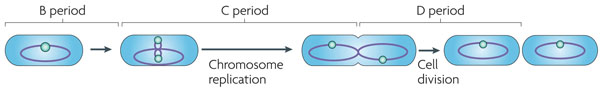
The bacterial cell cycle is traditionally divided into three stages: the period between division (cell ‘birth’) and the initiation of chromosome replication (known as the B period); the period required for replication (known as the C period); and the time between the end of replication and completion of division (known as the D period) (FIG. 1). In the enteric organism Escherichia coli and the spore former Bacillus subtilis, DNA replication begins at a single origin (oriC) on a single circular chromosome. Replication proceeds bidirectionally around the circumference of the chromosome, terminating at a region opposite oriC. During replication the chromosome remains in a condensed, highly ordered structure that is known as the nucleoid (see REF. 3 for a review of chromosome replication). Division is initiated near the end of chromosome segregation by the formation of a cytokinetic ring at the nascent division site. The tubulin-like protein FtsZ serves as the foundation for assembly of this ring and is required for recruitment of the division machinery (see REF. 4 for a review of bacterial cell division). Nutrient availability and growth rate could potentially affect any of the above steps.
Figure 1. The bacterial cell cycle.
The bacterial cell cycle is traditionally divided into three stages: the period between division (birth) and the initiation of chromosome replication (the B period); the period required for chromosome replication (the C period); and the time between the completion of chromosome replication and the completion of cell division (the D period). The bacterial cells (in this case, Escherichia coli) are outlined in black and contain highly schematic chromosomes (purple ovals) with oriC regions shown as green circles.
Our understanding of the bacterial cell cycle under different growth conditions derives largely from early physiological studies of B. subtilis and E. coli5. These studies indicated that, at constant temperature, mass doubling time decreases in response to increases in nutrient availability; however, both the C period and the D period remain essentially constant. Consequently, under nutrient-rich conditions, both E. coli and B. subtilis reach growth rates at which the period required for chromosome replication and segregation is greater than the mass doubling time. To resolve this paradox, rapidly growing cells initiate new rounds of chromosome replication before completing the previous round, a situation that results in two, four or even eight rounds of replication proceeding simultaneously. This phenomenon, which was first discovered in B. subtilis and termed ‘multifork replication’ (REF. 6), was formalized and further investigated by Cooper and Helmstetter in their influential 1968 paper5 (BOX 1). Notably, Cooper and Helmstetter’s work illuminated how cells balance largely constant rates of replication fork progression with nutrient-dependent changes in mass doubling time, by initiating replication and dividing more frequently when growing faster.
Box 1. Cooper and Helmstetter’s model.
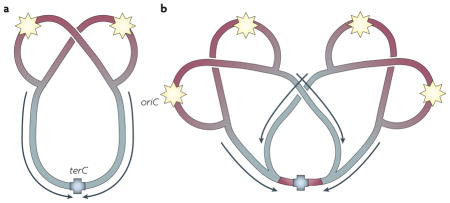
Replication during slow growth
In slow-growing bacterial cells (with a mass doubling time >C + D period), there is a single round of replication per division cycle. This type of growth resembles that of eukaryotes in that there is a gap (the B period), a period in which DNA replication takes place (the C period) and finally a period of chromosome segregation and cell division (the D period). During replication each cell has only two copies of the origin region (oriC) and one copy of the terminus (terC) (see the figure, part a).
Replication during fast growth (multifork replication)
In rapidly growing cells (with a mass doubling time ≤C + D period), each chromosome re-initiates a new round of replication before the first round has terminated, although only one round is initiated per cell division. Multifork replication ensures that at least one round of replication is finished before cytokinesis, to guarantee that each daughter cell receives at least one complete genome. During multifork replication cells can have four or more copies of the region proximal to oriC and one copy of the region proximal to terC (see the figure, part b). This imbalance has implications for gene expression levels as well as for the activity of the initiator protein DnaA.
Although arguably one of the most important insights in the field in the past 40 years, Cooper and Helmstetter’s model is limited in that it views the cell cycle as a single process, in which replication initiation is the triggering event that determines the timing of all subsequent steps in replication and cell division. This view does not take into consideration the effects of nutrients and metabolic status on events that occur after replication initiation, nor does it explain how cell cycle events are coordinated with mass doubling to ensure that new rounds of replication are initiated only once per division cycle and cell size homeostasis is maintained. Recent work suggests that, instead of being a single process, the bacterial cell cycle is a set of coordinated but independent events (see REFS 7,8 for an eloquent presentation of this model). This more nuanced view is the model to which we subscribe.
Multifork replication is not a universal feature of the bacterial life cycle: the aquatic bacterium Caulobacter crescentus has temporally compartmentalized cell cycle stages, a situation analogous to the eukaryotic cell cycle9. For simplicity, however, in this Review we treat B. subtilis and E. coli as representative Gram-positive and Gram-negative bacteria, respectively. In addition, we use the term ‘division cycle’ instead of cell cycle to refer to the period of time between the birth of a cell and its own subsequent division. This Review focuses first on chromosome replication and then on cell division, but these should not be regarded as independent processes.
Chromosome replication
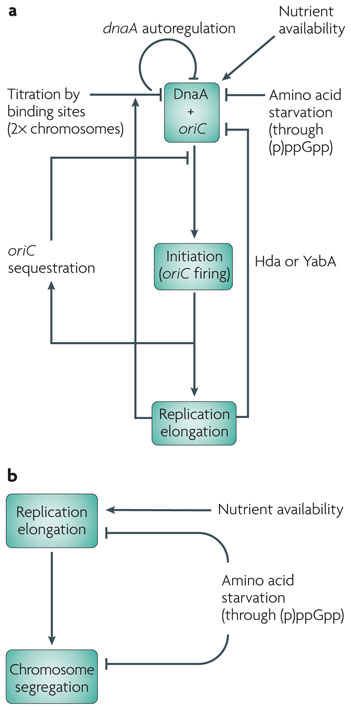
Chromosome replication is coordinated with cell growth to ensure that: at each origin, replication initiates once and only once per division cycle; at least one round of replication is completed and the nucleoids have segregated before the completion of cell division; and there are sufficient nutrients to support these processes. Not surprisingly, both the initiation of replication and the replication process itself (referred to here as elongation) are subject to metabolic controls (FIG. 2).
Figure 2. Nutrient availability, DNA replication and chromosome segregation.
Green boxes represent the relevant events in the cell cycle. a| Nutrient availability is a key determinant for replication initiation. The production of the initiation protein DnaA and other essential components of the replication machinery is proportional to carbon availability and growth rate. Amino acid starvation directly inhibits replication initiation through the production of guanosine tetraphosphate and guanosine pentaphosphate (collectively known as (p)ppGpp). Growth rate might also affect initiation indirectly, potentially through the additional feedback mechanisms that transmit the cell cycle and replication status to the replication machinery, to prevent overinitiation. For example, DnaA represses the transcription of the dnaA operon. In addition, DnaA binding to loci other than the origin of replication (oriC), such as datA in Escherichia coli, titrates DnaA and thereby inhibits replication initiation; replication doubles the copy number of datA, further titrating DnaA. In E. coli, newly replicated oriC contains hemimethylated GATC sites, which are sequestered to prevent reinitiation. Finally, replication elongation inhibits re-initiation through Hda in E. coli or YabA in Bacillus subtilis. The beta clamp, encoded by dnaN, is essential for mediating this control. b| Nutrient availability and amino acid starvation also affect replication elongation and chromosome segregation.
DnaA and origin firing: the entry point for metabolic control
The initiation of chromosome replication is the entry point for metabolic control of the bacterial division cycle10. Replication is coupled to growth rate primarily through nutrient-dependent changes in the synthesis and activity of the protein DnaA (FIG. 2a). A member of the AAA+ family of ATPases, DnaA binds to specific sites at the origin of replication and causes the AT-rich DNA in this region to unwind, which permits the replisome to load11,12. DnaA is rate limiting for the initiation of replication. Artificially increasing DnaA synthesis leads to overinitiation13–15, whereas blocking DnaA synthesis inhibits initiation16. In addition to its role in replication initiation, DnaA is a transcription factor that regulates its own expression and that of several other genes17,18.
Nutrient availability has a dramatic effect on DnaA accumulation and, hence, the initiation of chromosome replication. The intracellular concentration of DnaA increases with the growth rate to ensure that origin firing is triggered more frequently in rapidly growing cells19. Conversely, starving cells of amino acids abolishes de novo DnaA synthesis and prevents new rounds of chromosome replication20.
It has been proposed that DnaA expression in E. coli is regulated by nutrient availability through the small nucleotides guanosine tetraphosphate and guanosine pentaphosphate (collectively referred to as (p)ppGpp)10. (p)ppGpp synthesis is induced by amino acid starvation, through the enzyme RelA, or by carbon starvation, through the enzyme SpoT21. (p)ppGpp directly affects the transcription of many genes by altering the stability of the transcription complex21,22. (p)ppGpp also inhibits transcription of dnaA, although it remains to be elucidated whether this effect is direct or indirect23. Induction of (p)ppGpp, either by starvation or by overexpressing a constitutive form of RelA, inhibits replication initiation24–26. Together, these data support a model in which (p)ppGpp is responsible for the nutrient-dependent control of DnaA expression and replication initiation (FIG. 2a). In C. crescentus, carbon starvation inhibits replication initiation through SpoT by a different mechanism to that in E. coli: the proteolysis of DnaA27,28
In addition to (p)ppGpp, several other mechanisms modulate DnaA synthesis and activity to ensure that the initiation of chromosome replication occurs only once per division cycle (FIG. 2a). These mechanisms include: autoregulation of dnaA transcription29,30; sequestration of newly replicated, hemimethylated origin DNA away from DnaA by the protein SeqA31; titration of free DnaA by DnaA-binding sites outside oriC32,33; and regulation of DnaA activity by proteins such as DNA initiator-associating protein (DiaA) in E. coli34 and Soj in B. subtilis35. It remains to be discovered whether any of these control mechanisms are subject to nutrient-dependent regulation.
Once DnaA has triggered initiation, elongation-dependent mechanisms inhibit DnaA activity, further preventing over-initiation (FIG. 2a). In E. coli, re-initiation is prevented in part by the interaction between DnaA, DnaN (the beta clamp that ensures DNA polymerase processivity) and Hda, a protein that converts initiation-active DnaA–ATP to the less active DnaA–ADP in an elongation-dependent manner36,37. In slow-growing B. subtilis cells, YabA (a protein unrelated to Hda) also inhibits reinitiation, apparently by tethering DnaA to DnaN38–40. YabA delays replication initiation under nutrient-poor conditions but not under nutrient-rich conditions, suggesting that it might be involved in the metabolic control of initiation41,42.
Cell growth and the initiation of chromosome replication
The achievement of a specific size or mass has been widely accepted as the primary mechanism that links cell growth to the initiation of chromosome replication. According to this model, a factor that is required for the initiation of replication, generally assumed to be DnaA, accumulates in a growth-rate-dependent manner, reaching threshold levels when cells attain a specific size (termed the ‘initiation mass’) to trigger replication. This concept stems from a 1968 paper by Donachie43, in which he plotted the E. coli replication results of Cooper and Helmstetter5 against the Salmonella enterica subsp. enterica serovar Typhimurium cell size data from Schaechter et al.2 and observed that cell mass per origin at initiation was constant regardless of growth rate. At growth rates between 1 and 2 mass doublings per hour, cells initiated replication at the same mass. At faster growth rates, when cells had on average twice the number of origins as their slow-growing counterparts, the mass at initiation was exactly twice that of the slow-growing cells43.
The idea that achievement of a specific mass is the signal that triggers the initiation of DNA replication is appealing. First, it provides a simple explanation for the link between cell growth and DNA replication. Second, a constant initiation mass means that short cells must delay replication until they reach the appropriate size, whereas longer cells would initiate DNA replication earlier. Therefore, coupling initiation to cell size provides a straightforward mechanism for maintaining cell size homeostasis in a given population.
Despite its appeal, more recent data suggest that mechanisms controlling the initiation of DNA replication are more complex than originally proposed. For example, the timing of replication initiation is normal in B. subtilis cells that are 35% shorter than wild type, indicating that replication can be uncoupled from cell mass in this organism44. Moreover, initiation mass increases 1.6- to 2-fold in slow-growing E. coli K-12 cells compared with the initiation mass in cells growing at a normal rate45–47.
Another appealing hypothesis is that division, rather than achievement of a specific mass, serves as a checkpoint for origin firing48. However, replication is not notably perturbed when cell division is blocked49. Therefore, it is unlikely that division has a major impact on the initiation of replication.
Notably, with the exception of the B. subtilis short-cell experiments, the studies described above were all carried out in wild-type strains. Although this approach allows for correlative analysis, it does not directly test any of the hypotheses about the factors governing the initiation of chromosome replication. Moreover, a wealth of data suggests that the factors triggering initiation differ depending on growth rate45,47,48. For these reasons, we favour an integrated model in which multiple factors control origin firing. each factor would be more or less important depending on the growth rate — for example, factor ‘X’ could be important at rapid growth rates that support multifork replication, whereas factor ‘Y’ could be active only under slow-growth conditions. This type of model not only accounts for growth-rate-dependent differences in initiation mass but also provides the flexibility that is necessary to accommodate rapid changes in nutrient availability.
Replication elongation and chromosome segregation
Although metabolism exerts its best known effect on the initiation of chromosome replication, downstream steps in replication are also affected by changes in nutrient status (FIG. 2b). The E. coli C period was originally approximated to be constant under varying growth conditions5, but this finding turns out to apply only to cells cultured under nutrient-rich conditions (with a mass doubling time of less than 1 hour). when mass doubling times are greater than 1 hour, the C period can increase more than twofold46,50. It is not apparent why replication rates are sensitive to growth rates in E. coli, but it is clear that nutrient availability has a substantial impact on the elongation of replication.
Elongation is a costly process that requires large amounts of cellular resources; in particular, deoxynucleoside 5′-triphosphates (dNTPs) are in high demand during replication. The dNTP pool needs to be constantly replenished and is hence highly susceptible to nutrient availability. Data from thymine-requiring bacteria indicate that varying the exogenous thymine levels dramatically affects the replication elongation rate51. Not surprisingly, the ribonucleotide reductase that synthesizes dNTPs is regulated, by factors including DnaA, to keep up with the demand of replication52.
In B. subtilis replication elongation is directly controlled by nutrient availability. In response to amino acid starvation, induction of (p)ppGpp leads to a global, non-disruptive replication arrest, most likely through inhibition of primase activity53. Inhibition of replication by (p)ppGpp might serve as a surveillance mechanism to keep DNA synthesis in ‘suspended animation’ when resources such as dNTP are depleted. In this way genome integrity is maintained until growth conditions improve and the inhibition is relieved.
Genetic data from B. subtilis also point to a link between sugar metabolism and replication elongation. Mutations in enzymes that are required for the terminal, three-carbon steps of glycolysis suppress conditional mutations in genes that encode the lagging strand DNA polymerase, the helicase and the primase54. The biochemical nature of this link is not known, but it has been proposed that metabolic signals might directly regulate the activity of the replication proteins. Collectively, these results suggest that bacteria have evolved several means of coordinating DNA replication elongation with metabolism, an important adaptation to life in an uncertain environment.
At the end of elongation, newly replicated chromosomal termini need to be segregated, a step that was recently found to be subject to metabolic control. In E. coli, increases in (p)ppGpp can lead to cell cycle arrest before terminus segregation. It was discovered that loss of either SeqA or DNA adenine methylase (Dam) abolishes (p)ppGpp-induced cell cycle arrest26. SeqA binds not only to hemimethylated DNA at oriC to regulate initiation but also beyond the origin to affect chromosome organization55. It is possible that the failure to segregate the termini is due to prolonged chromosome cohesion, promoted by SeqA, or failure to decatenate interlinked chromosomes26.
The research described above has led to a more holistic view of the role of metabolism in replication control that extends well beyond origin firing. Nutrient availability and metabolic status seem to affect every step, from initiation, through elongation to chromosome segregation. Although we are just beginning to identify the components involved, there seems to be an extensive regulatory network that ensures that replication, an essential, demanding but vulnerable process, is coordinated with growth. This network is crucial in the wild, as it allows cells to maintain genome integrity even in the face of rapidly changing environmental conditions.
Cell division
Like replication, division must be coupled to growth to ensure that average cell size is maintained under a given growth condition. Cells that divided before they doubled in mass would, after several generations, become unsustainably small. Conversely, a population of cells that routinely divided a substantial time after they had doubled in mass would ultimately grow into filaments that are no longer viable. In addition to this temporal form of control, a second regulatory network ensures that cell size increases under carbon-rich conditions, allowing cells to maintain a constant DNA to cell mass ratio during multifork replication2,44,56,57.
The temporal control of cell division
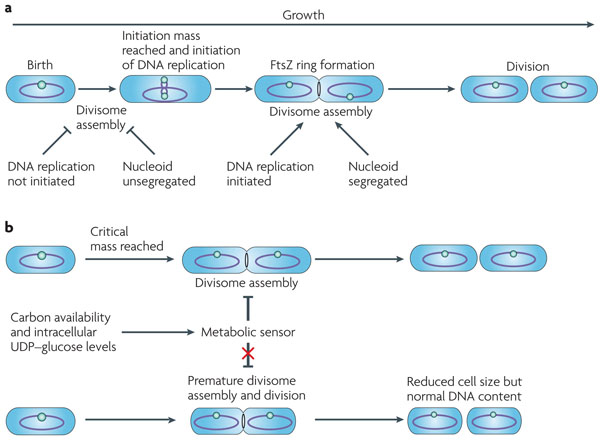
Precisely how division is coordinated with mass doubling is not clear. One possibility is that a factor analogous to DnaA accumulates in newborn cells, reaching the critical concentration only when cells have doubled in mass. To ensure that division is coordinated with mass doubling, the accumulation of such a factor would need to be dependent on nutrient availability and growth rate. Potential candidates include essential components of the division apparatus. However, none of the cell division genes identified to date, including ftsZ, seems to be regulated by growth rate in either B. subtilis or E. coli58,59.
Alternatively, it has been proposed that replication — a process that is itself dependent on growth rate — serves as a checkpoint for division (FIG. 3a). In support of this idea, inhibiting the initiation of replication in B. subtilis through the use of conditional mutants leads to the formation of elongated cells with acentric cytokinetic rings adjacent to centrally positioned nucleoids60,61. These cells experience a notable division delay due to DnaA-dependent repression of FtsL17, a crucial component of the division machinery. FtsL is highly unstable62 and therefore DnaA-dependent transcriptional inhibition quickly leads to a decrease in FtsL levels and a severe division delay.
Figure 3. Spatial and temporal regulation of cell division is achieved through multiple layers of control.
a| FtsZ assembly is coordinated with the initiation of DNA replication and nucleoid segregation to ensure that daughter cells receive complete copies of the bacterial genome. b| In Bacillus subtilis, the glucolipid biosynthesis pathway serves as a metabolic sensor to transmit information about carbon availability and growth rate, through the intracellular UDP–glucose concentration, to the division apparatus. When this sensor is functional (top), division is coupled to the achievement of a specific size (termed ‘critical mass’) as well as to mass doubling time. When this sensor is defective (bottom), division is uncoupled from the achievement of critical mass but remains sensitive to mass doubling time, resulting in reduced cell size.
Chromosome replication, segregation and the positional control of division
The acentric position of the FtsZ ring in cells that are blocked for initiation suggests that replication plays a part in the positional regulation of cell division. Consistent with this, allowing cells to assemble the replication initiation complex (DnaA and the primosome) but preventing DNA synthesis, either by thymine starvation of a thymine auxotroph or by the addition of the nucleotide analogue 6-(p-hydroxyphenylazo)-uracil, leads to the formation of a medially positioned FtsZ ring63. This observation suggests that the initiation of chromosome replication displaces the origin region from mid-cell, freeing an assembly point for the division machinery at the same position. In E. coli, medial FtsZ assembly and division have been observed in anucleate cells64,65, suggesting that replication is dispensable for cytokinesis in this organism.
Nucleoid segregation, a process that is partially dependent on metabolic status and cell growth, also seems to play a part in coordinating division with the rest of the cell cycle (FIG. 3a). Wild-type bacteria rarely divide across unsegregated nucleoids, and the vast majority of cells with FtsZ rings have two separate nucleoids4,44. Although it is possible for FtsZ rings to form across unsegregated nucleoids44,61,63, this situation is prevented in part by the actions of DNA-binding proteins, SlmA in E. coli and nucleoid occlusion protein (Noc) in B. subtilis66,67
Coordinating cell size with nutrient availability
Carbon availability is the primary determinant of cell size for rapidly growing bacteria2,57 (FIG. 3b). In B. subtilis information about carbon availability is transmitted directly to the division apparatus by accumulation of the nucleotide sugar UDP–glucose44. UDP–glucose inhibits division through its interaction with the bifunctional diacylglycerol glucosyltransferase, UgtP. Under conditions in which UDP–glucose is high, such as during growth in carbon-rich medium, UgtP inhibits FtsZ assembly and delays maturation of the cytokinetic ring until cells have reached the appropriate length. Under these conditions UgtP is distributed uniformly throughout the cytoplasm and localizes to the cytokinetic ring in an FtsZ-dependent manner, consistent with its role as a division inhibitor. Conversely, under conditions in which UDP–glucose levels are low, such as during growth in carbon-poor medium, the intracellular concentration of UgtP drops and the remaining protein is sequestered away from mid-cell in small, randomly positioned foci. The exact mechanism underlying this change in UgtP concentration and localization remains to be determined. UgtP inhibits FtsZ assembly in vitro, indicating that it interacts directly with FtsZ to inhibit division. The molecular mechanism by which UgtP prevents FtsZ assembly has yet to be determined; possibilities include capping the growing polymers or preventing the formation of stabilizing lateral interactions.
Carbon-dependent increases in cell size ensure that cells have sufficient room to accommodate the extra DNA that is generated by multifork replication. Defects in UDP–glucose biosynthesis reduce cell size by ~30% under carbon-rich conditions (when cells have a mass doubling time of ~25 minutes) and lead to a 3.5-fold increase in the frequency of FtsZ rings that are assembled across unsegregated nucleoids; Noc does not seem to inhibit FtsZ assembly across unsegregated nucleoids in these cells. However, under carbon-poor conditions (when cells have a mass doubling time of ~80 minutes), defects in UDP–glucose biosynthesis have little effect on cell morphology, and FtsZ ring formation takes place across segregated or partially segregated nucleoids44. Despite the reduction in cell size, the timing of FtsZ ring formation and division is still precisely coordinated with mass doubling in UDP–glucose-deficient cells. This observation implies that a second, UDP–glucose-independent pathway ensures that FtsZ assembly and division are coupled to cell growth.
E. coli cells also increase in size under carbon-rich conditions68, and cells shifted from carbon-poor to carbon-rich medium rapidly increase their growth rate but delay division until they reach the appropriate size for the new conditions69. Although UgtP is not conserved in E. coli, the accumulation of UDP–glucose does seem to be important for coordinating cell size with carbon availability. Studies indicate that mutations in two genes that are required for UDP–glucose biosynthesis, phosphoglucomutase (pgm) and glucose-1-phosphate uridylyltransferase (galU), reduce E. coli cell size by ~30% without substantially affecting mass doubling time70 (N.S. Hill and R.B. Weart, unpublished observations) (FIG. 3b). In the absence of a UgtP homologue, it is not clear what the UDP–glucose-sensitive effector is in E. coli, but on the basis of the data from B. subtilis it is reasonable to speculate that it is a UDP–glucose-binding protein. Systematic analysis of genes encoding such proteins should identify the effector responsible for coordinating nutrient availability with cell size in this Gram-negative organism.
Future directions
Cooper and Helmstetter’s work set the stage for our current understanding of the factors that couple cell cycle progression to nutrient availability and metabolic status in bacteria. Recent discoveries, many of which are discussed above, have enriched our knowledge and led to a nuanced view of the bacterial cell cycle as a set of coordinated but independent processes.
Nonetheless, this field is still in its infancy. It was only recently that we were able to determine the molecular basis of long-standing observations such as the sensitivity of chromosome replication to starvation and the increase in cell size that accompanies multifork replication. These baby steps provide a foundation for unravelling the precisely orchestrated signals that govern the bacterial cell cycle. Together with advances in systems biology that combine modelling with experimentation and advances in techniques that allow cell cycle processes to be followed in single cells, this foundation will allow the identification of the molecular mechanisms that are responsible for coupling cell growth and metabolic status to chromosome replication and division. Future work will clarify whether replication and division are programmed, consecutive steps in a linear pathway, as suggested by Cooper and Helmstetter5, or independent events that are linked together by their shared dependence on nutrient availability and metabolic status, as first proposed by Nordstrom and Boye6 and supported by recent work.
Acknowledgments
We thank D. Bates, A. Goranov and B. Hill for comments. Work in the Wang laboratory is supported by the Welch Foundation (Q-1698) and Public Health Service grants (GM084003 and DP2OD004433) from the US National Institutes of Health. Work in the Levin laboratory is supported by Public Health Services grant (GM64671) from the US National Institutes of Health and a National Science Foundation CAREER award (MCB-0448186).
Footnotes
DATABASES
Entrez Genome Project: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genomeprj
Bacillus subtilis | Caulobacter crescentus | Escherichia coli | Salmonella enterica subsp. enterica serovar Typhimurium
UniProtKB: http://www.uniprot.org
Dam | DiaA | DnaA | DnaN | FtsL | FtsZ | Hda | Noc | RelA | SeqA | SlmA | Soj | SpoT | UgtP | YabA
FURTHER INFORMATION
Jue D. Wang’s homepage: http://www.bcm.edu/genetics/?pmid=11048
Petra A. Levin’s homepage: http://www.biology.wustl.edu/levin/
Contributor Information
Jue D. Wang, Email: jdwang@bcm.edu, The Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas 77030, USA
Petra A. Levin, Email: plevin@biology.wustl.edu, The Department of Biology, Washington University, St. Louis, Missouri 63130, USA
References
- 1.Fantes P, Nurse P. Control of cell size at division in fission yeast by a growth-modulated size control over nuclear division. Exp Cell Res. 1977;107:377–386. doi: 10.1016/0014-4827(77)90359-7. [DOI] [PubMed] [Google Scholar]
- 2.Schaechter M, Maaloe O, Kjeldgaard NO. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 3.Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 4.Harry E, Monahan L, Thompson L. Bacterial cell division: the mechanism and its precison. Int Rev Cytol. 2006;253:27–94. doi: 10.1016/S0074-7696(06)53002-5. [DOI] [PubMed] [Google Scholar]
- 5.Cooper S, Helmstetter CE. Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol. 1968;31:519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- 6.Yoshikawa H, O’Sullivan A, Sueoka N. Sequential replication of the Bacillus subtilis chromosome, III. Regulation of initiation. Proc Natl Acad Sci USA. 1964;52:973–980. doi: 10.1073/pnas.52.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nordstrom K, Bernander R, Dasgupta S. The Escherichia coli cell cycle: one cycle or multiple independent processes that are co-ordinated? Mol Microbiol. 1991;5:769–774. doi: 10.1111/j.1365-2958.1991.tb00747.x. [DOI] [PubMed] [Google Scholar]
- 8.Boye E, Nordstrom K. Coupling the cell cycle to cell growth. EMBO Rep. 2003;4:757–760. doi: 10.1038/sj.embor.embor895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laub MT, Shapiro L, McAdams HH. Systems biology of Caulobacter. Annu Rev Genet. 2007;41:429–441. doi: 10.1146/annurev.genet.41.110306.130346. [DOI] [PubMed] [Google Scholar]
- 10.Zyskind JW, Smith DW. DNA replication, the bacterial cell cycle, and cell growth. Cell. 1992;69:5–8. doi: 10.1016/0092-8674(92)90112-p. [DOI] [PubMed] [Google Scholar]
- 11.Kaguni JM. DnaA: controlling the initiation of bacterial DNA replication and more. Annu Rev Microbiol. 2006;60:351–375. doi: 10.1146/annurev.micro.60.080805.142111. [DOI] [PubMed] [Google Scholar]
- 12.Mott ML, Berger JM. DNA replication initiation: mechanisms and regulation in bacteria. Nature Rev Microbiol. 2007;5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- 13.Xu YC, Bremer H. Chromosome replication in Escherichia coli induced by oversupply of DnaA. Mol Gen Genet. 1988;211:138–142. doi: 10.1007/BF00338404. [DOI] [PubMed] [Google Scholar]
- 14.Skarstad K, Lobner-Olesen A, Atlung T, von Meyenburg K, Boye E. Initiation of DNA replication in Escherichia coli after overproduction of the DnaA protein. Mol Gen Genet. 1989;218:50–56. doi: 10.1007/BF00330564. [DOI] [PubMed] [Google Scholar]
- 15.Ogura Y, Imai Y, Ogasawara N, Moriya S. Autoregulation of the dnaA-dnaN operon and effects of DnaA protein levels on replication initiation in Bacillus subtilis. J Bacteriol. 2001;183:3833–3841. doi: 10.1128/JB.183.13.3833-3841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaus N, O’Day K, Peters W, Wright A. Isolation and characterization of amber mutations in gene dnaA of Escherichia coli K-12. J Bacteriol. 1981;145:904–913. doi: 10.1128/jb.145.2.904-913.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goranov AI, Katz L, Breier AM, Burge CB, Grossman AD. A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proc Natl Acad Sci USA. 2005;102:12932–12937. doi: 10.1073/pnas.0506174102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gon S, et al. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 2006;25:1137–1147. doi: 10.1038/sj.emboj.7600990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiaramello AE, Zyskind JW. Expression of Escherichia coli dnaA and mioC genes as a function of growth rate. J Bacteriol. 1989;171:4272–4280. doi: 10.1128/jb.171.8.4272-4280.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanawalt PC, Maaloe O, Cummings DJ, Schaechter M. The normal DNA replication cycle. II J Mol Biol. 1961;3:156–165. doi: 10.1016/s0022-2836(61)80042-9. [DOI] [PubMed] [Google Scholar]
- 21.Cashel M, Gentry DR, Hernandez VH, Vinella D. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt FC, et al., editors. ASM; Washington DC: 1996. [Google Scholar]
- 22.Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 23.Chiaramello AE, Zyskind JW. Coupling of DNA replication to growth rate in Escherichia coli: a possible role for guanosine tetraphosphate. J Bacteriol. 1990;172:2013–2019. doi: 10.1128/jb.172.4.2013-2019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schreiber G, Ron EZ, Glaser G. ppGpp-mediated regulation of DNA replication and cell division in Escherichia coli. Curr Microbiol. 1995;30:27–32. doi: 10.1007/BF00294520. [DOI] [PubMed] [Google Scholar]
- 25.Levine A, Vannier F, Dehbi M, Henckes G, Seror SJ. The stringent response blocks DNA replication outside the ori region in Bacillus subtilis and at the origin in Escherichia coli. J Mol Biol. 1991;219:605–613. doi: 10.1016/0022-2836(91)90657-r. [DOI] [PubMed] [Google Scholar]
- 26.Ferullo DJ, Lovett ST. The stringent response and cell cycle arrest in Escherichia coli. PLoS Genet. 2008;4:e1000300. doi: 10.1371/journal.pgen.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lesley JA, Shapiro L. SpoT regulates DnaA stability and initiation of DNA replication in carbon-starved Caulobacter crescentus. J Bacteriol. 2008;190:6867–6880. doi: 10.1128/JB.00700-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorbatyuk B, Marczynski GT. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol Microbiol. 2005;55:1233–1245. doi: 10.1111/j.1365-2958.2004.04459.x. [DOI] [PubMed] [Google Scholar]
- 29.Atlung T, Clausen ES, Hansen FG. Autoregulation of the dnaA gene of Escherichia coli K12. Mol Gen Genet. 1985;200:442–450. doi: 10.1007/BF00425729. [DOI] [PubMed] [Google Scholar]
- 30.Braun RE, O’Day K, Wright A. Autoregulation of the DNA replication gene dnaA in E. coli K-12. Cell. 1985;40:159–169. doi: 10.1016/0092-8674(85)90319-8. [DOI] [PubMed] [Google Scholar]
- 31.Lu M, Campbell JL, Boye E, Kleckner N. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 32.Hansen FG, Christensen BB, Atlung T. The initiator titration model: computer simulation of chromosome and minichromosome control. Res Microbiol. 1991;142:161–167. doi: 10.1016/0923-2508(91)90025-6. [DOI] [PubMed] [Google Scholar]
- 33.Nozaki S, Yamada Y, Ogawa T. Initiator titration complex formed at datA with the aid of IHF regulates replication timing in Escherichia coli. Genes Cells. 2009;14:329–341. doi: 10.1111/j.1365-2443.2008.01269.x. [DOI] [PubMed] [Google Scholar]
- 34.Keyamura K, et al. The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes. Genes Dev. 2007;21:2083–2099. doi: 10.1101/gad.1561207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray H, Errington J. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell. 2008;135:74–84. doi: 10.1016/j.cell.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 36.Riber L, et al. Hda-mediated inactivation of the DnaA protein and dnaA gene autoregulation act in concert to ensure homeostatic maintenance of the Escherichia coli chromosome. Genes Dev. 2006;20:2121–2134. doi: 10.1101/gad.379506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kato J, Katayama T. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. Embo J. 2001;20:4253–4262. doi: 10.1093/emboj/20.15.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noirot-Gros MF, et al. Functional dissection of YabA, a negative regulator of DNA replication initiation in Bacillus subtilis. Proc Natl Acad Sci USA. 2006;103:2368–2373. doi: 10.1073/pnas.0506914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho E, Ogasawara N, Ishikawa S. The functional analysis of YabA, which interacts with DnaA and regulates initiation of chromosome replication in Bacillus subtils. Genes Genet Syst. 2008;83:111–125. doi: 10.1266/ggs.83.111. [DOI] [PubMed] [Google Scholar]
- 40.Soufo CD, et al. Cell-cycle-dependent spatial sequestration of the DnaA replication initiator protein in Bacillus subtilis. Dev Cell. 2008;15:935–941. doi: 10.1016/j.devcel.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 41.Hayashi M, Ogura Y, Harry EJ, Ogasawara N, Moriya S. Bacillus subtilis YabA is involved in determining the timing and synchrony of replication initiation. FEMS Microbiol Lett. 2005;247:73–79. doi: 10.1016/j.femsle.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 42.Noirot-Gros MF, et al. An expanded view of bacterial DNA replication. Proc Natl Acad Sci USA. 2002;99:8342–8347. doi: 10.1073/pnas.122040799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donachie WD. Relationship between cell size and time of initiation of DNA replication. Nature. 1968;219:1077–1079. doi: 10.1038/2191077a0. [DOI] [PubMed] [Google Scholar]
- 44.Weart RB, et al. A metabolic sensor governing cell size in bacteria. Cell. 2007;130:335–347. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wold S, Skarstad K, Steen HB, Stokke T, Boye E. The initiation mass for DNA replication in Escherichia coli K-12 is dependent on growth rate. EMBO J. 1994;13:2097–2102. doi: 10.1002/j.1460-2075.1994.tb06485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bipatnath M, Dennis PP, Bremer H. Initiation and velocity of chromosome replication in Escherichia coli B/r and K-12. J Bacteriol. 1998;180:265–273. doi: 10.1128/jb.180.2.265-273.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Churchward G, Estiva E, Bremer H. Growth rate-dependent control of chromosome replication initiation in Escherichia coli. J Bacteriol. 1981;145:1232–1238. doi: 10.1128/jb.145.3.1232-1238.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bates D, Kleckner N. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donachie WD. Co-ordinate regulation of the Escherichia coli cell cycle or The cloud of unknowing. Mol Microbiol. 2001;40:779–785. doi: 10.1046/j.1365-2958.2001.02439.x. [DOI] [PubMed] [Google Scholar]
- 50.Michelsen O, Teixeira de Mattos MJ, Jensen PR, Hansen FG. Precise determinations of C and D periods by flow cytometry in Escherichia coli K-12 and B/r. Microbiology. 2003;149:1001–1010. doi: 10.1099/mic.0.26058-0. [DOI] [PubMed] [Google Scholar]
- 51.Churchward G, Bremer H. Determination of deoxyribonucleic acid replication time in exponentially growing Escherichia coli B/r. J Bacteriol. 1977;130:1206–1213. doi: 10.1128/jb.130.3.1206-1213.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herrick J, Sclavi B. Ribonucleotide reductase and the regulation of DNA replication: an old story and an ancient heritage. Mol Microbiol. 2007;63:22–34. doi: 10.1111/j.1365-2958.2006.05493.x. [DOI] [PubMed] [Google Scholar]
- 53.Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Janniere L, et al. Genetic evidence for a link between glycolysis and DNA replication. PLoS ONE. 2007;2:e447. doi: 10.1371/journal.pone.0000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waldminghaus T, Skarstad K. The Escherichia coli SeqA protein. Plasmid. 2009;61:141–150. doi: 10.1016/j.plasmid.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 56.Sharpe ME, Hauser PM, Sharpe RG, Errington J. Bacillus subtilis cell cycle as studied by fluorescence microscopy: constancy of cell length at initiation of DNA replication and evidence for active nucleoid partitioning. J Bacteriol. 1998;180:547–555. doi: 10.1128/jb.180.3.547-555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sargent MG. Control of cell length in Bacillus subtilis. J Bacteriol. 1975;123:7–19. doi: 10.1128/jb.123.1.7-19.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weart RB, Levin PA. Growth rate-dependent regulation of medial FtsZ ring formation. J Bacteriol. 2003;185:2826–2834. doi: 10.1128/JB.185.9.2826-2834.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rueda S, Vicente M, Mingorance J. Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. J Bacteriol. 2003;185:3344–3351. doi: 10.1128/JB.185.11.3344-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rowland SL, Katis VL, Partridge SR, Wake RG. DivIB, FtsZ and cell division in Bacillus subtilis. Mol Microbiol. 1997;23:295–302. doi: 10.1046/j.1365-2958.1997.2141580.x. [DOI] [PubMed] [Google Scholar]
- 61.Harry EJ, Rodwell J, Wake RG. Co-ordinating DNA replication with cell division in bacteria: a link between the early stages of a round of replication and mid-cell Z ring assembly. Mol Microbiol. 1999;33:33–40. doi: 10.1046/j.1365-2958.1999.01439.x. [DOI] [PubMed] [Google Scholar]
- 62.Bramkamp M, Weston L, Daniel RA, Errington J. Regulated intramembrane proteolysis of FtsL protein and the control of cell division in Bacillus subtilis. Mol Microbiol. 2006;62:580–591. doi: 10.1111/j.1365-2958.2006.05402.x. [DOI] [PubMed] [Google Scholar]
- 63.Regamey A, Harry EJ, Wake RG. Mid-cell Z ring assembly in the absence of entry into the elongation phase of the round of replication in bacteria: co-ordinating chromosome replication with cell division. Mol Microbiol. 2000;38:423–434. doi: 10.1046/j.1365-2958.2000.02130.x. [DOI] [PubMed] [Google Scholar]
- 64.Jaffe A, D’Ari R, Norris V. SOS-independent coupling between DNA replication and cell division in Escherichia coli. J Bacteriol. 1986;165:66–71. doi: 10.1128/jb.165.1.66-71.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun Q, Yu XC, Margolin W. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol Microbiol. 1998;29:491–503. doi: 10.1046/j.1365-2958.1998.00942.x. [DOI] [PubMed] [Google Scholar]
- 66.Bernhardt TG, de Boer PA. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu LJ, Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 68.Donachie WD, Begg KJ. Cell length, nucleoid separation, and cell division of rod-shaped and spherical cells of Escherichia coli. J Bacteriol. 1989;171:4633–4639. doi: 10.1128/jb.171.9.4633-4639.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cooper S. Cell division and DNA replication following a shift to a richer medium. J Mol Biol. 1969;43:1–11. doi: 10.1016/0022-2836(69)90074-6. [DOI] [PubMed] [Google Scholar]
- 70.Lu M, Kleckner N. Molecular cloning and characterization of the pgm gene encoding phosphoglucomutase of Escherichia coli. J Bacteriol. 1994;176:5847–5851. doi: 10.1128/jb.176.18.5847-5851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]