Abstract
Genetic mutations in the mitotic regulatory kinase BUBR1 are associated with the cancer susceptible disorder mosaic variegated aneuploidy (MVA). In patients with biallelic mutations, a missense mutation pairs with a truncating mutation. Here we show that cell lines derived from MVA patients with biallelic mutations have an impaired mitotic checkpoint, chromosome alignment defects, and low overall BUBR1 abundance. Ectopic expression of BUBR1 restored mitotic checkpoint activity, proving that BUBR1 dysfunction causes chromosome segregation errors in the patients. Combined analysis of patient cells and functional protein replacement demonstrates that all MVA mutations fall in two distinct classes: those that impose specific defects in checkpoint activity or microtubule attachment and those that lower BUBR1 protein abundance. Low protein abundance is the direct result of the absence of transcripts from truncating mutants combined with high protein turnover of missense mutants. In this group of missense mutants, the amino acid change consistently occurs in or near the BUBR1 kinase domain. Our findings provide a molecular explanation for chromosomal instability in patients with biallelic genetic mutations in BUBR1.
Keywords: Mitosis, Cancer, BUBR1, Aneuploidy, Mitotic Checkpoint
Introduction
Mosaic Variegated Aneuploidy (MVA, OMIM: 257300), also referred to as Premature Chromatid Separation (PCS) Syndrome, an autosomal recessive syndrome characterized by constitutional aneuploidy and very early onset cancer predisposition. Typically, individuals with MVA display microcephaly, growth- and mental retardation, as well as other, milder, physical anomalies. In addition, 37% of patients develop cancers including rhadomyosarcoma, Wilms tumor and leukemia, mostly within the first 3 years of life, not seldom even in utero (see 1 of table (1) and (2, 3)). The MVA syndrome has been linked to mono- or biallelic mutations in the BUB1B locus, encoding the predicted serine/threonine kinase BUBR1 (BUBR1 is the accepted alias name for the BUB1B protein, and therefore used throughout this study)(2, 3). As indicated by the name, mosaic aneuploidies are found in cells of various tissues from MVA patients, suggesting underlying defects in the fidelity of chromosome segregation during development. Consistent with this, BUBR1 is critical for several processes that govern chromosome segregation during cell divisions. Error-free chromosome segregation requires that each sister of a duplicated chromosome is attached via their kinetochores to spindle microtubules from two opposing spindle poles (4). Onset of cell division before each an every kinetochore is attached to the mitotic spindle is normally prevented by the mitotic checkpoint (5). One of the essential components of this checkpoint is BUBR1 (6-9). BUBR1 directly inhibits the E3 ubiquitin ligase Anaphase Promoting Complex/Cyclosome (APC/C) that promotes chromosome segregation by targeting essential cell cycle regulators such as Cyclin B and Securin/PTTG for destruction (10, 11). This inhibitory property of BUBR1 resides in the highly conserved amino-terminal 450 amino acids and does not absolutely require the carboxy-terminal kinase domain (12-14). Consistent with the presence of BUBR1 mutations as a cause for aneuploidy in MVA patients, cells from a Japanese patient had an impaired ability to respond to the microtubule poison colcemid (3, 15). In addition to a role in the mitotic checkpoint, BUBR1 is required for the establishment of stable interactions between kinetochores and spindle microtubules (16, 17).
The high incidence of tumors in MVA patients suggests a causal link between aneuploidy and tumor formation. In sporadic cancers, chromosomal instability (CIN), the frequent missegregation of whole chromosomes, has also been proposed to be a contributing force in carcinogenesis (18, 19). The causes of CIN in human tumors are unknown but likely involve dysfunction of some machineries that normally promote error-free chromosome segregation. Defects in attachment error-correction mechanisms (16, 20), centrosome duplication, cytokinesis or the mitotic checkpoint have all been postulated to promote CIN in tumors (19, 21). In rare cases, mutations in regulators of chromosome segregation in sporadic human tumors have been reported, including in BUBR1 and the homologous kinase BUB1 (18), but no clear functional link between these mutations and chromosome segregation errors in such tumors has been established.
The mutations in BUBR1 associated with MVA fall in two classes: missense or frameshift mutations that result in truncated protein products (hereafter referred to as ‘truncation’), and missense mutations that cause single amino acid substitutions (hereafter referred to as ‘substitution’). In four families with individuals that carried biallelic mutations, a truncating mutation was combined with one amino acid substitution, often in the kinase domain (2). In 8 other families, one, predominantly truncating, mutation was found (3). We set out to examine the molecular causes of chromosome segregation errors in MVA patients. Our results present a rationale for why specific combinations of biallelic mutations cause aneuploidy in MVA patients.
Materials & Methods
Plasmids and shRNA-Based Protein Replacement
The pSuper-based shRNA plasmids used in this study: Mock (22), BUBR1 (23). LAP-BUBR1-WT has been created by cloning the RNAi resistant allele from pcDNA3-myc-BUBR1ΔsiRNA (23) to pIC58 (24). Mutants were obtained by site-directed mutagenesis and the LAP-mock control was created by mutating the second codon after the LAP-tag of LAP-BUBR1-WT to a stop codon.
Cells were cotransfected with a marker plasmid along with pSuper-BUBR1 or pSuper-mock and shRNA-insensitive LAP-BUBR1-WT or mutants in a 1:8:5 ratio (U2OS) or 1:10:5 (HeLa). This ratio was based on the functional rescue by wild-type in relation to the shRNA. Marker plasmids were pSpectrin-GFP for flow cytometry and pEYFP-H2B for live cell imaging and immunofluoresence. pBabe-puro was used for expression level studies in U2OS and HeLa, transfected cells were selected by treatment with puromycin.
Cell culture
HeLa and U2OS cells were grown in DMEM supplemented with 8%FBS and pen/strep (50μg/ml). EBV-transformed lymphoblastoid cell lines were obtained from cell repositories (European Collection of Cell Cultures (ECACC), Salisbury, UK, or Coriell Institute for Medical Research, New Jersey, USA). Fibroblasts were obtained as primary cell lines and were subsequently SV40-transformed using standard procedures. All samples were obtained with informed consent from the family and under multicenter ethics approval (MREC05/02/17).
SV40 transformed fibroblasts and EBV transformed lymphoblastoids were grown in DMEM and RPMI respectively supplemented with 10%FBS and pen/strep (50μg/ml). HTR34 cells were created by infection of HeLa cells stably expressing Tet repressor (a gift of M. Timmers) with retrovirus carrying pSuperior-retro-puro-BUBR1 and grown on medium with TetSystem-approved FBS (Clontech).
Detailed Materials and Methods can be found in the Supplemental material.
Results & Discussion
MVA patient cell lines are checkpoint deficient and have chromosome alignment defects
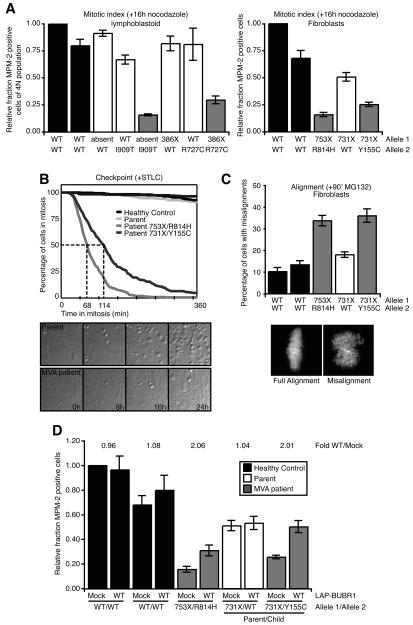
Mono- and biallelic mutations in BUB1B were identified in British and Japanese MVA patients (2, 3). Previous studies had shown that centromere cohesion was lost in colcemid-treated MVA fibroblasts (3, 25), and that colcemid addition to the media of fibroblasts from Japanese MVA patients did not cause an accumulation in mitosis and increased the amount of cells with micronuclei (3). To examine the fidelity of chromosome segregation in MVA patients, we established lymphoblastoid and fibroblast cell lines of MVA patients as well as of their respective parents or unrelated healthy individuals (an overview is presented in Supplementary Table 1). Flow cytometric analysis combined with time-lapse imaging revealed that the MVA patient cells failed to accumulate in mitosis in checkpoint dependent manner after treatment with the spindle depolymerizing drug nocodazole (Fig. 1A) or EG5 inhibitor S-trityl-L-cysteine (STLC) (26, 27) (Fig. 1B). Whereas control STLC treated cells were delayed in mitosis for the duration of the experiment, 50% of the population of MVA patient cell lines exited within 68 (753X/R814H) and 114 min (731X/Y155C). Furthermore MVA patient cells, treated with proteasome inhibitor MG132 to prevent mitotic exit, showed a 2-3 fold increase in the amount of chromosome misalignment compared to control lines (Fig. 1C).
Fig. 1. Checkpoint deficiency and chromosome alignment defects in MVA patient cell lines are directly due to dysfunctional BUBR1.
(A) Flow cytometric analysis of MPM-2 positivity of MVA lymphoblastoid (left) and fibroblast (right) cell lines, treated with nocodazole for 16 hours. Graphs represent the fraction of MPM-2 positive cells of 4N population relative to healthy control cells (average of at least 3 (left) or 5 (right) experiments, ± s.e.m.). (B) Analysis of mitotic delay in MVA fibroblast cell lines treated with STLC and imaged by differential interference contrast (DIC) microscopy. Graph indicates the percentage of cells in mitosis in time (total of 3 experiments, total of 300 cells). (C) Chromosome alignment in MVA fibroblast cell lines treated with MG132 for 90 minutes. Graph indicates the fraction of mitotic cells with full alignment relative to healthy control cells (average of 4 experiments, at least 70 cells counted per experiment, ± s.e.m.). (D) Flow cytometric analysis of MPM-2 positivity of MVA fibroblast cell lines, transfected with control or LAP-BUBR1 and treated with nocodazole for 16 hours. Graphs represent the fraction of MPM-2 positive cells of 4N population relative to healthy control cells (average of 5 experiments, ± s.e.m.).
Strikingly, restoration of high levels of BUBR1 by transient expression of Localization and Affinity Purification (LAP)-tagged BUB1B cDNA (23), induced a 2-fold increase in the response to nocodazole in patient cells (Fig. 1D). The amount of mitotic cells was however unchanged in LAP-BUBR1 expressing control cells (Fig. 1D). Importantly, comparison of the relative activity of the mitotic checkpoint of cells from one of the MVA patients (731X/Y155C) to that of cells of the relevant parental control (731X/WT) showed that mitotic checkpoint activity was almost fully restored to parental levels upon re-expression of BUBR1 in the patient cells (Fig. 1D). Together, these findings show that is the two mitotic processes in which BUBR1 participates, mitotic checkpoint signaling and chromosome alignment, are severely impaired in cells from MVA patients.
All kinase domain-localized mutations in BUBR1 reduce overall BUBR1 protein abundance
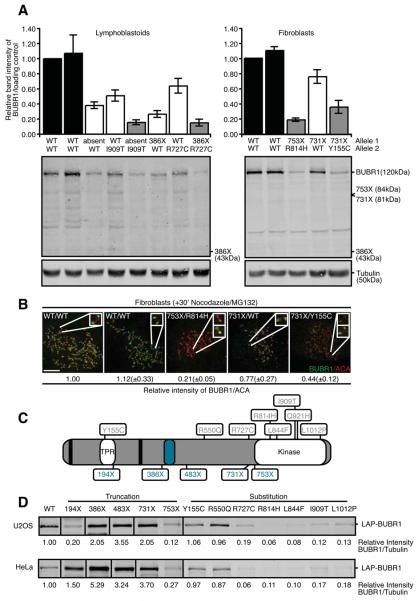
Most BUBR1 substitution mutations identified in British MVA patients are located in or in close proximity to the region encoding the kinase domain of BUBR1. This has inspired the hypothesis that BUBR1 kinase activity and/or substrate recognition are important for chromosome segregation and are affected by the MVA mutations. Although an intriguing hypothesis, analysis of BUBR1 protein expression showed that abundance of full-length substitution mutant BUBR1 protein in all patient lines (absent/I909T, 386X/R727C, 753X/R814H or 731X/Y155C) was decreased 2-6 fold compared to wild-type alleles (Fig. 2A). Interestingly, BUBR1 protein abundance was most severely decreased when mutations occurred in or near the kinase domain, but less by a mutation in the N-terminal TPR domain (Fig. 2A). Importantly, levels of full-length BUBR1 in the parent cell lines, carrying one mutant allele, were consistently in between those of patients and unrelated healthy controls (Fig. 2A). Low overall protein abundance translated to low levels of BUBR1 on unattached kinetochores (Fig. 2B). Enhancing contrast settings, however, showed that the residual protein could be recruited to kinetochores, indicative of at least partial functionality of mutant BUBR1 (Fig. 2B lower insets). Similar results were obtained by expression of wild-type and mutant LAP-BUBR1 in human tissue culture cells. This approach allowed analysis of all MVA mutations including those of which no patient derived cell lines were available (a schematic representation of the mutants is shown in Fig. 2C). Please note that fusing the LAP-tag to the amino terminus of BUBR1 did not affect its function or subcellular localization (see below). All mutant proteins with substitution mutations in or near the kinase domain showed a 5-10 fold decrease in levels compared to wild-type BUBR1 in HeLa and U2OS cells (Fig. 2D and S1). These data show that a wide range of mutations in or near the kinase domain can destabilize BUBR1. Importantly, the two substitution mutations located away from the kinase domain, Y155C and R550Q, did not affect protein levels when compared to wild-type BUBR1 (Fig. 2D).
Fig. 2. Low BUBR1 protein abundance in MVA patient cells.
(A) Quantitative immunoblot of lysates of immortalized MVA patient-derived lymphoblastoid (left) and fibroblast (right) cell lines. Antibodies can recognize truncated proteins if overexpressed in HeLa. Graphs indicate the band intensity of BUBR1/Tubulin relative to healthy control cells (average of 4 experiments, ± s.e.m.). (B) Immortalized fibroblasts treated with nocodazole and MG132 for 30 minutes, immunostained for BUBR1 and centromeres (ACA). Upper insets show representative kinetochore pairs, lower insets show the same kinetochore pairs with increased intensity. Quantification of BUBR1/ACA on kinetochores relative to healthy control cells is shown (average of 3 experiments, 25 kinetochores were quantified per cell for 5 cells per experiment, ± s.e.m.). (C) Schematic representation of BUBR1. Biallelic mutations found in MVA patients, KEN-box motives (black stripes) and BUB3-binding domain (blue oval) are indicated. (D) Quantitative immunoblot of lysates of HeLa and U2OS cells transfected with wild-type or mutant LAP-BUBR1 together with pBabe-Puro and selected for transfection by puromycin. Band intensity of BUBR1/Tubulin relative to wild-type is indicated (average of at least 3 experiments, bands were aligned for visualization purposes, full blot is presented in supplemental Figures (S1A)).
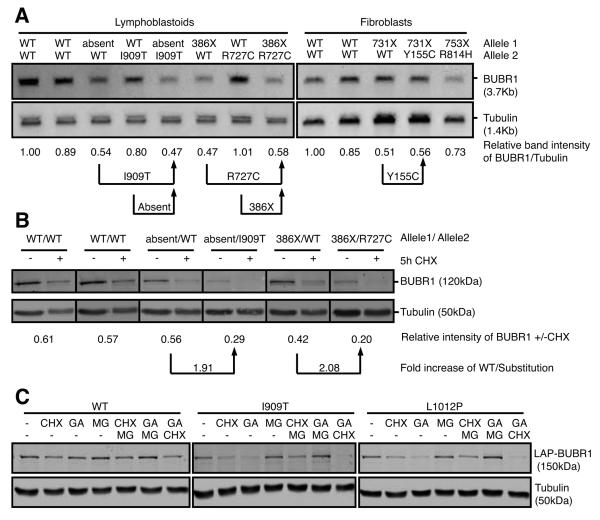
Substitution mutations affect BUBR1 protein stability
To investigate the cause of low BUBR1 protein abundance, mRNA levels and protein turnover were measured in patient cells for which relevant parental control cells were available to allow analysis of specific single gene products. For instance, comparison of mRNA from 386X/R727C cells to that from 386X/WT cells provides information on the level of R727C transcript relative to wild-type. As shown by Northern blotting, mRNA expression of three substitution alleles, I909T, R727C and Y155C, was unaffected (Fig. 3A). We next addressed whether the substitution mutations, rather than affecting mRNA levels, promote protein turnover. Treatment of the lymphoblastoid cell lines with the translation inhibitor cycloheximide for 5 hours showed that the protein turnover of the substitution mutant proteins I909T and R727C was increased ~2-fold compared to wild-type protein (Fig. 3B). Additionally, U2OS cells stably expressing LAP-tagged wild-type, I909T or L1012P mutant BUBR1 were treated with inhibitors to three major pathways that control protein abundance (protein translation (cycloheximide, CHX), folding (geldanamycin, GA) and degradation (MG132)). As expected from data of the patient cells, treatment with CHX reduced mutant levels more severely than wild-type (Fig. 3C). Furthermore, whereas levels of wild-type BUBR1 showed only minor changes, levels of both mutant proteins were severely decreased after 5h treatments with the HSP90 inhibitor (Fig. 3C, GA). Importantly, combined treatments of CHX and GA removed virtually all mutant BUBR1 protein (Fig. 3C). This shows that stability of these substitution mutants but not wild-type relied on heat-shock proteins and suggested that the mutations cause protein misfolding. In support of this, inhibition of proteasomal degradation with MG132 (Fig. 3C, MG) prevented the enhanced protein turnover caused by HSP90 inhibition (Fig. 3C, MG+GA). Thus, HSP90 activity is needed for folding of BUBR1 substitution mutants and for preventing their clearance via proteasomal degradation. Thus low abundance of substitution mutants is, at least in part, caused by decreased protein stability.
Fig. 3. All kinase-domain-localized MVA mutations affect overall BUBR1 protein abundance.
(A) Northern blot of mRNA isolated from immortalized MVA lymphoblastoid (left) and fibroblast (right) cell lines. Band intensity of BUBR1/Tubulin relative to healthy control cells is shown (representative of 2 experiments). (B) Quantitative immunoblot of lysates of immortalized MVA lymphoblastoid cell lines. Band intensity of BUBR1/Tubulin of cells treated cells with cycloheximide for 5 hours relative to untreated cells is shown (average of 3 experiments). (C) Quantitative immunoblot of lysates of U2OS cells stably expressing wild-type or mutant LAP-BUBR1 together with BUBR1 shRNA, treated with DMSO (−), cycloheximide (CHX), geldanamycin (GA), MG132 (MG) or combinations for 5 hours (representative experiment is shown).
None of the truncated proteins, 386X, 731X and 753X, could be detected in lysates of MVA patient cell lines or in parental controls (Fig. 2A). However most truncations, except LAP-194X and LAP-753X, showed expression levels comparable to or higher than wild-type BUBR1 in U2OS and HeLa cells (Fig. 2D and S1). This indicates that truncations also affected BUBR1 protein levels, although not necessarily by decreased protein stability. In patient cells, mRNA from the 386X allele could not be detected and although the relevant parental controls are missing, the 2-fold reduction in mRNA levels of 731X/WT relative to healthy controls suggested that transcripts from the 731X allele were also absent (Fig. 3A). These findings suggested that the premature STOP codons decreased transcript stability and thereby contributed to reducing BUBR1 levels. This was in agreement with a previous suggestion that expression of truncated BUBR1 proteins may be prevented by nonsense-mediated mRNA decay (NMD, see for review (28)) (3), and supported by the finding that both 386X and 731X are expressed when NMD is circumvented by plasmid-driven expression in HeLa or U2OS cells. It is, however, unclear what the status of 194X, 483X or 753X is in patient cells. Plasmid-driven expression in cancer cells of 194X and 753X shows levels that are nonetheless lower than wild-type BUBR1, suggesting that the truncations may also affect protein stability, however BUBR1 abundance of truncation mutants is mainly affected on mRNA level.
MVA-associated mutations in the kinase domain of BUBR1 cause chromosome segregation defects primarily by lowering protein abundance
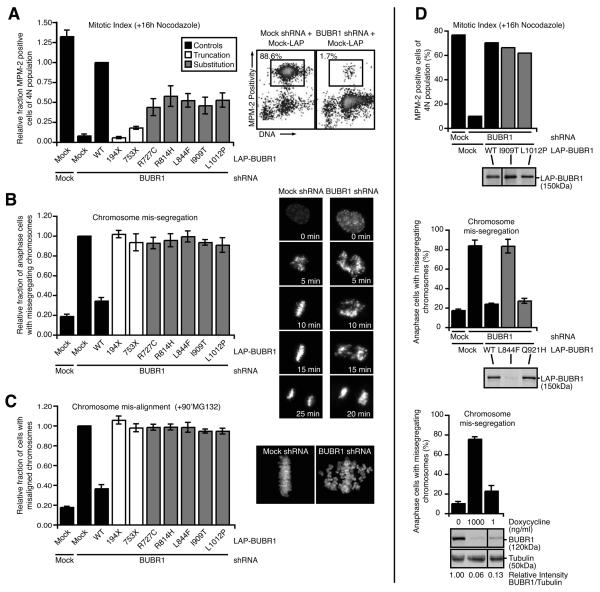
The previous analyses showed that the MVA-associated substitution and truncation mutations in BUBR1 could be divided in two categories: those that affect its protein abundance, and those that don't. To examine if mutants from the ‘low abundance’ category compromise BUBR1 function, various aspects of chromosome segregation were examined in cells in which endogenous BUBR1 was transiently replaced with exogenous epitope-tagged mutant BUBR1 (23). As a consequence of checkpoint inactivation, cells depleted of BUBR1 were unable to delay mitosis in the presence of nocodazole (Fig. 4A), and over 80% of the cells showed massive chromosome missegregations as determined by time-lapse microscopy (Fig. 4B). Checkpoint function was restored by expression of wild-type, shRNA-insensitive LAP-BUBR1, but the truncated proteins LAP-194X and LAP-753X as well as the five kinase-domain-localized substitution mutants were unable to sustain a mitotic delay in response to spindle depolymerization and caused severe chromosome missegregations (Fig. 4B). In addition, these mutants also affected the ability of chromosomes to form stable microtubule attachments: While wild-type LAP-BUBR1 restored efficient chromosome alignment in BUBR1-depleted cells, neither of the unstable truncations nor the five substitution mutants could restore chromosome alignment in BUBR1-depleted cells (Fig. 4C).
Fig. 4. Low BUBR1 levels is the primary cause of BUBR1 dysfunction by MVA Mutations in the kinase domain.
(A) Flow cytometric analysis of MPM-2 positivity of U2OS cells transfected with control (Mock) or BUBR1 shRNA in combination with control or RNAi insensitive LAP-BUBR1 cDNA, treated with nocodazole for 16 hours. Boxes indicate percentage of cells positive for MPM-2. Graph represents the fraction of MPM-2 positive cells of 4N population relative to the LAP-BUBR1 WT control (average of at least 3 experiments, ± s.e.m.). (B) Analysis of chromosome segregation by live imaging of U2OS expressing H2B-EYFP and transfected as in 4A. Graph indicates the fraction of cells with chromosome mis-segregations relative to BUBR1 depleted cells (average of 3 experiments, a total of at least 238 cells, ± s.e.m.). (C) HeLa cells, transfected as in 4A, were treated with MG132 for 90 minutes and analyzed for chromosome alignment. Graph indicates the fraction of cells with misaligned chromosomes relative to BUBR1 depleted cells (average of 3 experiments, at least 90 cells counted per experiment, ± s.e.m.). (D, upper) Flow cytometric analysis of MPM-2 positivity of U2OS cells transfected control (Mock) or BUBR1 shRNA in combination with control or RNAi insensitive LAP-BUBR1 cDNA, substitution mutants were overexpressed to levels comparable to wild-type, and treated with nocodazole for 16 hours. Graph represents the percentage of MPM-2 positive cells of 4N population. Immunoblot shows the expression of wild-type and mutant LAP-BUBR1 in lysates from the same experiment (1 representative experiment). (D, middle) Analysis of chromosome segregation by live imaging of U2OS expressing H2B-EYFP and transfected as in 4A. Graph indicates the percentage of cells with chromosome mis-segregations (average of 3 experiments, a total of at least 270 cells, ± s.e.m.). Immunoblot shows the expression of wild-type and mutant LAP-BUBR1 in lysates from a representative experiment. (D, lower) Analysis of chromosome segregation by live imaging of HeLa cells expressing inducible BUBR1 shRNA and H2B-EYFP treated with 0, 1 or 1000 ng/ml doxycycline. Graph indicates the percentage of cells with chromosome mis-segregations (average of 3 experiments, a total of at least 350 cells, ± s.e.m.). Quantitative immunoblot shows the expression of BUBR1 and Tubulin in lysates from a representative experiment, band intensity of BUBR1/Tubulin relative to untreated control is indicated (average of 3 experiments)
The inability of the ‘low abundance’ mutants to restore BUBR1 functionality on our assays raised the possibility that these disease-associated mutations affected chromosome segregation primarily by lowering BUBR1 protein abundance without affecting BUBR1 function directly. Three lines of evidence strongly support this. First, forced overexpression of the poorly expressed substitution mutants I909T and L1012P to levels comparable to wild-type BUBR1 fully restored the response to nocodazole (Fig. 4D, upper graph). This showed that these mutations do not impose significant constraints on BUBR1 function other than affecting overall BUBR1 protein abundance. Second, of the two substitution mutations that were reported to occur within one allele (L844F and Q921H) (2), L844F was poorly expressed and unable to restore any of the defects in BUBR1-depleted cells (Fig. 4D middle graph, S2A and S2B). In striking contrast, the Q921H mutation was indistinguishable from wild-type LAP-BUBR1, both on protein level and functionality. This suggests that the L844F substitution is the likely cause of mitotic defects in the patient carrying the doubly mutated allele. Third, the amount of segregation errors is highly sensitive to the amount of BUBR1 present in the cell. BUBR1 abundance was reduced to different levels by titration of doxycyline to a HeLa cell line stably carrying inducible expression of an shRNA to BUBR1. Whereas saturating amounts of doxycyline reduced BUBR1 protein to ~6% and caused chromosome mis-segregation in the majority of cells, low amounts of doxycyline, while still significantly reducing BUBR1 levels to ~13%, had virtually no impact on the fidelity of chromosome segregation (Fig. 4D, lower graph). These findings support the hypothesis that mitotic checkpoint deficiency in MVA patient cells carrying mutations in the BUBR1 kinase domain is caused by low BUBR1 levels. Interestingly, of the two newly identified substitution mutations reported here (Y155C and R727C), only the mutation located near the kinase domain, R727C, confers a similar level of protein instability. As there is no information on structural properties of BUBR1, it is at present unclear why these specific mutations affect protein abundance. Interestingly, however, mapping of the orthologous residues in the recently published crystal structure of the highly similar BUB1 kinase (29) suggests that these residues are needed for the overall integrity of the BUBR1 kinase domain (unpublished observation). From these comparisons, it is to be expected that substitution of these amino acids for their respective disease-causing counterparts will disturb kinase domain folding and stability of the protein. In support of this, we have observed a two-fold increase in the turnover of BUBR1 as a result of the substitution mutations. Furthermore, inhibition of HSP90, a chaperone that aids protein folding, further decreased protein levels of substitution mutants. This finding indicates a strong tendency of kinase domain-located substitution mutants to misfolding, which raises the interesting possibility that this region of BUBR1 is highly sensitive to amino-acid substitutions. Detailed insights into BUBR1 structure will be crucial to further investigate this.
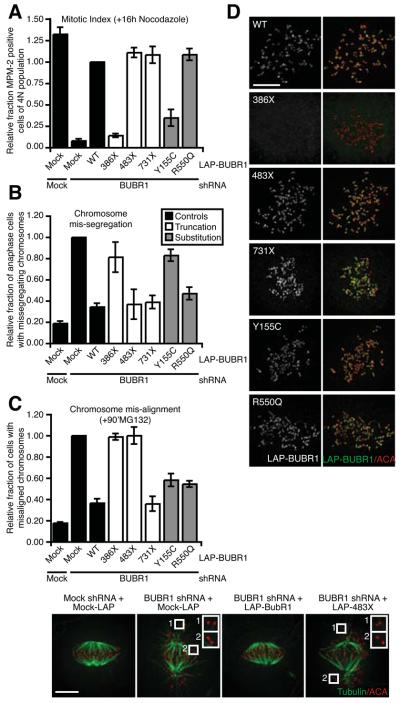
Stable MVA mutants cause direct functional BUBR1 defects and reveal novel aspects of BUBR1's roles in mitosis
We next examined functionality of the MVA mutations in BUBR1 that have no significant effect on BUBR1 protein levels. Again, all mutants were assayed on the capability to rescue mitotic checkpoint defects and chromosome misalignment in cells depleted of endogenous BUBR1 (as in Fig. 4A-4C). Analysis of the two stable substitution mutants revealed expression of LAP-R550Q rescued both functions of BUBR1 (Fig. 5A-5C). Interestingly, however, the Y155C substitution mutation that is located in the amino-terminal TPR domain was a separation-of-function allele: LAP-Y155C restored chromosome alignment to BUBR1-depleted cells (Fig. 5C) but failed to reconstitute mitotic checkpoint activity (Fig. 5A and 5B). The TPR domain of BUBR1 is essential for BUBR1 functionality in checkpoint signaling via interaction with KNL-1/Blinkin (30, 31). Similar to previously reported BUBR1 TPR mutants, LAP-Y155C properly localized to unattached kinetochores (Fig. 5D).
Fig. 5. The MVA-associated BUBR1 mutants that do not affect protein stability directly compromise BUBR1 function.
(A) Flow cytometric analysis of MPM-2 positivity of U2OS cells transfected as in 4A, treated with nocodazole for 16 hours. Graph represents the fraction of MPM-2 positive cells of 4N population relative to the LAP-BUBR1 WT control (average of at least 3 experiments, ± s.e.m.). (B) Analysis of chromosome segregation by live imaging of U2OS expressing H2B-EYFP and transfected as in 4A. Graph indicates the fraction of cells with chromosome mis-segregations relative to BUBR1 depleted cells (average of 3 experiments, a total of at least 268 cells, ± s.e.m.). (C) HeLa cells, transfected as in 4A, were treated with MG132 for 90 minutes and analyzed for chromosome alignment (left) or cold-stable microtubules (right). Graph indicates the fraction of cells with misaligned chromosomes relative to BUBR1 depleted cells (average of 3 experiments, at least 100 cells counted per experiment, ± s.e.m.). Insets show representative unattached kinetochore pairs, scale bar is 5μm. (D) HeLa cells transfected as in 4A, treated with nocodazole and MG132 for 30 minutes, immunostained for LAP-BUBR1 and centromeres (ACA), scale bar is 5μm.
Expression of LAP-386X, a protein that lacks the kinase domain, the BUB3-interaction domain and a CDC20-binding region, could not restore checkpoint activity nor chromosome alignment (Fig. 5A-5C). As expected from the absence of the BUB3-interaction domain, LAP-386X could not localize to unattached kinetochores (Fig. 5D). In contrast, the slightly longer protein LAP-483X that has the BUB3-binding region included, could be recruited to unattached kinetochores (Fig. 5D) and was fully functional in restoring mitotic checkpoint activity (Fig. 5A and 5B). This indicated that the extended amino-terminus of BUBR1 is sufficient for sustained mitotic checkpoint activity. LAP-483X, however, could not rescue chromosome misalignments in BUBR1-depleted cells, and as a result LAP-483X-reconstituted cells had a significantly extended mitosis (Fig. S3A and S3B), indicative of a sustained mitotic checkpoint response to unattached chromosomes. Thus, amino acids 483-1050 of human BUBR1 were indispensable for attachment of chromosomes to the mitotic spindle (Fig. 5C). This is in agreement with studies in Saccharomyces cerevisiae and mice describing two conserved KEN motives, both of which are intact in 483X, that are essential for the budding yeast and murine checkpoints (12-14). However unlike murine BUBR1, which can act as functional checkpoint component without localizing to unattached kinetochores (14), amino acid 1-386 of human BUBR1 lacking the BUB3-binding domain, essential for kinetochore localization is not sufficient.
Surprisingly, the kinase domain did not contribute to chromosome alignment; since LAP-731X has no kinase domain yet is fully capable of supporting the microtubule attachment function of BUBR1 (Fig. 5C). Furthermore, LAP-731X restored the response to nocodazole as well as proper chromosome segregation in unperturbed mitosis (Fig. 5A and 5B), suggesting that, at least in our assays and in these cells, the kinase domain of human BUBR1 is not required for the mitotic checkpoint nor for stable microtubule attachments. Combined, these findings have two important implications: First, neither the kinase activity nor the 265 amino-acid kinase domain are required for establishing mitotic checkpoint activity or stable microtubule attachments. Second, stable kinetochore-microtubule attachments rely on the central part of human BUBR1 (484-715 (note that 731X contains only 715 amino acids of BUBR1 sequence)). If 731X restores all functions of BUBR1, what is then the role, if any, of the kinase domain? Although careful live cell analyses of 731X did not reveal subtle defects in our experiments, it cannot be excluded that such defects would become apparent under certain conditions. However our demonstration that BUBR1 is destabilized by all kinase-domain-localized MVA mutations, offers an alternative explanation for the role of the BUBR1 kinase domain. Our data show that the integrity of the kinase domain needs to be preserved in order for chromosome segregation to be free of errors. This opens the possibility that the kinase domain is involved in regulating BUBR1 protein stability in a non-catalytic fashion, perhaps through ATP binding or cofactor association (Fig. S4).
A model for aneuploidy by biallelic mutations in BUBR1
The appearance of aneuploidy in MVA patients carrying the specific combination of biallelic mutations is explained by a combination of a reduction in the efficiency of chromosome attachment with an impaired mitotic checkpoint response. Our molecular analyses provide a rationale for how the specific combinations of mutations in each patient can cause these defects. In one group of patients, both mutations in a biallelic combination reduce BUBR1 protein levels to such an extent that both chromosome attachment and the mitotic checkpoint are significantly impaired, while in other patients the same outcome is reached by a reduction in protein combined with direct functional impairment (Fig. S5). In conclusion, we propose that chromosomal instability in MVA patients carrying BUB1B mutations is a result of low BUBR1 protein abundance. It will be of interest to examine whether genetic or epigenetic alterations of BUBR1 protein expression may also underlie other pathologies.
Supplementary Material
Acknowledgements
The authors thank the families that provided samples and the clinicians that recruited them to the research, including A. Kumar, N. Gregerson, A. Kidd and A. Plaja. We thank Helmut Hanenberg for undertaking the SV40 transformation of fibroblasts from MVA patients, Stephen Taylor and Marc Timmers for reagents, Livio Kleij and Hetty van Teeffelen for technical support, Holger Rehmann for helpful input, Rene Medema and Susanne Lens for critically reading the manuscript and the Lens, Medema and Kops labs for insights and discussion. This work was supported by grants awarded to GK (UU-2006-3664, VIDI-91776336), SH and NR are supported by Cancer Research UK and the Institute of Cancer Research (UK).
References
- 1.Micale M, Schran D, Emch S, Kurczynski T, Rahman N, Van Dyke D. Mosaic variegated aneuploidy without microcephaly: Implications for cytogenetic diagnosis. Am J Med Genet A. 2007;143A:1890–3. doi: 10.1002/ajmg.a.31848. [DOI] [PubMed] [Google Scholar]
- 2.Hanks S, Coleman K, Reid S, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–61. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 3.Matsuura S, Matsumoto Y, Morishima K, et al. Monoallelic BUB1B mutations and defective mitotic-spindle checkpoint in seven families with premature chromatid separation (PCS) syndrome. Am J Med Genet A. 2006;140:358–67. doi: 10.1002/ajmg.a.31069. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka TU, Desai A. Kinetochore-microtubule interactions: the means to the end. Curr Opin Cell Biol. 2008;20:53–63. doi: 10.1016/j.ceb.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–93. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 6.Taylor SS, Ha E, McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–36. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan GK, Jablonski SA, Sudakin V, Hittle JC, Yen TJ. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J Cell Biol. 1999;146:941–54. doi: 10.1083/jcb.146.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell. 2009;16:105–17. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudakin V, Ganoth D, Dahan A, et al. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–97. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–88. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 12.Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21:655–67. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King EM, van der Sar SJ, Hardwick KG. Mad3 KEN boxes mediate both Cdc20 and Mad3 turnover, and are critical for the spindle checkpoint. PLoS ONE. 2007;2:e342. doi: 10.1371/journal.pone.0000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malureanu LA, Jeganathan KB, Hamada M, Wasilewski L, Davenport J, van Deursen JM. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev Cell. 2009;16:118–31. doi: 10.1016/j.devcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuura S, Ito E, Tauchi H, Komatsu K, Ikeuchi T, Kajii T. Chromosomal instability syndrome of total premature chromatid separation with mosaic variegated aneuploidy is defective in mitotic-spindle checkpoint. Am J Hum Genet. 2000;67:483–6. doi: 10.1086/303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ditchfield C, Johnson VL, Tighe A, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–80. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lampson MA, Kapoor TM. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat Cell Biol. 2004;7:93–8. doi: 10.1038/ncb1208. [DOI] [PubMed] [Google Scholar]
- 18.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–85. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 19.Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–67. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–27. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. J Cell Sci. 2008;121:3859–66. doi: 10.1242/jcs.039537. [DOI] [PubMed] [Google Scholar]
- 22.Jelluma N, Brenkman AB, van den Broek NJ, et al. Mps1 Phosphorylates Borealin to Control Aurora B Activity and Chromosome Alignment. Cell. 2008;132:233–46. doi: 10.1016/j.cell.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 23.Kops GJ, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci USA. 2004;101:8699–704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheeseman IM, Desai A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Science's STKE. 2005;2005:pl1. doi: 10.1126/stke.2662005pl1. [DOI] [PubMed] [Google Scholar]
- 25.Callier P, Faivre L, Cusin V, et al. Microcephaly is not mandatory for the diagnosis of mosaic variegated aneuploidy syndrome. Am J Med Genet A. 2005;137A:204–7. doi: 10.1002/ajmg.a.30783. [DOI] [PubMed] [Google Scholar]
- 26.Skoufias DA, DeBonis S, Saoudi Y, et al. S-trityl-L-cysteine is a reversible, tight binding inhibitor of the human kinesin Eg5 that specifically blocks mitotic progression. J Biol Chem. 2006;281:17559–69. doi: 10.1074/jbc.M511735200. [DOI] [PubMed] [Google Scholar]
- 27.Hauf S, Cole RW, LaTerra S, et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–94. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fasken M, Corbett A. Process or perish: quality control in mRNA biogenesis. Nat Struct Mol Biol. 2005;12:482–8. doi: 10.1038/nsmb945. [DOI] [PubMed] [Google Scholar]
- 29.Kang J, Yang M, Li B, et al. Structure and Substrate Recruitment of the Human Spindle Checkpoint Kinase Bub1. Mol Cell. 2008;32:394–405. doi: 10.1016/j.molcel.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiyomitsu T, Obuse C, Yanagida M. Human Blinkin/AF15q14 Is Required for Chromosome Alignment and the Mitotic Checkpoint through Direct Interaction with Bub1 and BubR1. Dev Cell. 2007;13:663–76. doi: 10.1016/j.devcel.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 31.D' Arcy S, Davies OR, Blundell TL, Bolanos-Garcia VM. Defining the molecular basis of BUBR1 kinetochore interactions and anaphase-promoting complex/cyclosome (APC/C)-CDC20 inhibition. Journal of Biological Chemistry. 2010:1–23. doi: 10.1074/jbc.M109.082016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.