Abstract
Background
The t(9;22)(q34;q11), generating the Philadelphia (Ph) chromosome, is found in more than 90% of patients with chronic myeloid leukemia (CML). As a result of the translocation, the 3' portion of the ABL1 oncogene is transposed from 9q34 to the 5' portion of the BCR gene on chromosome 22 to form the BCR/ABL1 fusion gene. At diagnosis, in 5-10% of CML patients the Ph chromosome is derived from variant translocations other than the standard t(9;22).
Results
We report a molecular cytogenetic study of 452 consecutive CML patients at diagnosis, that revealed 50 cases identifying three main subgroups: i) cases with variant chromosomal rearrangements other than the classic t(9;22)(q34;q11) (9.5%); ii) cases with cryptic insertions of ABL1 into BCR, or vice versa (1.3%); iii) cases bearing additional chromosomal rearrangements concomitant to the t(9;22) (1.1%). For each cytogenetic group, the mechanism at the basis of the rearrangement is discussed.
All breakpoints on other chromosomes involved in variant t(9;22) and in additional rearrangements have been characterized for the first time by Fluorescence In Situ Hybridization (FISH) experiments and bioinformatic analyses. This study revealed a high content of Alu repeats, genes density, GC frequency, and miRNAs in the great majority of the analyzed breakpoints.
Conclusions
Taken together with literature data about CML with variant t(9;22), our findings identified several new cytogenetic breakpoints as hotspots for recombination, demonstrating that the involvement of chromosomes other than 9 and 22 is not a random event but could depend on specific genomic features. The presence of several genes and/or miRNAs at the identified breakpoints suggests their potential involvement in the CML pathogenesis.
Background
Chronic myeloid leukemia (CML) is characterized by the constitutive expression of the 5'BCR/3'ABL1 fusion gene resulting from the t(9;22)(q34;q11); this translocation is evident in more than 90% of patients and produces the Philadelphia chromosome (Ph)[1].
In 5-10% of CML patients, the 5'BCR/3'ABL1 fusion gene arises from complex variant rearrangements which may involve one or more chromosomes in addition to 9 and 22 [2,3]. In some variant t(9;22) cases, additional material is transferred onto the Ph chromosome, resulting in a "masked" Ph whereas other CML patients show a classic Ph and an atypical der(9) chromosome as a consequence of a rearrangement between the der(9)t(9;22) and another chromosome [4,5]. Serial translocations or a single simultaneous event are alternative hypotheses proposed to justify the occurrence of these complex rearrangements [6].
In a subset of CML patients, cryptic rearrangements have been postulated to induce the chimeric gene formation, such as a nonreciprocal insertion between chromosomes 9 and 22 or two sequential translocations restoring the partner chromosomes morphology [7-11].
Microdeletions on the der(9) chromosome next to the t(9;22) breakpoint have been described in patients with classic and variant Ph translocations, and appear to be a valuable prognostic factor [12-17]. Recently, the frequency of such deletions has been investigated in the subgroup of CML patients with a masked Ph chromosome [18]. Additional genomic deletions on the third derivative chromosome have also been described in CML cases with variant translocations [19,20].
To our knowledge, an accurate breakpoints identification and bioinformatic analysis of other chromosomes involved in variant t(9;22) or in concomitant chromosomal rearrangements apart from the t(9;22) has never been performed in CML.
In this paper, a detailed molecular cytogenetic characterization of 50 (11.1%) out of 452 chronic phase (CP) CML patients was carried out to define the precise breakpoints on chromosomes other than 9 and 22. Bioinformatic analysis of breakpoint regions was performed to investigate the presence of repeated elements (Alu, LINE), GC content, Segmental Duplications (SDs), miRNAs, and known genes. Our findings, taken together with a review of literature data, allowed us to identify new cytogenetic hotspots in CML cases with variant t(9;22).
Methods
Patients
The study included 452 CP-CML patients. All of them were newly diagnosed at our hospital between 1990 and 2009. As a consequence of the long time span for sample accrual, several therapeutic regimens (hydroxyurea, interferon-α, imatinib, nilotinib, and dasatinib) were adopted.
Out of the 452 CP-CML cases, 50 showed variant t(9;22) or additional chromosomal rearrangements, 9 of which have been characterized in previous reports by our group [19-23].
Conventional cytogenetics
Conventional cytogenetic analysis of a 24-48 hour culture was performed at diagnosis of CML on bone marrow cells by standard techniques and evaluated by Giemsa-Trypsin-Giemsa (GTG) banding at about the 400-band level according to the ISCN [24]. At least 25 metaphases were analyzed for each case.
Identification of cytogenetic hotspots
To identify new cytogenetic hotspots, an estimate of the Haploid Autosomal Length (HAL) of the bands involved in variant t(9;22) cases was performed [25,26]. We calculated the number of breaks expected (E) in any band, given the null hypothesis of a random distribution of all breaks across the genome. Reviewing large series of CML patients with variant t(9;22) we assessed the number of breaks observed (O) in each band and divided this value by the expected (E) value to determine an O/E ratio. An O/E ratio >1 identified new cytogenetic hotspots.
FISH analysis
FISH analysis was performed on bone marrow samples of all CP-CML patients at diagnosis using "home-brew" FISH probes specific for ABL1 and BCR genes, validated in previous papers [13,16,27]. Breakpoints characterization and deletions size definition were carried out with additional bacterial artificial chromosome (BAC) and Phage P1-derived artificial chromosome (PAC) probes. All clones were selected according to the University of California Santa Cruz (UCSC http://genome.ucsc.edu/index.html; March 2006 release) database [28]; the mapping of each clone was first tested on normal human metaphases. Chromosome preparations were hybridized in situ with probes labeled with biotin by nick translation [29]. Briefly, 500 ng of labeled probe were used for FISH experiments; hybridization was performed at 37°C in 2× standard saline citrate (SSC), 50% (vol/vol) formamide, 10% (wt/vol) dextran sulphate, 5 μg COT1 DNA (Bethesda Research Laboratories, Gaithersburg, MD), and 3 μg sonicated salmon sperm DNA in a volume of 10 μL. Post-hybridization washing was done at 60°C 0.1× SSC. Biotin-labeled DNA was detected with Cy3-conjugated avidin. In cohybridization experiments, other probes were directly labeled with fluorescein. Chromosomes were identified by 4',6-diamidino-2-phenylindole (DAPI) staining. Digital images were obtained using a Leica DMRXA epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments, Boston, MA). Cy3 (red; New England Nuclear, NJ), fluorescein (green; NEN Life Science Products, Boston, MA), and DAPI (blue) fluorescence signals, which were detected using specific filters, were recorded separately as gray-scale images. Pseudocoloring and merging of images were performed with Adobe Photoshop software.
Bioinformatic analysis
Breakpoint regions on other chromosomes involved in variant t(9;22) and additional rearrangements were included in 250 Kb size intervals, according to the resolution limit of the BAC clones used for breakpoints definition. Each interval was checked for the presence of interspersed repeats classes (Alu and LINE repeats), SDs, GC content, and gene density. The UCSC Table Browser [28] was queried for summary analysis about the items belonging to the tracks "RepeatMasker", "Segmental Dups", "GC Percent", and "RefSeq Genes". For SDs and RefSeq gene analysis, both "Item count" and "Item Bases" values were considered, to assess their number and the bases percentage involved in SDs or coding sequences, respectively. For each genomic feature, the obtained value was normalized to the mean value for the examined chromosome. For example, in case 1, the breakpoint mapped in 1q32.1 (chr1:203,949,120-204,199,120) showed an Alu frequency of 13.47%. As the mean Alu content inside chromosome 1 is estimated to be 11.9%, the normalized value will be 1.13 (i.e. 13.47/11.90). Therefore, greater or lesser values than 1 correspond to regions with a richer or poorer content of a specific genomic feature than those observed along the entire chromosome.
In view of the known low miRNAs density in the human genome, regions spanning 2 Mb proximally and distally to breakpoints were investigated by querying the UCSC database at the track "sno/miRNA". For each chromosome the expected miRNA density within a 4 Mb interval was established according to the following formula: number of miRNA along the entire chromosome/size in bp of the chromosome × 4000000 bp. The identification of the predicted miRNAs target genes was performed by querying the miRGen database http://www.diana.pcbi.upenn.edu/cgi-bin/miRGen/v3/Targets.cgi. Intersection data from the three widely used target prediction programs (miRanda, PicTar, TargetScan) were considered. The definition of target genes function as oncogenes or tumor suppressor genes (TSGs) was made according to the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/gene/) database.
Results
FISH data
Cytogenetic analysis and FISH experiments with specific BAC/PAC probes for the ABL1 and BCR genes allowed us to detect 50 (11.1%) out of 452 cases, that identify 3 main subgroups of CML patients showing variant t(9;22) rearrangements, the occurrence of cryptic insertions of the ABL1 in the BCR region (or vice versa), and the presence of additional chromosomal abnormalities, respectively (Table 1).
Table 1.
Cytogenetic groups identified in a large series of CML patients at diagnosis
| Cases | Chromosomal rearrangements | Ph chromosome | Deletions |
5'BCR/3'ABL location |
5'ABL/3'BCR location |
|---|---|---|---|---|---|
| Variant t(9;22) | |||||
| #1 | t(1;9;22)(q32.1;q34;q11) | Ph+ | del(1)(q32), del(9)(q34), del(22)(q11) | Ph | 5'ABL deleted, 3'BCR on der(1) |
| #2 | t(1;9;22)(p34.2;q34;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(1) |
| #3 | der(9)t(9;22)(q34;q11), der(1)ins(1;22) (p36;q11q13), der(22)t(9;22) |
Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(1) |
| #4 | t(2;9;22)(q37.1;q34;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(2) |
| #5 | t(3;9;22)(p21.31;q34;q11) | Ph+ | del(22)(q11) | Ph | 5'ABL on der(9), 3'BCR on der(3) |
| #6 | t(3;9;22)(p21.31;q34;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(3) |
| #7 | t(3;9;22)(p21.31;q34;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(3) |
| #8 | t(4;9;22)(p16.3;q34;q11) | Ph+ | del(9)(q34) | Ph | 5'ABL on der(4), 3'BCR on der(4) |
| #9 | t(4;9;22)(p16;q34;q11) | Ph+ | del(9)(q34), del(22)(q11) | Ph | 5'ABL on der(4), 3'BCR deleted |
| #10 | t(4;9;22)(q12;q34;q11) | Ph+ | del(4)(q12), del(22)(q11) | Ph | 5'ABL deleted, 3'BCR on der(4) |
| #11 | t(4;9;22)(p16.2;q34;q11) | Ph+ | del(4)(p16), del(9)(q34), del(22)(q11) | Ph | 5'ABL deleted, 3'BCR deleted |
| #12 | t(6;9;22)(p12.3;q34;q11) | Ph+ | del(6)(p12), del(9)(q34) | Ph | 5'ABL deleted, 3'BCR on der(6) |
| #13 | t(6;9;22)(p21.31;q34;q11) | Ph+ | del(6)(p21p21), del(9)(q34q34), del(22)(q11q11) |
Ph | 5'ABL deleted, 3'BCR on der(6) |
| #14 | t(6;9;22)(q14.1;q34;q11) | Ph+(×2) | - | Ph | 5'ABL on der(9), 3'BCR on der(6) |
| #15 | t(7;9;22)(p14.3;q34;q11) | Ph+ | del(7)(p14), del(9)(q34), del(22)(q11) | Ph | 5'ABL deleted, 3'BCR deleted |
| #16 | t(7;9;22)(p22;q34;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(7) |
| #17 | der(9)t(9;22)(q34;q11), der(9)ins(9;9) (q34;q22q34) |
Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(9) |
| #18 | t(9;22;9;14)(q34;q11;p12;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(9) |
| #19 | t(9;11;22)(q34;q13.1;q11) | Ph+ | del(9)(q34), del(11)(q13), del(22)(q11) | Ph | 5'ABL deleted, 3'BCR deleted |
| #20 | t(9;11;22)(q34;q21;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(11) |
| #21 | t(9;12;22)(q34;q23.3;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(12) |
| #22 | t(9;12;22)(q34;q13.2;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(9) |
| #23 | t(9;12;22)(q34;q24.31;q11) | Ph+ | del(9)(q34), del(12)(q24), del(22)(q11) | Ph | 5'ABL deleted, 3'BCR on der(12) |
| #24 | t(9;12;22)(q34;q24.21;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(12) |
| #25 | t(9;13;22)(q34;q14.12;q11) | Ph+ | del(9)(q33q34), del(13)(q14), del(22)(q11) | Ph | 5'ABL on der(13), 3'BCR deleted |
| #26 | der(9)t(9;22)(q34;q11), der(14)ins(14;9)(q32;q34q34), der(22)t(9;22) |
Ph+ | del(9)(q34), del(22)(q11) | Ph | 5'ABL on der(14), 3'BCR on der(14) |
| #27 | t(9;14;15;22)(q34;q24.2;?;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(14) |
| #28 | t(9;15;22)(q34;q24.3;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(15) |
| #29 | t(9;16;22) (q34;p11.2;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(16) |
| #30 | t(9;16;17;22)(q34;q24.3;p13.1;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(17) |
| #31 | t(9;17;22)(q34;q21.2;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(9) |
| #32 | t(9;17;22) (q34;q25.3;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(17) |
| #33 | t(9;17;22) (q34;p13.3;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(17) |
| #34 | t(9;19;22)(q34;q13.32;q11) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(19) |
| #35 | t(9;21;22)(q34;q22.13;q11) | Ph+ | del(9)(q34), del(21)(q22), del(22)(q11) | Ph | 5'ABL deleted, 3'BCR deleted |
| #36 | t(9;X;22)(q34;q13.1;q11) | Ph+ | del(9)(q33q34) | Ph | 5'ABL deleted, 3'BCR on der(X) |
| #37 | t(7;9;22)(q22.2;q34;q11) | masked Ph | - | der(7) | 5'ABL on der(9), 3'BCR on der(9) |
| #38 | t(8;9;22)(p12;q34;q11) | masked Ph | - | der(8) | 5'ABL on der(9), 3'BCR on der(9) |
| #39 | t(9;10;22)(q34;p11.22;q11) | masked Ph | del(9)(q34), del(22)(q11) | der(10) | 5'ABL deleted, 3'BCR deleted |
| #40 | t(9;15;22)(q34;q24.1;q11) | masked Ph | - | der(22) | 5'ABL on der(9), 3'BCR on der(9) |
| #41 | t(6;9;12;22)(p22.1;q34;q13.13;q11) | masked Ph | del(9)(q34) | der(22) | 5'ABL on der(9), 3'BCR on der(6) |
| #42* | der(9)ins(9;22)(q34;q11q11), der(22)t(20;22)(q13.33;q11) |
masked Ph | - | der(9) | 5'ABL on der(9), 3'BCR on der(20) |
| #43* | der(22)t(12;22)(p13;q11)ins(22;9)(q11;q34q34) | masked Ph | del(9)(q34) | der(22) | 5'ABL deleted, 3'BCR on der(12) |
| Cryptic insertions | |||||
| #44 | ins(9;22)(q34;q11q11) | Ph- | del(9)(q34), del(22)(q11q12) | der(9) | 5'ABL deleted, 3'BCR on der(22) |
| #45 | der(22)t(9;22)(q34;q11)ins(22;22)(q11;q11q12) | Ph- | - | der(22) | 5'ABL on der(9), 3'BCR on der(22) |
| #46* | der(9)ins(22;9)(q11;q34q34)t(9;11)(p22.3;p15.4) | Ph- | - | der(22) | 5'ABL on der(9), 3'BCR on der(22) |
| #47* | ins(9;22)(q34;q11q11), t(1;20;21) | Ph- | - | der(9) | 5'ABL on der(9), 3'BCR on der(22) |
| #42* | der(9)ins(9;22)(q34;q11q11), der(22)t(20;22)(q13.33;q11) |
masked Ph | - | der(9) | 5'ABL on der(9), 3'BCR on der(20) |
| #43* | der(22)t(12;22)(p13;q11)ins(22;9)(q11;q34q34) | masked Ph | del(9)(q34) | der(22) | 5'ABL deleted, 3'BCR on der(12) |
| Additional chromosomal rearrangements | |||||
| #48 | t(9;22)(q34;q11), del(11)(q14q24) | Ph+ | del(11)(q14q24) | Ph | 5'ABL on der(9), 3'BCR on der(9) |
| #49 | t(9;22)(q34;q11), t(14;15)(q32.31;q24.1) | Ph+ | del(14)(q32), del(22)(q11) | Ph | 5'ABL on der(9), 3'BCR on der(9) |
| #50 | der(9)t(9;22)(q34;q11)t(9;11)(p24.1;q12.1) | Ph+ | - | Ph | 5'ABL on der(9), 3'BCR on der(9) |
| #46* | der(9)ins(22;9)(q11;q34q34)t(9;11)(p22.3;p15.4) | Ph- | - | der(22) | 5'ABL on der(9), 3'BCR on der(22) |
| #47* | ins(9;22)(q34;q11q11), t(1;20;21) | Ph- | - | der(9) | 5'ABL on der(9), 3'BCR on der(22) |
Three subgroups of CML patients have been identified according to the chromosomal rearrangements detected by FISH experiments: cases showing variant t(9;22) rearrangements, patients characterized by cryptic insertions of the ABL1 into the BCR region (or vice versa), and cases bearing additional chromosomal abnormalities. Cases #1- #36 showed a Ph+ chromosome together with a masked der(9) whereas a masked Ph in association with a classic der(9) was revealed in patients #37- #43. For each subgroup, the chromosomal rearrangements identified, the appearance of the der(22) chromosome, the occurrence of microdeletions next to chromosomal breakpoints, the location of the 5'BCR/3'ABL1 and of the 3'ABL1/5'BCR fusion genes are reported.
* indicates cases included in several cytogenetic subgroups.
1. Variant t(9;22) rearrangements
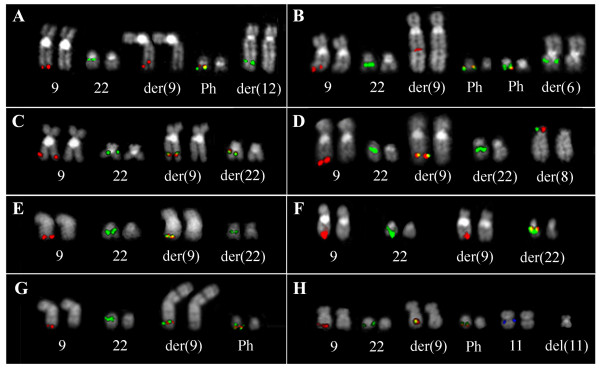
Forty-three (9.5%) out of 452 CML patients showed the involvement of one (90.7%) or more chromosomes (9.3%) in addition to 9 and 22. These complex variant translocations generated a classic Ph together with a masked der(9) in 36 out of 43 cases (83.7%) and a masked Ph in association with a classic der(9) chromosome in 7 patients (16.3%) (Table 1). Cases with a masked der(9) showed the presence of additional material belonging to partner chromosomes other than chromosome 22 (Fig. 1A, B). Several chromosomes were involved in these variant translocations, with a prevalence of chromosomes 4, 6, 12, and 17 (Table 1). The 5'BCR/3'ABL1 fusion gene was localized on the Ph chromosome in all these cases, whereas the 5'ABL1/3'BCR gene was retained on the der(9) only in 4 (11.1%) out of 36 patients (Table 1). In the remaining 32 (88.9%) cases, the 5'ABL1/3'BCR gene was not detected on der(9) due to deletions and/or 3'BCR transfer onto partner chromosomes (Table 1; Fig. 1A, B). Molecular cytogenetic characterization performed to verify the presence of microdeletions at the level of the rearrangements breakpoints revealed sequences loss in 18 out of the 43 (42%) cases. Among these 18 patients, 10 (55.6%) showed microdeletions of sequences belonging to the third partner chromosome, revealing a high incidence of this kind of deletions in t(9;22) variant rearrangement cases (Table 1).
Figure 1.
FISH pattern observed with specific probes for the ABL1 and BCR genes on bone marrow metaphases from the analyzed CML patients. Examples from each of the identified cytogenetic groups are shown: "masked der(9)" (A, B), "masked Ph" (C, D), "cryptic insertions" (E, F), and "concomitant rearrangements" (G, H).
Among 7 CML cases with a "masked Ph" chromosome, 3 showed the 5'BCR/3'ABL1 fusion signal on 22q11, the second breakpoint on the derivative chromosome 22 mapping inside chromosome 9 sequences distal to the ABL1 gene (Table 1; Fig. 1C). In 3 cases the fusion gene was detected on the third partner chromosome, the second chromosome 22 breakpoint being localized centromerically to the BCR gene (Table 1; Fig. 1D). In patient #42 the insertion of 5' BCR into the ABL1 gene caused the 5'BCR/3'ABL1 localization on der(9). The 5'ABL1/3'BCR gene was detected on the der(9) in 3 cases with masked Ph, was deleted in case #39 whereas in the remaining patients the 5'ABL1 gene was retained on the der(9) and the 3'BCR gene was transferred onto other derivative chromosomes (Table 1). Chromosome 9 sequences loss next to the rearrangement breakpoint was observed in case #43 and an unusual loss of a region of about 400 Kb localized telomerically to the ABL1 gene was detected in case #41 [22] (Table 1).
2. Cryptic insertions
Six (1.3%) out of the 452 CML cases showed cryptic insertions of ABL1 into BCR, or vice versa, as the cause of the 5'BCR/3'ABL1 fusion gene generation (Fig. 1E, F). Four (66.7%) of these cases are indicated as "Ph negative" (Ph-), with chromosome 22 appearing normal without the presence of additional genomic material (Table 1). Two (33.3%) out of these 6 cases were also included in the previous group as they showed variant rearrangements generating a masked Ph (Table 1). The 5'BCR/3'ABL1 gene was detected on the der(9) or on the der(22) at a ratio of 1:1 as a consequence of 5' BCR insertion in 9q34 or 3' ABL1 insertion in 22q11, respectively (Table 1).
3. Chromosomal rearrangements concomitant to the presence of 5'BCR/3'ABL1
Conventional and molecular cytogenetic analysis showed 5 (1.1%) out of 452 CML cases bearing additional chromosomal rearrangements concomitant to the generation of the 5'BCR/3'ABL1 fusion gene (Table 1; Fig. 1G, H). Cases #46 and #47 were also included in the previous patients group as they showed cryptic insertions; the remaining 3 CML cases carried a classic Ph chromosome.
Bioinformatic analysis of breakpoints on other chromosomes involved in variant t(9;22) or in concomitant chromosomal rearrangements
FISH experiments with BAC clones specific for other chromosomes involved in variant or additional chromosomal rearrangements revealed a total number of 58 breakpoints. These breakpoints were mapped within a single BAC clone or in the region between two overlapping or adjacent clones (Table 2). In cases with sequences loss, two different breakpoints were defined at the level of the deleted regions boundaries.
Table 2.
Bioinformatic analysis of the analyzed breakpoints
| Cytogenetic Band | Case | Molecular Breakpoint | 250 Kb interval | ALU | LINE | REF SEQ (IB) | REF SEQ (IC) | SD (IC) | SD (IB) | GC |
|---|---|---|---|---|---|---|---|---|---|---|
| 1p34.2 | 2 | RP11-318G20_RP11-632A13 | chr1:40,158,249-40,408,249 | 2,27 | 0,77 | 0,71 | 2,16 | 0,67 | 0,21 | 1,06 |
| 1q32.1 | 1 | RP11-219P13_RP11-1089F13 | chr1:203,949,120-204,199,120 | 1,13 | 0,78 | 1,11 | 3,09 | 0,44 | 0,27 | 1,07 |
| 1q32.1 | 1 | RP11-145I13_RP11-57I17 | chr1:205,496,738-205,746,738 | 0,52 | 1,55 | 0,76 | 1,85 | 0,22 | 0,15 | 0,91 |
| 2q37.1 | 4 | RP11-332L11_RP11-94I20 | chr2:234,468,641-234,718,641 | 0,69 | 0,94 | 1,3 | 1,01 | 0 | 0 | 1,05 |
| 3p21.31 | 5 | RP11-804H15_RP11-3B7 | chr3:49,112,155-49,393,337 | 4,17 | 0,16 | 1,7 | 7,05 | 0 | 0 | 1,24 |
| 3p21.31 | 6 | RP11-352L13_RP11-419G15 | chr3: 48,395,535-48,645,535 | 2,37 | 0,38 | 1,84 | 9,88 | 0,77 | 0,44 | 1,32 |
| 3p21.31 | 7 | RP11-316 M24 | chr3:48,787,955-49,037,955 | 4,6 | 0,45 | 2,02 | 6,58 | 0 | 0 | 1,18 |
| 4p16.2 | 11 | RP11-323F5_RP11-341O1 | chr4:4,566,405-4,816,405 | 1,22 | 0,54 | 0,33 | 0,65 | 0 | 0 | 1,18 |
| 4p16.3 | 8 | RP11-919N24 | chr4:2,277,155-2,527,155 | 3,37 | 0,56 | 1,9 | 1,31 | 0,38 | 0,33 | 1,34 |
| 4q12 | 10 | RP11-680 M13_RP11-167A8 | hr4:58,262,438-58,512,438 | 0,39 | 1,34 | 0 | 0 | 0 | 0 | 0,94 |
| 4q12 | 10 | RP11-622J1_RP11-793H8 | chr4:58,796,340-59,046,340 | 0,5 | 1,28 | 0 | 0 | 0 | 0 | 0,95 |
| 6p12.3 | 12 | RP3-347E1_RP11-446F17 | chr6: 46,562,151-46,812,151 | 0,67 | 1,1 | 1,76 | 1,72 | 0 | 0 | 0,95 |
| 6p12.3 | 12 | RP1-142O9_RP11-28H17 | chr6:49,380,518-49,630,518 | 0,82 | 1,04 | 0,93 | 1,72 | 0 | 0 | 0,92 |
| 6p21.31 | 13 | RP11-666K4_RP11-652G7 | chr6: 36,095,599-36,345,599 | 1,52 | 0,82 | 1,63 | 4,32 | 0 | 0 | 1,09 |
| 6p22.1 | 41 | RP11-635O11 | chr6:27,927,951-28,177,951 | 1,2 | 0,78 | 0,21 | 4,75 | 1,05 | 0,49 | 0,98 |
| 6q14.1 | 14 | RP11-1063N1_RP11-422O8 | chr6:83,741,988-83,991,988 | 1,21 | 0,86 | 2,37 | 2,16 | 0 | 0 | 0,95 |
| 7p14.3 | 15 | RP11-803J20_RP11-350H1 | chr7:32,789,214-33,039,214 | 1,91 | 0,65 | 1,06 | 2,54 | 0,92 | 5,42 | 1,05 |
| 7q22.2 | 37 | RP11-251G23 | chr7:104,889,282-105,139,282 | 2,55 | 0,5 | 1,95 | 2,12 | 0,61 | 0,07 | 1,09 |
| 8p12 | 38 | RP11-346L1_RP11-113G10 | chr8:37,576,531-37,826,531 | 2,13 | 0,44 | 1,01 | 4,09 | 0 | 0 | 1,2 |
| 9p22.3 | 46 | RP11-307K19_RP11-518K17 | chr9:15,456,627-15,734,271 | 1,47 | 1,44 | 2,16 | 1,87 | 0 | 0 | 0,93 |
| 9p24.1 | 50 | RP11-1084A8 | chr9:6,611,140-6,861,140 | 3,44 | 0,34 | 1,78 | 2,34 | 0,16 | 0,05 | 1,07 |
| 9q22.31 | 17 | RP11-412A12 | chr9:93,506,167-93,756,167 | 1,18 | 0,78 | 2,32 | 0,46 | 0 | 0 | 1,12 |
| 10p11.22 | 39 | RP11-241I20 | chr10:32,250,540-32,500,540 | 2,18 | 0,68 | 0,49 | 0,82 | 0,47 | 0,15 | 1,02 |
| 11p15.4 | 46 | RP11-120E20 | chr11:3,312,588-3,573,461 | 1,56 | 0,64 | 0,43 | 1,1 | 40,59 | 18,97 | 1,1 |
| 11q12.1 | 50 | RP11-624G17 | chr11:56,933,830-57,183,830 | 2,55 | 0,44 | 1,1 | 3,85 | 0 | 0 | 1,17 |
| 11q13.1 | 19 | RP11-665N17 | chr11:64,231,470-64,481,470 | 2,03 | 0,24 | 1,73 | 6,88 | 0 | 0 | 1,32 |
| 11q13.1 | 19 | RP11-821O7_RP11-755F10 | chr11:65,514,670-65,764,670 | 1,28 | 1,4 | 2,01 | 2,2 | 0 | 0 | 1,11 |
| 11q14.2 | 48 | RP11-185J12 | chr11: 85,624,712-85,874,712 | 1,45 | 1,09 | 1,6 | 3,02 | 0 | 0 | 0,96 |
| 11q21 | 20 | RP11-8N17 | chr11:95,707,670-95,957,670 | 0,45 | 1,6 | 0,49 | 0,82 | 0 | 0 | 0,9 |
| 11q24.2 | 48 | RP11-417F7 | chr11:124,646,245-124,896,245 | 1,21 | 0,52 | 2 | 0,82 | 0 | 0 | 1,13 |
| 12p13.32 | 43 | RP11-319E16 | chr12:5,129,118-5,379,118 | 0,41 | 0,94 | 0 | 0 | 0 | 0 | 1,04 |
| 12q13.13 | 41 | RP11-199A1_RP11-714I16 | chr12:50,354,699-50,604,699 | 1,17 | 0,52 | 1,47 | 2 | 4,8 | 0,33 | 1,14 |
| 12q13.2 | 22 | RP11-559I11_RP11-973D8 | chr12:54,434,528-54,684,528 | 2,86 | 0,38 | 1,65 | 6,35 | 0,48 | 0,48 | 1,11 |
| 12q23.3 | 21 | RP11-643D8_RP11-711H11 | chr12:103,184,790-103,434,790 | 1,41 | 0,58 | 1,33 | 2,33 | 0 | 0 | 1,07 |
| 12q24.21 | 24 | RP11-812F13_RP11-379F8 | chr12:114,736,981-114,986,981 | 0,91 | 0,86 | 0,98 | 0,33 | 0 | 0 | 1,01 |
| 12q24.31 | 23 | RP11-463O12_RP11-197N18 | chr12:121,846,884-122,096,884 | 2,14 | 0,57 | 1,91 | 4,67 | 0 | 0 | 1,25 |
| 12q24.31 | 23 | RP11-338K17_RP11-380L11 | chr12:122,702,493-122,952,493 | 2,21 | 0,72 | 2,18 | 1,67 | 0 | 0 | 1,1 |
| 13q14.12 | 25 | RP11-106H11 | chr13:45,408,774-45,658,774 | 0,98 | 0,88 | 2,43 | 3,5 | 0 | 0 | 0,98 |
| 13q14.12 | 25 | RP11-24B19 | chr13:50,678,660-50,928,660 | 0,9 | 0,65 | 2,09 | 4,38 | 0 | 0 | 1,06 |
| 14q11.2 | 18 | RP11-749G5_RP11-298I3 | chr14:22,280,500-22,530,500 | 3,01 | 0,39 | 1,38 | 10,02 | 0 | 0 | 1,16 |
| 14q24.2 | 27 | RP11-667E7 | chr14:71,197,805-71,447,805 | 1,05 | 1,2 | 0,81 | 0,41 | 0 | 0 | 1,01 |
| 14q32.31 | 49 | RP11-796G6_RP11-350L3 | chr14:101,148,480-101,398,480 | 1 | 1,41 | 1,07 | 2,5 | 0,44 | 0,44 | 1,07 |
| 14q32.31 | 49 | RP11-114H15_RP11-356L8 | chr14:101,630,161-101,880,161 | 2,78 | 0,54 | 2,38 | 2,92 | 0 | 0 | 1,14 |
| 15q22.2 | 27 | RP11-74K1 | chr15:58,405,774-58,655,774 | 1,06 | 0,44 | 1,7 | 3,7 | 0,41 | 0,14 | 0,96 |
| 15q24.1 | 40 | RP11-247C2 | chr15:72,080,168-72,330,168 | 1,06 | 0,56 | 1 | 8,53 | 3,28 | 2,71 | 1,19 |
| 15q24.1 | 49 | RP11-247C2 | chr15:72,080,168-72,330,168 | 1,06 | 0,56 | 1 | 8,53 | 3,28 | 2,71 | 1,19 |
| 15q24.3 | 28 | RP11-20 M10 | chr15:75921863-76171863 | 0,88 | 0,47 | 0,82 | 1,48 | 6,78 | 5,58 | 1,2 |
| 16p11.2 | 29 | RP11-779B17 | chr16:31,765,790-32,015,790 | 2,15 | 0,56 | 0,39 | 0,28 | 6,11 | 5,91 | 0,93 |
| 16q24.3 | 30 | RP11-79A1 | chr16:88,199,243-88,449,243 | 1,92 | 0,21 | 1,86 | 5,1 | 0 | 0 | 1,21 |
| 17p13.1 | 30 | RP11-89D11 | chr17:7,395,110-7,645,110 | 1,66 | 0,64 | 1,56 | 6,66 | 0,29 | 0 | 1,1 |
| 17p13.3 | 33 | RP5-1029F21 | chr17:267,052-517,052 | 1,11 | 1,18 | 1,49 | 0,52 | 0,58 | 0,23 | 1,02 |
| 17q21.2 | 31 | RP11-156A24 | chr17:36,799,760-37,049,760 | 0,39 | 0,52 | 0,47 | 2,45 | 1,59 | 1,92 | 1,1 |
| 17q25.3 | 32 | RP11-46E14 | chr17:75,272,119-75,522,119 | 0,83 | 0,63 | 0,22 | 0,87 | 0,29 | 0,93 | 1,2 |
| 19q13.32 | 34 | RP11-846 M4 | chr19:51,871,627-52,121,627 | 1,41 | 0,64 | 0,93 | 1,77 | 0 | 0 | 1,03 |
| 20q13.33 | 42 | RP4-591C20_RP11-266K16 | chr20:62,058,835-62,308,835 | 0,95 | 0,29 | 1,47 | 4,29 | 1,87 | 0,91 | 1,27 |
| 21q22.13 | 35 | RP11-315B15_RP11-777J19 | chr21:37,536,287-37,809,765 | 2,2 | 0,89 | 1,77 | 2,67 | 0 | 0 | 0,98 |
| 21q22.13 | 35 | RP11-105O24_RP11-1021I19 | chr21:37,747,110-37,997,110 | 0,98 | 1,02 | 1,57 | 2,67 | 0 | 0 | 1,04 |
| Xq13.1 | 36 | RP11-69L22_RP11-237F13 | chrX:68,788,268-69,038,268 | 0,66 | 1,58 | 3,32 | 1,95 | 0 | 0 | 0,97 |
The 250 Kb size intervals covering the molecular breakpoints were analyzed for the presence of Alu, LINE, RefSeq Genes, SDs, and GC. The reported values were normalized to the mean value for each chromosome. In cases characterized by sequences deletions or the involvement of several chromosomes, more than one molecular breakpoints was identified.
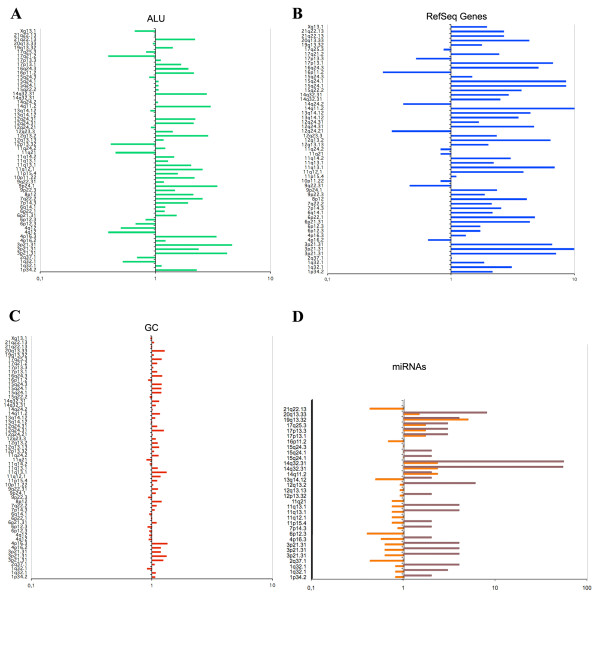
Interestingly, the majority of breakpoints on chromosomes involved in variant or additional chromosomal rearrangements showed a high frequency of Alu repeats (Table 2; Fig. 2A). In fact, 41 out of 58 (71%) breakpoints showed an Alu content of more than one whereas the remaining 17 out of 58 (29%) had a content of less than one. Instead, the LINE content was lower than one in 44 out of 58 (76%) breakpoints (Table 2). Thirty-five out of 41 breakpoints (85%) with Alu >1 showed a LINE amount < 1 (Table 2).
Figure 2.
Histograms representing the frequency distribution of genomic features in the breakpoint regions. All values shown for Alu (A), RefSeq Genes Item count (B), GC (C), and miRNAs (D) are reported in logarithmic scale. Chromosomal bands with a null value were excluded from the analysis. The observed distribution of miRNAs within 4 Mb intervals (brown) was compared to the expected mean value for each chromosome (orange).
Most of the analyzed breakpoints map within gene-rich regions as a RefSeq Genes Item count of more than one was observed in 45 out of 58 (78%) breakpoints (Table 2; Fig. 2B). Moreover, 40 out of 58 (69%) breakpoints showed a RefSeq Genes Item bases value of more than 1 (Table 2). It is worthy of note that 34 out of 41 bp (83%) with Alu >1 showed a RefSeq Genes Item count >1 (Table 2). The number of known genes localized at breakpoints and their function as oncogenes and/or TSGs are reported in Table 3.
Table 3.
Genes with known function identified in breakpoint regions
| Cytogenetic Band | Case | Molecular Breakpoint | Breakpoint region | Deleted region | Number of known genes | TSG/Oncogenes |
|---|---|---|---|---|---|---|
| 1p34.2 | 2 | RP11-318G20_RP11-632A13 | chr1:40,270,704-40,309,712 | 1 | ||
| 1q32.1 | 1 | RP11-1089F13_RP11-145I13 | chr1:203,989,316-205,431,690 | 26 | IKBKE, RASSF5, IL10, IL24 | |
| 2q37.1 | 4 | RP11-332L11_RP11-94I20 | chr2:234,593,254-234,594,028 | 0 | ||
| 3p21.31 | 5 | RP11-804H15_RP11-3B7 | chr3:49,112,155-49,222,551 | 8 | ||
| 3p21.31 | 6 | RP11-352L13_RP11-419G15 | chr3:48,468,265-48,572,806 | 4 | ||
| 3p21.31 | 7 | RP11-316 M24 | chr3:48,859,939-48,965,972 | 4 | ||
| 4p16.2 | 11 | 4ptel_RP11-341O1 | chr4:0-4,673,371 | 68 | GAK | |
| 4p16.3 | 8 | RP11-919N24 | chr4:2,317,528-2,486,782 | 2 | ||
| 4q12 | 10 | RP11-167A8_RP11-622J1 | chr4:58,389,737-58,916,984 | 0 | ||
| 6p12.3 | 12 | RP11-446F17_RP1-142O9 | chr6: 46,562,151-46,812,151 | 14 | ||
| 6p21.31 | 13 | RP11-666K4_RP11-652G7 | chr6:36,174,067-36,267,130 | chr6:46,715,042-49,457,682 | 2 | |
| 6p22.1 | 41 | RP11-635O11 | chr6:27,960,335-28,145,568 | 5 | ||
| 6q14.1 | 14 | RP11-1063N1_RP11-422O8 | chr6:83,875,190-83,886,368 | 1 | ||
| 7p14.3 | 15 | RP11-803J20_RP11-350H1 | chr7:32,469,044-33,359,385 | 10 | ||
| 7q22.2 | 37 | RP11-251G23 | chr7:104,957,075-105,071,488 | 3 | RINT1 | |
| 8p12 | 38 | RP11-346L1_RP11-113G10 | chr8:37,687,186-37,715,876 | 1 | ||
| 9p22.3 | 46 | RP11-307K19_RP11-518K17 | chr9:15,455,865-15,456,627 | 1 | ||
| 9p24.1 | 50 | RP11-1084A8 | chr9:6,611,140-6,861,140 | 2 | KDM4C | |
| 9q22.31 | 17 | RP11-412A12 | chr9:93,553,640-93,708,694 | 1 | ||
| 10p11.22 | 39 | RP11-241I20 | chr10:32,303,829-32,447,252 | 1 | ||
| 11p15.4 | 46 | RP11-120E20 | chr11:3,573,461-3,758,006 | 5 | ART1, NUP98 | |
| 11q12.1 | 50 | RP11-624G17 | chr11:56,953,550-57,164,109 | 10 | ||
| 11q13.1 | 19 | RP11-665N17_RP11-821O7 | chr11:64,481,470-65,571,636 | 57 | FAU, SYVN1, RELA, CFL1, CST6 | |
| 11q14.2 | 48 | RP11-185J12_RP11-417F7 | chr11: 85,874,712-124,646,245 | 297 | TSG11, CADM1, FAT3, MLL, ATM, YAP1, ZBTB16, THY1, SDHD, ARHGEF12, BTG4, PPP2R1B, NOX4, POU2F3, TBRG1, TAGLN, CBL | |
| 11q21 | 20 | RP11-8N17 | chr11:95,755,457-95,909,884 | 2 | ||
| 12p13.32 | 43 | RP11-319E16 | chr12:5,163,930-5,344,301 | 0 | ||
| 12q13.13 | 41 | RP11-199A1_RP11-714I16 | chr12:50,423,209-50,425,214 | 1 | ||
| 12q13.2 | 22 | RP11-559I11_RP11-973D8 | chr12:54,583,954-54,618,511 | 2 | ||
| 12q23.3 | 21 | RP11-643D8_RP11-711H11 | chr12:103,271,502-103,364,761 | 0 | ||
| 12q24.21 | 24 | RP11-812F13_RP11-379F8 | chr12:114,705,782-115,018,181 | 1 | ||
| 12q24.31 | 23 | RP11-197N18_RP11-338K17 | chr12:121,978,125-122,877,008 | 19 | ||
| 13q14.12 | 25 | RP11-106H11_RP11-24B19 | chr13:45,658,774-50,678,660 | 33 | RB1, ARL11, TRIM13, FAM10A4 | |
| 14q11.2 | 18 | RP11-749G5_RP11-298I3 | chr14:22,375,744-22,435,254 | 3 | ||
| 14q24.2 | 27 | RP11-667E7 | chr14:71,230,541-71,415,068 | 1 | ||
| 14q32.31 | 49 | RP11-350L3_RP11-114H15 | chr14:101,273,490-101,716,645 | HSP90AA1 | ||
| 15q22.2 | 27 | RP11-74K1 | chr15:58,530,516-58,531,032 | 1 | ||
| 15q24.1 | 40 | RP11-247C2 | chr15:72,158,366-72,251,969 | 3 | ||
| 15q24.1 | 49 | RP11-247C2 | chr15:72,158,366-72,251,969 | 3 | ||
| 15q24.3 | 28 | RP11-20 M10 | chr15:75,965,658-76,128,067 | 3 | ||
| 16p11.2 | 29 | RP11-779B17 | chr16:31,786,062-31,995,517 | 1 | ||
| 16q24.3 | 30 | RP11-79A1 | chr16:88,125,792-88,522,693 | 9 | ||
| 17p13.1 | 30 | RP11-89D11 | chr17:7,436,436-7,603,767 | 18 | TP53 | |
| 17p13.3 | 33 | RP5-1029F21 | chr17:343,377-440,727 | 1 | VPS53 | |
| 17q21.2 | 31 | RP11-156A24 | chr17:36,841,610-37,007,910 | 9 | ||
| 17q25.3 | 32 | RP11-46E14 | chr17:75,316,874-75,477,363 | 4 | ||
| 19q13.32 | 34 | RP11-846 M4 | chr19:51,889,293-52,103,962 | 6 | ||
| 20q13.33 | 42 | RP4-591C20_RP11-266K16 | chr20:62,100,354-62,267,316 | 9 | ||
| 21q22.13 | 35 | RP11-777J19_RP11-105O24 | chr21:37,641,259-37,872,927 | 1 | ||
| Xq13.1 | 36 | RP11-69L22_RP11-237F13 | chrX:68,857,148-68,969,387 | 1 |
The number of the genes with known function mapping in the breakpoint regions or located inside the deleted regions was reported for each case. Known oncogenes and TSGs have been identified according to the NCBI.
In the search for SDs, 49 out of 58 (84%) and 51 out of 58 (88%) breakpoints revealed SDs Item count and SDs Item bases of less than one, respectively (Table 2). In cases showing the presence of SDs within breakpoint regions no specific association with chromosomes 9 and 22 was detected, as the duplicated elements recognized several chromosomal regions.
Finally, a GC content >1 was detected in 43 out of 58 (74%) breakpoints (Table 2; Fig. 2C). A GC content of more than one was identified in 34 out of 41 (83%) breakpoints with Alu >1 and in 34 out of 45 (76%) with a RefSeq Genes Item count >1 (Table 2).
The search for miRNAs revealed a different density from the expected value in 32 out of 58 (55%) breakpoint regions (Fig. 2D). In detail, in 30 (94%) and 2 out of 32 (6%) breakpoints a higher or lower number of miRNA than the expected value was identified, respectively (Fig. 2D). In the remaining 26 out of 58 (45%) breakpoints no miRNA was revealed in the 4 Mb analyzed intervals. It is noteworthy that in case #49 with an additional t(14;15)(q32;q24) a miRNA cluster of 54 elements was revealed in the 14q32 breakpoint region. In this patient a microdeletion of about 450 Kb was detected on 14q32, resulting in the loss of almost the entire miRNA cluster. The list of miRNAs found at the breakpoints is reported in Table 4; in addition to the 14q32 miRNA cluster a total number of 63 known miRNA was identified, 8 (13%) of which show involvement in hematological malignancies. Moreover, querying the miRGen database (the intersection data from the miRanda, PicTar, and TargetScan programs) allowed the identification of the predicted target genes in 19 out of 63 (30%) analyzed miRNAs (see Additional File 1). Among the identified target genes, several play a role as oncogenes or TSGs (see Additional File 1). Noteworthy, some miRNAs share the same target oncogenes or TSGs; for example, PPM1D (protein phosphatase, Mg2+/Mn2+ dependent, 1D) and AKT3 (v-akt murine thymoma viral oncogene homolog 3) genes are the most frequent miRNAs targets (see Additional File 1).
Table 4.
Known miRNA mapped at the breakpoint regions.
| Cytogenetic Band | miRNA | miRNA position | Hematologic Malignancies involvement |
|---|---|---|---|
| 1p34.2 | mir-30e | chr1:40,992,614-40,992,705 | no |
| mir-30c-1 | chr1:40,995,543-40,995,631 | no | |
| 1q32.1 | mir-135b | chr1:203,684,053-203,684,149 | no |
| mir-29c | chr1:206,041,820-206,041,907 | CLL36,37, ALL37 | |
| mir-29b-2 | chr1:206,042,411-206,042,491 | AML38, ALL38 | |
| mir-205 | chr1:207,672,101-207,672,210 | no | |
| 2q37.1 | mir-562 | chr2:232,745,607-232,745,701 | no |
| 3p21.31 | mir-1226 | chr3:47,866,049-47,866,123 | no |
| mir-425 | chr3:49,032,585-49,032,671 | no | |
| mir-191 | chr3:49,032,805-49,033,396 | AML39, ALL40 | |
| mir-566 | chr3:50,185,763-50,185,856 | no | |
| 4p16.3 | mir-943 | chr4:1,957,909-1,958,002 | no |
| mir-571 | chr4:333,946-334,041 | no | |
| 6p12.3 | mir-586 | chr6:45,273,389-45,273,485 | no |
| 7p14.3 | mir-550-2 | chr7:32,739,118-32,739,214 | no |
| mir-548n | chr7:34,946,897-34,946,971 | no | |
| 11p15.4 | mir-675 | chr11:1,974,565-1,974,637 | no |
| mir-483 | chr11:2,111,940-2,112,015 | no | |
| 11q12.1 | mir-130a | chr11:57,164,997-57,165,585 | no |
| 11q13.1 | mir-1237 | chr11:63,892,650-63,892,751 | no |
| mir-192 | chr11:64,414,935-64,415,544 | no | |
| mir-194-2 | chr11:64,415,153-64,415,737 | no | |
| mir-612 | chr11:64,968,505-64,968,604 | no | |
| 11q21 | mir-548l | chr11:93,839,309-93,839,394 | no |
| 12p13.32 | mir-200c | chr12:6,942,873-6,943,440 | no |
| mir-141 | chr12:6,943,271-6,943,865 | no | |
| 12q13.13 | mir-1293 | chr12:48,914,192-48,914,262 | no |
| 12q13.2 | mir-196a-2 | chr12:52,671,789-52,671,898 | no |
| mir-615 | chr12:52,714,001-52,714,096 | no | |
| mir-148b | chr12:53,017,267-53,017,365 | no | |
| mir-1228 | chr12:55,874,554-55,874,626 | no | |
| mir-616 | chr12:56,199,213-56,199,309 | no | |
| mir-26a-2 | chr12:56,504,409-56,504,992 | no | |
| 13q14.12 | mir-16-1 | chr13:49,521,110-49,521,198 | CLL41, MDS42 |
| mir-15a | chr13:49,521,006-49,521,588 | CLL43, MM44 | |
| 14q11.2 | mir-208a | chr14:22,927,645-22,927,715 | no |
| mir-208b | chr14:22,957,036-22,957,112 | no | |
| 14q32.31 | miRNA cluster 1 | chr14:99,645,745--101,096,512 | AML45, B-cell malignancies46 |
| 14q32.31 | miRNA cluster 2 | chr14:99,645,745-103,653,604 | AML45, B-cell malignancies46 |
| 15q24.1 | mir-630 | chr15:70,666,612-70,666,708 | no |
| mir-631 | chr15:73,433,005-73,433,079 | B cell lymphomas47 | |
| 15q24.3 | mir-184 | chr15:77,289,185-77,289,268 | no |
| 16p11.2 | mir-1826 | chr16:33,873,009-33,873,093 | no |
| 17p13.1 | mir-195 | chr17:6,861,408-6,861,994 | CLL37, ALL37 |
| mir-497 | chr17:6,861,954-6,862,065 | no | |
| mir-324 | chr17:7,067,340-7,067,422 | no | |
| 17p13.3 | mir-22 | chr17:1,563,947-1,564,031 | no |
| mir-132 | chr17:1,899,952-1,900,052 | T-cell leukemia48 | |
| mir-212 | chr17:1,900,315-1,900,424 | no | |
| 17q25.3 | mir-657 | chr17:76,713,671-76,713,768 | no |
| mir-338 | chr17:76,714,278-76,714,344 | no | |
| mir-1250 | chr17:76,721,591-76,721,703 | no | |
| 19q13.32 | mir-330 | chr19:50,834,092-50,834,185 | no |
| mir-642 | chr19:50,870,026-50,870,122 | no | |
| mir-769 | chr19:51,214,030-51,214,147 | no | |
| mir-220c | chr19:53,755,341-53,755,423 | no | |
| 20q13.33 | mir-1-1 | chr20:60,561,708-60,562,278 | no |
| mir-133a-2 | chr20:60,572,314-60,572,915 | no | |
| mir-124-3 | chr20:61,280,297-61,280,383 | no | |
| mir-941-1 | chr20:62,021,238-62,021,326 | no | |
| mir-941-2 | chr20:62,021,545-62,021,633 | no | |
| mir-941-3 | chr20:62,021,657-62,021,745 | no | |
| mir-1914 | chr20:62,043,262-62,043,341 | no | |
| mir-647 | chr20:62,044,428-62,044,523 | no | |
| 21q22.13 | mir-802 | chr21:36,014,883-36,014,976 | no |
For each cytogenetic band identified in this study are reported known miRNAs, the mapping position derived from the UCSC database querying, and their implication in hematological malignancies according to literature data.
Identification of cytogenetic hotspots
Our study revealed 46 cytogenetic breakpoints on other chromosomes involved in variant t(9;22) rearrangements (see Additional File 2). The assessment of the O/E ratio for each breakpoint allowed us to identify 24 hotspots, 12 of which have been previously described in literature [26] (see Additional File 2). Notably, 4 out of 12 new hotspots showed a ratio >2 involving the chromosomal bands 4q12, 9p11, 11q21 and 21q22 (see Additional File 2).
To investigate the breakpoints distribution in the genome, a review of literature data about variant t(9;22) following the study by Fisher et al. was carried out [4,30-33]. In total, 60 new hotspots were identified, 18 of which have already been reported [26]. However, 10 previously published hotspots were not supported by our literature review (see Additional File 2). Among the 60 new hotspots, 27 showed an O/E ratio > 2.
Treatment response
Data on the response to treatment in the analyzed CML patients were only available for about 50% of the cases; a summary is shown in Table 5. All the cases evaluable for the response to interferon-α therapy were non responders whereas 11 out of 17 (65%) cases treated with imatinib achieved cytogenetic response. Among patients resistant to imatinib, 3 (75%) treated with dasatinib achieved CCyR.
Table 5.
Response to treatment of 50 CML patients included in the study
| Case | Hydroxyurea | Interferon-α | Imatinib | Nilotinib | Dasatinib |
|---|---|---|---|---|---|
| #1 | NE | NE | NR | NR | CCyR |
| #2 | NE | NE | CCyR | NE | NE |
| #3 | NA | NA | NA | NA | NA |
| #4 | NA | NA | NA | NA | NA |
| #5 | NE | NE | CCyR | NE | NE |
| #6 | NE | NE | NR | NE | NE |
| #7 | NA | NA | NA | NA | NA |
| #8 | NA | NA | NA | NA | NA |
| #9 | NA | NA | NA | NA | NA |
| #10 | NA | NA | NA | NA | NA |
| #11 | NE | NR | NE | NE | NE |
| #12 | NR* | NE | NE | NE | NE |
| #13 | NE | NE | CCyR | NE | NE |
| #14 | NA | NA | NA | NA | NA |
| #15 | NA | NA | NA | NA | NA |
| #16 | NA | NA | NA | NA | NA |
| #17 | NE | NE | CCyR | NE | NE |
| #18 | NA | NA | NA | NA | NA |
| #19 | NE | NE | CCyR | NE | NE |
| #20 | NA | NA | NA | NA | NA |
| #21 | NE | NE | CCyR | NE | NE |
| #22 | NA | NA | NA | NA | NA |
| #23 | NA | NA | NA | NA | NA |
| #24 | NA | NA | NA | NA | NA |
| #25 | NE | NR | NR | NE | CyCR |
| #26 | NE | NR | CCYR | NE | NE |
| #27 | NA | NA | NA | NA | NA |
| #28 | NA | NA | NA | NA | NA |
| #29 | NA | NA | NA | NA | NA |
| #30 | NE | NE | CCyR | NE | NE |
| #31 | NE | NE | NR | NE | NE |
| #32 | NA | NA | NA | NA | NA |
| #33 | NA | NA | NR | NE | NR |
| #34 | NA | NA | NA | NA | NA |
| #35 | NE | NE | CCyR | NE | NE |
| #36 | NA | NA | NA | NA | NA |
| #37 | NR* | NE | NE | NE | NE |
| #38 | NE | NR | NE | NE | NE |
| #39 | NA | NA | NA | NA | NA |
| #40 | NE | NR | NE | NE | NE |
| #41 | NE | NR | PCyR | NE | NE |
| #42 | NA | NA | NA | NA | NA |
| #43 | NE | NR | CCyR | NE | NE |
| #44 | NA | NA | NA | NA | NA |
| #45 | NA | NA | NA | NA | NA |
| #46 | NE | CHR | NE | NE | NE |
| #47 | NA | NA | NA | NA | NA |
| #48 | NE | NR | NE | NE | NE |
| #49 | NE | NR | NR | NE | CCyR |
| #50 | NA | NA | NA | NA | NA |
NE = Not Evaluable; NA = Not Available; NR = Non Responder; CCyR = Complete Cytogenetic Response; CHR = Complete Hematologic Response; PcyR = Partial Cytogenetic Response; * = patient who died in the pre imatinib era, of blast crisis.
Discussion
Literature data indicate that breakpoints on additional chromosomes involved in CML cases with variant t(9;22) are not distributed randomly in the genome but show hotspots [26]. Several genomic features such as the density of CpG islands, genes, Alu repeats, recombination events, openness of the chromatin structure and transcription activity have been correlated to the occurrence of breakpoints in variant t(9;22) cases [26,33,34].
In this study, we have performed for the first time a precise molecular cytogenetic characterization of breakpoints involved in variant t(9;22) or in additional rearrangements, in 50 CML cases. To identify genomic elements with a role in the occurrence of chromosomal translocations, bioinformatic analysis was carried out to investigate the distribution and density of several genomic features, such as Alu, LINE, GC, SDs, miRNAs, and genes at breakpoint regions. To date, according to the miRBase database http://www.mirbase.org[35] the total number of known miRNAs is very low (about 720) as compared to the human genome size (3.1 × 109 bp). In this study the miRNAs density within the 4 Mb analyzed intervals resulted higher than the expected value in 32 out of 58 (55%) breakpoint regions. These findings suggest a potential role for miRNAs in the pathogenesis of CML cases with variant or additional chromosomal rearrangements. Few miRNAs located at breakpoint regions have previously been described in several hematological malignancies [36-48]. However, none of them was involved in CML. It is worth noting the presence of the miRNA cluster next to the breakpoint region in 14q32 (case #49). miRNAs in this region are organized in an imprinted domain regulated by a differentially methylated region located upstream of the miRNA cluster. It has been reported that these miRNAs act as tumor suppressor genes and that changes in their methylation status could promote tumor development [49]. Querying of miRGen and NCBI databases showed the involvement of interesting target oncogenes or TSGs implicated in a wide variety of biological processes including cell proliferation, differentiation, apoptosis, and tumorigenesis.
Increasing evidence shows a high density of interspersed repetitive elements, such as Alu and LINE, at some chromosomal translocation breakpoints, suggesting a mediator role of some recurrent rearrangements in tumors [50]. Because a much higher density of Alu repeats has been observed in the DNA sequences flanking the ABL1 and BCR genes, it has been hypothesized that Alu elements provide hotspots for non allelic homologous recombination and mediate chromosomal translocation in CML [34,50]. Our data, supported by bioinformatic evidence, suggest that the high density of Alu repeats could increase the propensity to undergo rearrangements also of other chromosomes involved in variant t(9;22). In our CML series, a high Alu density was detected in 71% of the analyzed breakpoints. Moreover, a rich content of Alu repeats was revealed also on breakpoint regions identified in chromosomal rearrangements concomitant to the t(9;22).
Literature data revealed a preferential breakpoints distribution in CML cases with variant t(9;22) within the CG-richest regions of the genome corresponding to the G-light banding karyotype [26,33]. Our data confirmed this association, as 83% of the identified cytogenetic breakpoints mapped inside G-light bands. Moreover, we report the first bioinformatic evidence of the association between GC-content and breaks in cases with variant t(9;22), as 73% of the molecular breakpoints showed a GC content >1. In addition, these data showed that CG richness was related to other genomic features such as Alu content and a greater gene density than the mean expected value.
The search for SDs revealed a low density in the majority of the analyzed breakpoints, without showing any specific association with chromosomes 9 and 22 regions, unlike what has been reported about the occurrence of the t(9;22) in CML [51].
Moreover, our study provided an outline of the frequency and molecular features of the most relevant cytogenetic groups identified in a very large series of CML patients at diagnosis. Three-way translocations were the most frequent among variant t(9;22) rearrangements, chromosomes 4, 6, 12, and 17 being common partners. However, no cytogenetic breakpoints clustering was revealed when the same partner chromosome was rearranged, except for the 3p21 band, that was involved in 3 CML cases with variant t(9;22).
As to the mechanisms involved in the formation of the variant t(9;22) rearrangements, our data indicated that the most probable mechanism, identified in cases with a "masked der(9)" chromosome, is a single event consisting of multiple simultaneous breaks and rejoins (one-step model). In fact, splitting of the 5'ABL1/3'BCR fusion gene signal was observed in the majority (27 out of 36, 75%) of analyzed cases. A two-step mechanism was hypothesized in about 11% of cases bearing a "masked der(9)" chromosome; the permanence of the 5'ABL1/3'BCR gene on the der(9) suggests that a second break occurred inside the chromosome 22 sequence telomeric to the BCR gene. On the contrary, in 71.4% of cases (#37 - #41) with a "masked Ph" chromosome a second break located proximally to BCR or distally to ABL1 was identified, suggesting the occurrence of a two-step mechanism in the majority of the CML patients included in this group.
In our study, FISH 'walking' with BAC/PAC contigs belonging to the chromosomes 9 and 22 next to the t(9;22) breakpoint regions allowed us to assess the frequency of deletions in three main cytogenetic subgroups of CML patients and the size of these microdeletions. Confirming the deletion frequency reported in literature [12], 12 out of the 36 (33%) cases with a "masked der(9)" chromosome showed chromosome 9 and/or 22 sequences loss. Moreover, in about 55% of these patients we found extensive genomic deletions on the third chromosome, in addition to deletions on der(9). Chromosome 9 sequences deletions were detected in 3 out of 6 (50%) cases with a masked Ph (#39, #41, and #43) and in 1 out of 4 (25%) Ph- cases (#44). These frequencies are higher than the value recently reported in literature [18].
The biological significance and the prognostic impact of the cytogenetic molecular heterogeneity occurring in the generation of the 5'BCR/3'ABL1 fusion gene remain to be clarified. However, the bioinformatic analysis performed in this study on a large number of breakpoints in CML cases with variant t(9;22) or additional chromosomal alterations revealed that the rearranged regions are characterized by an elevated content of miRNAs, Alu repeats, GC and known genes.
In conclusion, this genomic analysis of breakpoint regions provides clues to a better understanding of the pathogenetic mechanisms that underlie CML onset. Further analyses will be needed to demonstrate the functional meaning of these genomic features.
List of abbreviations
(CP-CML): Chronic Myeloid Leukemia; (FISH): Fluorescence In Situ Hybridization; (CML): Chronic myeloid leukemia; (Ph): Philadelphia chromosome; (SDs): Segmental Duplications; (GTG): Giemsa-Trypsin-Giemsa; (HAL): Haploid Autosomal Length; (E): Number of breaks expected; (O): Number of breaks observed; (BAC): Bacterial artificial chromosome; (PAC): Phage P1-derived artificial chromosome; (UCSC): University of California Santa Cruz; (Ph-): Ph negative.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
FA, LA, and AZ were involved in the design and execution of the experiments, wrote the manuscript and contributed to the overall experimental design. NC conducted most of the FISH experiments. PC and LV performed conventional cytogenetic analysis; ARR contributed to clinical data collection. VL, MR and GS participated in the design of the study and supervised the manuscript preparation. All authors have read and approved the final manuscript.
Supplementary Material
Oncogenes and TSGs regulated by the analyzed miRNAs. The number of the predicted target genes for each analyzed miRNA was reported according to the miRGen database. Target genes with a role as oncogenes or TSGs were identified by querying the NCBI database.
Variant t(9;22) Breakpoints. The chromosomal bands involved in variant t(9;22) are shown according to our study and recent large series of CML patients reported in literature. The number of total breaks observed in each band is shown, together with the observed/expected (O/E) ratio. In bold are indicated cytogenetic hotspots, O/E ratio being > 1. The symbol * represents hotspots previously reported [26].
Contributor Information
Francesco Albano, Email: f.albano@ematba.uniba.it.
Luisa Anelli, Email: luisaanelli@yahoo.it.
Antonella Zagaria, Email: antonellazagaria@hotmail.com.
Nicoletta Coccaro, Email: n.coccaro@biologia.uniba.it.
Paola Casieri, Email: p.casieri@libero.it.
Antonella Russo Rossi, Email: antorussorossi@libero.it.
Laura Vicari, Email: l.vicari@libero.it.
Vincenzo Liso, Email: v.liso@ematba.uniba.it.
Mariano Rocchi, Email: rocchi@biologia.uniba.it.
Giorgina Specchia, Email: g.specchia@ematba.uniba.it.
Acknowledgements
The authors thank Ms MVC Pragnell, B.A. for language revision of the manuscript. The financial support of Associazione Italiana contro le Leucemie (AIL)-BARI is gratefully acknowledged.
References
- Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7:441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- Huret JL. Complex translocations, simple variant translocations and Ph-negative cases in chronic myelogenous leukaemia. Hum Genet. 1990;85:565–568. doi: 10.1007/BF00193575. [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F. The Mitelman database of chromosome aberrations in cancer. http://www.cgap.nci.nih.gov/Chromosomes/Mitelman
- Gorusu M, Benn P, Li Z, Fang M. On the genesis and prognosis of variant translocations in chronic myeloid leukemia. Cancer Genet Cytogenet. 2007;173:97–106. doi: 10.1016/j.cancergencyto.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Virgili A, Brazma D, Reid AG, Howard-Reeves J, Valgañón M, Chanalaris A, De Melo VA, Marin D, Apperley JF, Grace C, Nacheva EP. FISH mapping of Philadelphia negative BCR/ABL1 positive CML. Mol Cytogenet. 2008;1:14. doi: 10.1186/1755-8166-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald PH, Morris CM. Complex chromosomal translocations in the Philadelphia chromosome leukemias: serial translocations or a concerted genomic rearrangement? Cancer Genet Cytogenet. 1991;57:143–151. doi: 10.1016/0165-4608(91)90145-K. [DOI] [PubMed] [Google Scholar]
- Plas DC van der, Grosveld G, Hagemeijer A. Review of clinical, cytogenetic, and molecular aspects of Ph-negative CML. Cancer Genet Cytogenet. 1991;52:143–156. doi: 10.1016/0165-4608(91)90457-6. [DOI] [PubMed] [Google Scholar]
- Chase A, Huntly BJP, Cross NCP. Cytogenetics of chronic myeloid leukemia. Best Pract Res Clin Haematol. 2001;14:553–571. doi: 10.1053/beha.2001.0154. [DOI] [PubMed] [Google Scholar]
- Haigh S, Cuthbert G. Fluorescence in situ hybridization characterization of different cryptic BCR-ABL rearrangements in chronic myeloid leukemia. Cancer Genet Cytogenet. 2004;155:132–137. doi: 10.1016/j.cancergencyto.2004.02.026. [DOI] [PubMed] [Google Scholar]
- Reddy KS, Grove B. A Philadelphia-negative chronic myeloid leukemia with a BCR/ABL fusion gene on chromosome 9. Cancer Genet Cytogenet. 1998;107:48–50. doi: 10.1016/S0165-4608(98)00064-8. [DOI] [PubMed] [Google Scholar]
- Storlazzi CT, Anelli L, Surace C, Lonoce A, Zagaria A, Nanni M, Curzi P, Rocchi M. Molecular cytogenetic characterization of a complex rearrangement involving chromosomes 9 and 22 in a case of Ph negative chronic myeloid leukemia. Cancer Genet Cytogenet. 2002;136:141–145. doi: 10.1016/S0165-4608(02)00530-7. [DOI] [PubMed] [Google Scholar]
- Huntly BJ, Reid AG, Bench AJ, Campbell LJ, Telford N, Shepherd P, Szer J, Prince HM, Turner P, Grace C, Nacheva EP, Green AR. Deletions of the derivative chromosome 9 occur at the time of the Philadelphia translocation and provide a powerful and independent prognostic indicator in chronic myeloid leukemia. Blood. 2001;98:1732–1738. doi: 10.1182/blood.V98.6.1732. [DOI] [PubMed] [Google Scholar]
- Storlazzi CT, Specchia G, Anelli L, Albano F, Pastore D, Zagaria A, Rocchi M, Liso V. Breakpoint characterization of der(9) deletions in CML patients. Genes Chromosomes Cancer. 2002;35:271–276. doi: 10.1002/gcc.10116. [DOI] [PubMed] [Google Scholar]
- Huntly BJ, Guilhot F, Reid AG, Vassiliou G, Hennig E, Franke C, Byrne J, Brizard A, Niederwieser D, Freeman-Edward J, Cuthbert G, Bown N, Clark RE, Nacheva EP, Green AR, Deininger MW. Imatinib improves but may not fully reverse the poor prognosis of patients with CML with derivative chromosome 9 deletions. Blood. 2003;102:2205–2212. doi: 10.1182/blood-2002-09-2763. [DOI] [PubMed] [Google Scholar]
- Reid AG, Huntly BJ, Grace C, Green AR, Nacheva EP. Survival implications of molecular heterogeneity in variant Philadelphia-positive chronic myeloid leukaemia. Br J Haematol. 2003;121:419–427. doi: 10.1046/j.1365-2141.2003.04291.x. [DOI] [PubMed] [Google Scholar]
- Specchia G, Albano F, Anelli L, Storlazzi CT, Zagaria A, Liso A, Pannunzio A, Pastore D, Mestice A, Greco G, Liso V, Rocchi M. Derivative chromosome 9 deletions in chronic myeloid leukemia are associated with loss of tumor suppressor genes. Leuk Lymphoma. 2004;45:689–694. doi: 10.1080/10428190310001623900. [DOI] [PubMed] [Google Scholar]
- Quintas-Cardama A, Kantarjian H, Talpaz M, O'brien S, Garcia-Manero G, Verstovsek S, Rios MB, Hayes K, Glassman A, Bekele BN, Zhou X, Cortes J. Imatinib mesylate therapy may overcome the poor prognostic significance of deletions of derivative chromosome 9 in patients with chronic myelogenous leukemia. Blood. 2005;105:2281–2286. doi: 10.1182/blood-2004-06-2208. [DOI] [PubMed] [Google Scholar]
- De Melo VA, Milojkovic D, Marin D, Apperley JF, Nacheva EP, Reid AG. Deletions adjacent to BCR and ABL1 breakpoints occur in a substantial minority of chronic myeloid leukemia patients with masked Philadelphia rearrangements. Cancer Genet Cytogenet. 2008;182:111–115. doi: 10.1016/j.cancergencyto.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Albano F, Specchia G, Anelli L, Zagaria A, Storlazzi CT, Buquicchio C, Roberti MG, Liso V, Rocchi M. Genomic deletions on other chromosomes involved in variant t(9;22) chronic myeloid leukemia cases. Genes Chromosomes Cancer. 2003;36:353–360. doi: 10.1002/gcc.10183. [DOI] [PubMed] [Google Scholar]
- Anelli L, Albano F, Zagaria A, Liso A, Roberti MG, Rocchi M, Specchia G. A chronic myelocytic leukemia case bearing deletions on the three chromosomes involved in a variant t(9;22;11) Cancer Genet Cytogenet. 2004;148:137–140. doi: 10.1016/S0165-4608(03)00246-2. [DOI] [PubMed] [Google Scholar]
- Zagaria A, Anelli L, Albano F, Storlazzi CT, Liso A, Roberti MG, Buquicchio C, Liso V, Rocchi M, Specchia G. A fluorescence in situ hybridization study of complex t(9;22) in two chronic myelocytic leukemia cases with a masked Philadelphia chromosome. Cancer Genet Cytogenet. 2004;150:81–85. doi: 10.1016/j.cancergencyto.2003.08.018. [DOI] [PubMed] [Google Scholar]
- Zagaria A, Anelli L, Albano F, Vicari L, Schiavone EM, Annunziata M, Pane F, Liso V, Rocchi M, Specchia G. Molecular cytogenetic characterization of deletions on der(9) in chronic myelocytic leukemia. Cancer Genet Cytogenet. 2006;167:97–102. doi: 10.1016/j.cancergencyto.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Albano F, Zagaria A, Anelli L, Pannunzio A, Manodoro F, Coccaro N, Russo Rossi A, Liso V, Rocchi M, Specchia G. The double deceit generated by an insertion mechanism in chronic myeloid leukemia with t(9;9;22) Ann Hematol. 2008;87:923–926. doi: 10.1007/s00277-008-0511-y. [DOI] [PubMed] [Google Scholar]
- Mitelman F. ISCN. An International System for Human Cytogenetic Nomenclature. Basel, Switzerland: Karger; 1995. [Google Scholar]
- Daniel A. Structural differences in reciprocal translocations. Potential for a model of risk in Rcp. Hum Genet. 1979;51:171–182. doi: 10.1007/BF00287173. [DOI] [PubMed] [Google Scholar]
- Fisher AM, Strike P, Scott C, Moorman AV. Breakpoints of variant 9;22 translocations in chronic myeloid leukemia locate preferentially in the CG-richest regions of the genome. Genes Chromosomes Cancer. 2005;43:383–389. doi: 10.1002/gcc.20196. [DOI] [PubMed] [Google Scholar]
- Albano F, Anelli L, Zagaria A, Archidiacono N, Liso V, Specchia G, Rocchi M. "Home-brew" FISH assay shows higher efficiency than BCR-ABL dual color, dual fusion probe in detecting microdeletions and complex rearrangements associated with t(9;22) in chronic myeloid leukemia. Cancer Genet Cytogenet. 2007;174:121–126. doi: 10.1016/j.cancergencyto.2006.09.025. [DOI] [PubMed] [Google Scholar]
- The University of California Santa Cruz database. http://genome.ucsc.edu/index.html
- Lichter P, Tang Chang CJ, Call K, Hermanson G, Evans GA, Housman D, Ward DC. High resolution mapping of human chromosomes 11 by in situ hybridization with cosmid clones. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- El-Zimaity MM, Kantarjian H, Talpaz M, O'Brien S, Giles F, Garcia-Manero G, Verstovsek S, Thomas D, Ferrajoli A, Hayes K, Nebiyou Bekele B, Zhou X, Rios MB, Glassman AB, Cortes JE. Results of imatinib mesylate therapy in chronic myelogenous leukaemia with variant Philadelphia chromosome. Br J Haematol. 2004;125:187–195. doi: 10.1111/j.1365-2141.2004.04899.x. [DOI] [PubMed] [Google Scholar]
- Costa D, Carrió A, Madrigal I, Arias A, Valera A, Colomer D, Aguilar JL, Teixido M, Camós M, Cervantes F, Campo E. Studies of complex Ph translocations in cases with chronic myelogenous leukemia and one with acute lymphoblastic leukemia. Cancer Genet Cytogenet. 2006;166:89–93. doi: 10.1016/j.cancergencyto.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Richebourg S, Eclache V, Perot C, Portnoi MF, Akker J Van den, Terré C, Maareck O, Soenen V, Viguié F, Laï JL, Andrieux J, Corm S, Roche-Lestienne C. Fi-LMC Group. Mechanisms of genesis of variant translocation in chronic myeloid leukemia are not correlated with ABL1 or BCR deletion status or response to imatinib therapy. Cancer Genet Cytogenet. 2008;182:95–102. doi: 10.1016/j.cancergencyto.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Bennour A, Sennana H, Laatiri MA, Elloumi M, Khelif A, Saad A. Molecular cytogenetic characterization of variant Philadelphia translocations in chronic myeloid leukemia: genesis and deletion of derivative chromosome 9. Cancer Genet Cytogenet. 2009;194:30–37. doi: 10.1016/j.cancergencyto.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Jeffs AR, Wells E, Morris CM. Nonrandom distribution of interspersed repeat elements in the BCR and ABL1 genes and its relation to breakpoint cluster regions. Genes Chromosomes Cancer. 2001;32:144–154. doi: 10.1002/gcc.1176. [DOI] [PubMed] [Google Scholar]
- The miRBase database. http://www.mirbase.org
- Visone R, Rassenti LZ, Veronese A, Taccioli C, Costinean S, Aguda BD, Volinia S, Ferracin M, Palatini J, Balatti V, Alder H, Negrini M, Kipps TJ, Croce CM. Karyotype-specific microRNA signature in chronic lymphocytic leukemia. Blood. 2009;114:3872–3879. doi: 10.1182/blood-2009-06-229211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanette DL, Rivadavia F, Molfetta GA, Barbuzano FG, Proto-Siqueira R, Silva WA Jr, Falcao RP, Zago MA. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz J Med Biol Res. 2007;40:1435–1440. doi: 10.1590/s0100-879x2007001100003. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li Z, He C, Wang D, Yuan X, Chen J, Jin J. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol Dis. 2010;44:191–197. doi: 10.1016/j.bcmd.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, Fabbri M, Coombes K, Alder H, Nakamura T, Flomenberg N, Marcucci G, Calin GA, Kornblau SM, Kantarjian H, Bloomfield CD, Andreeff M, Croce CM. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulci V, Colombo T, Chiaretti S, Messina M, Citarella F, Tavolaro S, Guarini A, Foa' R, Macino G. Characterization of B- and T-lineage acute lymphoblastic leukemia by integrated analysis of MicroRNA and mRNA expression profiles. Genes Chromosomes Cancer. 2009;48:1069–1082. doi: 10.1002/gcc.20709. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–1529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons A, Nomdedeu B, Navarro A, Gaya A, Gel B, Diaz T, Valera S, Rozman M, Belkaid M, Montserrat E, Monzo M. Hematopoiesis-related microRNA expression in myelodysplastic syndromes. Leuk Lymphoma. 2009;50:1854–1859. doi: 10.3109/10428190903147645. [DOI] [PubMed] [Google Scholar]
- Hanlon K, Rudin CE, Harries LW. Investigating the targets of MIR-15a and MIR-16-1 in patients with chronic lymphocytic leukemia (CLL) PLoS One. 2009;4:e7169. doi: 10.1371/journal.pone.0007169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corthals SL, Jongen-Lavrencic M, de Knegt Y, Peeters JK, Beverloo HB, Lokhorst HM, Sonneveld P. Micro-RNA-15a and micro-RNA-16 expression and chromosome 13 deletions in multiple myeloma. Leuk Res. 2010;34:677–681. doi: 10.1016/j.leukres.2009.10.026. [DOI] [PubMed] [Google Scholar]
- Dixon-McIver A, East P, Mein CA, Cazier JB, Molloy G, Chaplin T, Andrew Lister T, Young BD, Debernardi S. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One. 2008;3:e2141. doi: 10.1371/journal.pone.0002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katrincsakova B, Takeda H, Urbankova H, Michaux L, Jarosova M, Vandenberghe P, Georges M, Charlier C, Wlodarska I. Methylation analysis of the imprinted DLK1-GTL2 domain supports the random parental origin of the IGH-involving del(14q) in B-cell malignancies. Epigenetics. 2009;4:469–475. doi: 10.4161/epi.4.7.9924. [DOI] [PubMed] [Google Scholar]
- Robbiani DF, Bunting S, Feldhahn N, Bothmer A, Camps J, Deroubaix S, McBride KM, Klein IA, Stone G, Eisenreich TR, Ried T, Nussenzweig A, Nussenzweig MC. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellon M, Lepelletier Y, Hermine O, Nicot C. Deregulation of microRNA involved in hematopoiesis and the immune response in HTLV-I adult T-cell leukemia. Blood. 2009;113:4914–4917. doi: 10.1182/blood-2008-11-189845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Kolomietz E, Meyn MS, Pandita A, Squire JA. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer. 2002;35:97–112. doi: 10.1002/gcc.10111. [DOI] [PubMed] [Google Scholar]
- Albano F, Anelli L, Zagaria A, Coccaro N, D'Addabbo P, Liso V, Rocchi M, Specchia G. Genomic segmental duplications on the basis of the t(9;22) rearrangement in Chronic Myeloid Leukemia. Oncogene. 2010;29:2509–2516. doi: 10.1038/onc.2009.524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oncogenes and TSGs regulated by the analyzed miRNAs. The number of the predicted target genes for each analyzed miRNA was reported according to the miRGen database. Target genes with a role as oncogenes or TSGs were identified by querying the NCBI database.
Variant t(9;22) Breakpoints. The chromosomal bands involved in variant t(9;22) are shown according to our study and recent large series of CML patients reported in literature. The number of total breaks observed in each band is shown, together with the observed/expected (O/E) ratio. In bold are indicated cytogenetic hotspots, O/E ratio being > 1. The symbol * represents hotspots previously reported [26].