Abstract
Monocytes and macrophages are critical effectors and regulators of inflammation and the innate immune response, the immediate, pre-programmed arm of the immune system. Dendritic cells initiate and regulate the highly pathogen-specific adaptive immune responses, and are central to the development of immunologic memory and tolerance. Recent in vivo experimental approaches in the mouse have unveiled new aspects of the developmental and lineage relationships among these cell populations. Despite this, the origin and differentiation cues for many tissue macrophages, monocytes, and dendritic cell subsets in mice, and the corresponding cell populations in humans, remain to be elucidated.
White blood cells or leukocytes are a diverse group of cell types that mediate the body's immune response. They circulate through the blood and lymphatic system and are recruited to sites of tissue damage and infection. Leukocyte subsets are distinguished by functional and physical characteristics. They have a common origin in hematopoietic stem cells and develop along distinct differentiation pathways in response to internal and external cues. The mononuclear phagocyte system represents a subgroup of leucocytes originally described as a population of bone marrow-derived myeloid cells that circulate in the blood as monocytes and populate tissues as macrophages in the steady state and during inflammation (1). In different tissues they can show significant heterogeneity with respect to phenotype, homeostatic turnover and function. The discovery of dendritic cells (DCs) as a distinct lineage of mononuclear phagocytes, specialized in antigen presentation to T cells and the initiation and control of immunity (2), revealed additional roles of these cells in shaping the immune response to pathogens, vaccines and tumors, as well as additional heterogeneity. Whereas a detailed map of the relationship between monocytes, DCs and their progenitors begins to emerge, other areas like the origin and renewal of tissue macrophage subsets remain less defined.
Monocytes (Fig. 1A) circulate in the blood, bone marrow, and spleen and do not proliferate in a steady state (3, 4). Monocytes represent immune effector cells, equipped with chemokine receptors and pathogen recognition receptors that mediate migration from blood to tissues during infection. They produce inflammatory cytokines and take up cells and toxic molecules. They can also differentiate into inflammatory DCs or macrophages during inflammation, and possibly, less efficiently, in the steady state. Migration to tissues and differentiation to inflammatory DC and macrophages is likely determined by the inflammatory milieu and pathogen associated pattern recognition receptors (5).
Fig. 1.

(A). Still frames from time-lapse intravital confocal microscopy of a crawling monocytes (arrow) and perivascular macrophages in the dermis (courtesy of F. Geissmann, for details see (52)) (B). Confocal microscopy image of the spleen from mice grafted with MDPs. DCs derived from the MDP graft are labeled in green (CD45.2 staining) and host-derived marginal zone metallophilic macrophages are labelled in red with CD169/Sialoadhesin (courtesy of F. Geissmann, for details see (24)). (C). Dividing LCs in the epidermis (courtesy of I. Chorro & F. Geissmann, for details see (35)). (D). Confocal micrograph of aortic whole mount from Cx3cr1GFP/GFPApoe-/- mouse, viewed from endothelial side (courtesy of K. Ley). (E). Intra-vital two photon microscopic image of intestinal villi of CD11c+ cells-depleted mice reconstituted by grafts of bone marrow monocytes, yielding red and green fluorescent lamina propria cells, respectively (courtesy S. Jung and G. Shakhar, for details see (26)).
Macrophages (Fig. 1, A and B) are resident phagocytic cells in lymphoid and non-lymphoid tissues, and are believed to be involved in steady-state tissue homeostasis via the clearance of apoptotic cells, and the production of growth factors. Macrophages are equipped with a broad range of pathogen recognition receptors that make them efficient at phagocytosis and induce production of inflammatory cytokines (6). The developmental origin and the function of tissue macrophage subsets, such as microglia (macrophages in the central nervous system), dermal macrophages (Fig. 1A), and splenic marginal zone and metallophilic macrophages (Fig. 1 B), remain insufficiently understood.
Classical DCs (cDCs) (Fig. 1, B and C) are specialized antigen-processing and presenting cells, equipped with high phagocytic activity as immature cells and high cytokine producing capacity as mature cells (7, 8). Although present in human circulation, cDCs are rare in mouse blood. cDCs are highly migratory cells that can move from tissues to the T-cell and B-cell zones of lymphoid organs via afferent lymphatics and high endothelial venules. cDCs regulate T cell responses both in the steady-state and during infection. They are generally short-lived and replaced by blood-borne precursors (Fig. 1B) (9, 10). Of note, they are distinct from Langerhans cells (LCs, DCs found in the epidermis) (Fig. 1C), which are not replaced by blood-borne cells at the steady state (11). Individual myeloid cell populations may share features of DC and macrophages and can be difficult to ascribe to one or the other cell type (Fig. 1 D and E).
Plasmacytoid DCs (PDCs) differ from cDCs in that they are relatively long lived and a proportion of them carry characteristic immunoglobulin rearrangements (12). They are present in the bone marrow and all peripheral organs. PDCs are specialized to respond to viral infection with a massive production of type I interferons (IFN), however, they also can act as antigen presenting cells and control T cell responses(13).
The development of the mononuclear phagocyte system is controlled by cytokines - small secreted proteins that promote cell-cell communication and can act as growth and differentiation factors. The generation of monocytes, macrophages and - to some extent - DCs is dependent on the cytokine and hematopoietic growth factor receptor Csf1r (c-fms, M-CSFR, CD115), expressed in monocytes, macrophages, and mononuclear phagocyte precursors (14-17). Characterization of op/op mice, a spontaneous mutant lacking a functional Csf1 gene, has revealed both the role of Csf1 in the development of mononuclear phagocytes, and also their broad functions. (17). The known ligands of Csf1r, Csf1/M-CSF (18)and interleukin (IL)-34 (19), are likely both important for the development of the mononuclear phagocyte lineage, as M-CSF-deficient mice have a milder phenotype than Csf1r-deficient mice. Another cytokine receptor closely related to Csf1r, fms-related tyrosine kinase 3 (FLT3 also known as Flk2) receptor, is critical for the development of cDCs and PDCs (10, 20-22).
Following on the early work on Csf1r, a large amount of work has been devoted in the past 20 years to investigating the origin and differentiation pathways of monocytes, macrophages, and DCs. This recently culminated in a series of studies that unveiled what is likely to be the main pathway for the development of DCs, monocytes and macrophages from bone marrow progenitors both in the steady state and during inflammation.
What are the precursors of monocytes, macrophages and DCs?
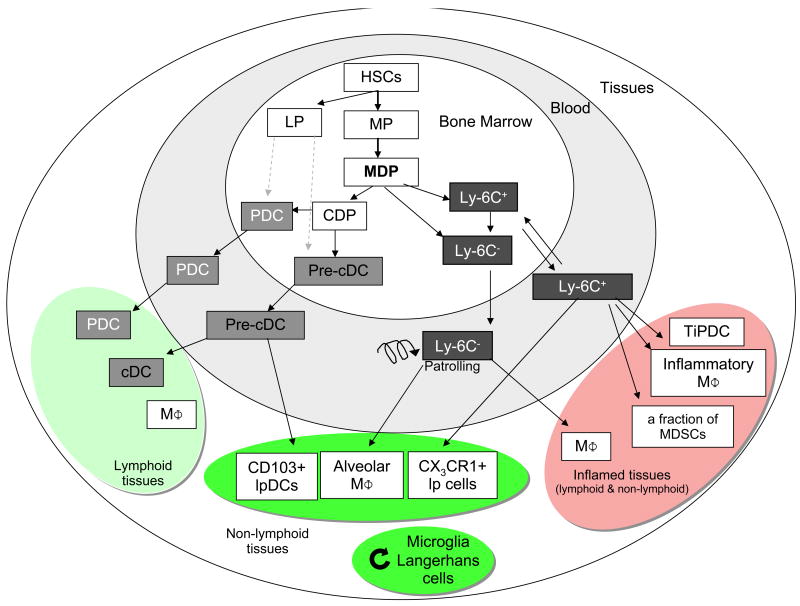
The use of multi-color fluorescence activated cell sorting has allowed idenfication of progenitors and differentiated cell populations on the basis of the expression of multiple cell surface proteins (Table S1). These cells can then be isolated from mouse bone marrow, blood or spleen, and grafted into recipient mice to determine lineage relationships. This methodology was recently used to delineate the DC developmental program in vivo (3, 9, 23-26) (see Fig. 1). Current models propose that blood monocytes, many macrophage subsets and most DCs originate in vivo from hematopoietic stem cell-derived progenitors with myeloid restricted differentiation potential (Fig. 2). Successive commitment steps in the bone marrow include common myeloid progenitors (CMPs), granulocyte-macrophage precursors (GMPs) and macrophage/DC progenitors (MDPs). MDPs are a subset of proliferating cells in the bone marrow that share phenotypic characteristics with myeloid precursor populations and give rise to many macrophages and DC subsets, but largely cannot differentiate into granulocytes, another cell type of the myeloid lineage. Within the bone marrow, MDPs differentiate to monocytes, and to the common DC precursors (CDPs).
Fig. 2.
Differentiation of DCs and macrophages in mice. In the bone marrow, hematopoietic stem cells (HSC) produce myeloid (MP) and lymphoid (LP) committed precursors. MP give rise to monocyte/macrophages and DC precursors (MDP). MDP give rise to monocytes, and to common DC precursor (CDP). Two monocyte subsets, Ly-6C+ and Ly-6C- leave the bone marrow to enter the blood. CDP give rise to pre-classical dendritic cells (pre-cDC) and plasmacytoid dendritic cells (PDC). Pre-cDC circulate in blood and enter lymphoid tissue, where they give rise to CD8α+ and CD8α- cDCs, and non-lymphoid tissues, where they may give rise to CD103+ lamina propria DC (lpDC). Under homeostatic conditions, Ly-6C- monocytes may contribute to alveolar macrophages (MΦ) and Ly-6C+ monocytes can become CX3CR1+ lpDCs in non-lymphoid tissues. During inflammation, Ly-6C+ monocytes give rise to monocyte-derived DCs, e.g. TNF and iNOS-producing dendritic cells (TipDC), inflammatory macrophages, and may contribute to myeloid-derived suppressor cells (MDSC) associated with tumors. They are also suspected to contribute to microglia and Langerhans cells in selected experimental conditions. Microglia and Langerhans cells can renew independently from the bone marrow (curved arrow). HSC can also leave their bone marrow niche and enter peripheral tissues, where they differentiate to myeloid cells during inflammation. It is unclear at this time if LP contribute significantly to PDC and cDCs (dashed arrow).
The CDPs are proliferating cells that differentiate into PDCs and the precursors for classical DCs (pre-cDCs) in the bone marrow, but have lost the potential to give rise to monocytes (9, 23, 27). At steady state, Pre-cDCs are found in the bone marrow, blood and spleen (9, 28). They enter lymph nodes from the blood through high endothelial venules and integrate into the DC network where they acquire a mature cDC surface phenotype and morphology. cDC development also involves cell division in lymphoid organs, which is controlled in part by regulatory T cells (9), Flt3 (10), and the lymphotoxin-β receptor (29). Pre-cDCs can also differentiate into CD103+ mucosal DCs in the lamina propria (26, 30).
Monocytes exit to the blood, and can enter tissues under inflammatory conditions. They give rise to subsets of macrophages and to inflammatory DCs that share many of the phenotypic features and functions of DCs, such as the ability to process and present antigen to T cells (3, 5, 26, 31, 32) (Fig. 1); however, it is now widely accepted that monocytes do not give rise to cDCs and PDCs (9, 24).
Are there exceptions to the classical developmental pathways?
Although many macrophages and DC subsets are renewed from bone marrow progenitors, there are notable exceptions. For example, neither microglia nor Langerhans cells (LCs) are dependent on the bone marrow for their renewal in the steady state, and possibly during inflammation (33, 34). LCs were recently shown to develop from an embryonic precursor that colonizes the epidermis before birth, differentiates in situ, and then proliferates during the first week of life to establish the LC network(35). Adult LC self renew in situ and can massively proliferate during inflammation (35). Thus, epidermal LCs appear to be their own progenitors. In contrast, hematopoietic stem cells and precursors recirculate between the periphery and the bone marrow in adults (36) and may contribute directly to the differentiation of certain myeloid cells in inflamed tissues (37). Lymphoid-restricted progenitors contribute little to peripheral lymphoid organs or tissue DCs, with the possible exception of the thymus (38, 39).
What are the origins of non-lymphoid tissue resident dendritic cells?
Recent studies have established the existence of two distinct DC populations in the lung and dermis (reviewed in (34)), which are characterized as CD103+ and CX3CR1+ DCs. Likewise, recent studies have revealed phenotypically definable CD11c+ CD103+ and CX3CR1+ subpopulations in the intestinal lamina propria (26, 30, 40) (Table S1), which are derived from from pre-cDCs or Ly6C+ monocytes, respectively (26). Mice reconstituted exclusively with CX3CR1+ lamina propria cells developed severe intestinal inflammation when challenged in an innate colitis model (26). Thus, the balance of lamina propria DC subsets is likely important for gut homeostasis. Of note, only CD103+ cells emigrate from the lamina propria to the mesenteric lymph nodes (30, 41). CX3CR1+ cells are poor T cell stimulators (41) and thus may represent lamina propria macrophages rather than DCs.
What is the developmental relationship between mature monocyte subsets?
Differentiation of MDP into monocytes and macrophages is less well understood than their differentiation into DCs. Among differentiated ‘mature’ monocytes in the bone marrow, blood and spleen, at least two phenotypic and functional subsets can be identified (reviewed in (3)), whose developmental relationship is still unclear. Adoptive transfer experiments demonstrate that bone marrow Gr1+/Ly-6Chigh monocytes can shuttle between the blood and the bone marrow, and lose Ly-6C expression (25, 42), suggesting that they give rise to Gr1-/Ly-6Clow monocytes. However, neither a genetic defect in or antibody-mediated depletion of Gr1+/Ly-6Chigh monocytes affect the generation of Gr1-/Ly-6Clow monocytes (43-46). When bone marrow is used a source of monocytes, current adoptive transfer protocols may not always distinguish mature Gr1+/Ly-6Chigh monocytes from their proliferating precursors because pre-monocytes are ill-defined. Therefore, a better characterization of monocyte precursors and new lineage tracking studies will be needed to establish the developmental relationship between monocyte subsets.
Are monocytes effector cells or circulating precursors?
Monocytes have long been considered as a developmental intermediate between bone marrow precursors and tissue macrophages. It is now clear, however, that many DCs and tissue macrophages do not originate from monocytes in a steady-state. Conversely, monocytes carry out specific effector functions during inflammation(3). “Inflammatory” Gr1+/Ly-6Chigh monocytes are referred to as such because they give rise to macrophages and DCs in a variety of infectious models. During infection with Listeria monocytogenes, Gr1+/Ly-6Chigh monocytes differentiate into DCs that produce inflammatory mediators such as tumor necrosis factor α, nitric oxide and reactive oxygen species (known as TipDCs) (5, 47). Gr1+/Ly-6Chigh monocytes are required for resistance to the pathogen Toxoplasma gondii, but in this case differentiate into mucosal macrophages, which differ in surface phenotype and inflammatory mediator production from TipDCs (48, 49). In some tumors, up to half of the myeloid-derived suppressor cells (cells that can protect tumors from immune attack (50)) are Gr1+/Ly-6Chigh. Upon spinal cord injury, recruited Gr1+/Ly-6Chigh monocytes differentiate into macrophages that critically contribute to the recovery (51). Thus, Gr1+/Ly-6Chigh monocytes can differentiate into a variety of macrophages and DCs subtypes that can both activate and inhibit the immune response, depending on local or systemic cues received and the nature of encountered pathogen.
Are Gr1-/Ly-6Clow monocytes a new cell subset with a role in tissue repair?
Gr1-/Ly-6Clow monocytes represent a functionally distinct monocyte subset (52). They are characterized by a smaller size, lack of expression of CCR2 and L-selectin, but higher expression of the chemokine receptor CX3CR1 and the integrin LFA-1 (31) (Table S1). Gr1-/Ly-6Clow monocytes were initially termed “resident” in mice because of their longer half-life in vivo and their localization to both resting and inflamed tissues after adoptive transfer. Intravital microscopy revealed that Gr1-/Ly-6Clow monocytes exhibit long-range crawling on the luminal side of the vascular endothelium and are ideally located to survey endothelial cells and surrounding tissues for damage or infection (52). Moreover, Gr1-/Ly-6Clow monocytes are recruited at a late phase to the ischemic myocardium, where they express vascular-endothelial growth factor (VEGF), and are proposed to promote healing via myofibroblast accumulation, angiogenesis and deposition of collagen (53). Transcriptional profiling of extravasated monocytes retrieved from the peritoneum after L. monocytogenes infection (3, 52) indicates that Gr1-/Ly-6Clow monocytes initiate a macrophage differentiation program that resembles the one described for “alternatively activated macrophages”, cells that are proposed to be involved in tissue repair (54) (also known as M2 macrophages). Conversely Gr1-/Ly-6Clow monocytes that entered the peritoneum at the same time initiate a more inflammatory “classical” transcriptional program that resembles TipDCs or M1 macrophage differentiation. Direct experimental evidence that monocytes are involved in tissue repair is missing, however, and the differential contributions of monocytes subsets to neoangiogenesis (55, 56) need further study.
What determines monocyte differentiation potential?
The emerging evidence that monocyte subsets exert distinct functions and have distinct fates, such as the differentiation into M1 and M2 macrophages, seemingly contradicts the notion of a general plasticity of monocytes/macrophages, which holds that monocytes respond to their environment by differentiating into a variety of macrophages and DC-like cells (reviewed in (57)). This apparent contradiction is likely based on the effects of cytokines on monocytes in vitro. Exposure to GM-CSF and IL-4 induces differentiation of human and mouse monocytes into DCs, irrespective of their subset (58, 59). Moreover, addition of transforming growth factor β1 confers the phenotype of LCs (60), whereas exposure to M-CSF induces monocytes to differentiate into macrophages. Addition of IFNγ (or lipopolysaccharide) to M-CSF induces the differentiation of M1-like macrophages whereas addition of IL-4 induces the differentiation of M2-like macrophages (54, 61, 62). M2 macrophages have been further subsetted depending on the culture conditions (62). The chemokine CXCL4 induces another macrophage phenotype with possible relevance to vascular diseases (63).These results suggest that monocytes are phenotypically polarized by the microenvironment to mount specific functional programs. M1 macrophages, whose prototypical activating stimuli are IFNγ and LPS, exhibit potent microbicidal properties and promote strong IL-12-mediated T helper 1 responses. In contrast, M2 macrophages support T helper 2-associated effector functions and may play a role in resolution of inflammation through endocytic clearance and trophic factor synthesis (54). In vitro studies may not, however, fully recapitulate in vivo differentiation, and thus heterogeneity of monocytes may influence their plasticity in vivo, with Gr1-/Ly-6Clow and Gr1+/Ly-6Chigh monocytes being committed to differentiate more readily into M2 cells or inflammatory DCs and M1 macrophages, respectively.
Transcriptional control of lineage commitment in the mononuclear phagocyte system
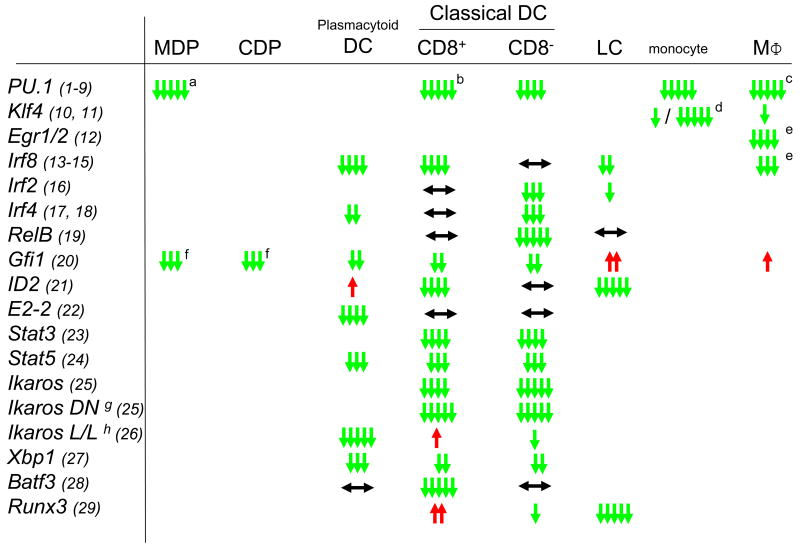
The successively more restricted lineage potential of MDPs, CDPs, pre-DCs, monocytes, macrophages and DCs involves the selection of specific gene expression programs. For some pathways the transcription factors responsible for these cell fate choices have recently begun to be elucidated. Although several transcription factor knockout mice show deficiencies in the mononuclear phagocyte system, they often display broad effects in multiple cell types (3, 34, 64) (Fig. 3). A few exceptions have been identified, such as the loss of PDCs in mice deficient for the basic helix-loop-helix transcription factor E2-2 (65) or the loss of CD8α+ cDCs in mice deficient for the basic leucine zipper transcription factor Batf3 (66). Similarly selective transcription factor requirements may be responsible for monocyte (44) and macrophage subtypes and will require further investigation.
Figure 3.
Phenotype of transcription factor knockout mice in different populations of the MPs. ↓ indicates a reduction in KO, ↑ an increase in KO and ↔ means unchanged numbers of the populations, suggesting a positive (green), negative (red) or no (black) effect on the development of the respective population. Number of arrows indicates the relative strength of the effect. Specific notes: (a) MDP have not been specifically analyzed but PU.1-deficient mice lack all myeloid progenitors of which MDP are a subpopulation, (b) in one study (SOM ref 7) CD8+ CD1 1c+ cells were detected in E16.5 embryos, (c) some macrophage subpopulations are present in the embryo, (d) LyC6-/LyC6+ respectively, (e) M-CSF dependent differentiation in culture, (f) MDP were not specifically studied, but the population of lin- ckit+ Flt3+ progenitors, which includes MDP was reduced; (g) dominant negative allele and (h) low level expression. References cited in this figure (1-29) are found in the SOM.
Broad effects on many lineages, however, do not necessarily exclude important functions in specific commitment events. An instructive example is the myeloid transcription factor PU.1. Although PU.1 is required for the earliest steps of myeloid lineage commitment in hemopoietic stem cells (67) and its absence results in general myeloid lineage deficiencies (3, 34), PU.1 has additional key selector gene functions at several branch points of myeloid lineage diversification, particularly during the macrophage versus DC choice of monocytes (68). The function of PU.1 at particular progenitor stages depends on its balance with antagonistic factors driving alternative fates. Intermediate expression of PU.1 in GMPs can overcome the neutrophil fate-inducing effects of the basic leucine zipper transcription factor C/EBPα and activate the macrophage-specifying zinc finger transcription factors Egr-1 and Egr-2 (3, 69)(3, 81). In contrast, high expression of PU.1 is required to induce a DC fate in monocytes and to antagonize the macrophage-inducing transcription factors c-Maf and MafB (3, 68). Of interest, the transcription factor balance specifying M2 or DC fate of monocytes in culture correlates with cell fate choice in vivo, as outlined above for the programs initiated by Ly6Clow and Ly6Chigh monocytes upon L. monocytogenes infection of the peritoneum (52, 68).
In general, gain-of-function experiments have been informative in defining transcription factor potential in driving particular cell fate choices. For example ectopic expression of the transcription factors MafB, c-Maf, Egr1, ICSBP/IRF8, KLF4 and PU.1 in early progenitors can drive monocyte/macrophage fates. PU.1 and RelB induce DC differentiation via monocytic intermediates and E-box proteins (likely mimicking endogenous E2-2) or SpiB can induce PDC fates (3). In most of these experiments, however, undefined or early progenitor-enriched populations were used. Given that the function of any factor will depend on other cooperating and antagonistic transcription factors expressed at each specific progenitor stage (70), this makes it difficult to precisely identify the affected commitment event. Such differentiation stage-specific manipulation of transcription factors will require models that faithfully reproduce defined differentiation stages in culture, the development of new genetic tools of inducible gene regulation in vivo or adoptive transfer of genetically marked, defined cell populations.
Another aspect of emerging importance is the coupling of cytokine receptor signaling with specific transcription factors in lineage commitment. For example, STAT3 is required for Flt3L but not GM-CSF mediated DC differentiation (21, 34, 64). Similarly, Id2 and Runx3 mediate TGFβ signals in LC differentiation (34, 64). Furthermore, MafB limits M-CSF signals and inhibits PU.1 activation in HSCs (67), whereas MafB and c-Maf together inhibit M-CSF dependent proliferative signals in mature monocytes and macrophages and assure their withdrawal from cell cycle (71).
Besides transcription factors, epigenetic modification with large-scale histone and DNA modifications like acetylation and methylation, and micro-RNAs have recently been reported to regulate important functions of mononuclear phagocytes (72, 73), and are also emerging as important determinants of lineage choice, but will not be discussed here.
Perspectives and future challenges
Although the studies discussed above have shed light on the in vivo development and homeostasis of the mononuclear phagocyte system, mechanisms that control replacement of many macrophage subsets and resident cells remain mysterious. Most organs contain resident tissue macrophages, many of which have long half-lives. After irradiation and other forms of tissue injury, bone marrow-derived cells frequently repopulate the injured organs. Whether bone marrow-derived cells under inflammatory conditions assume the same phenotype and function as resident cells that populate tissues under steady state conditions remains largely unclear. Fate mapping experiments investigating steady state developmental potential will therefore have to avoid experimental inflammation. Towards this end, Cre recombinase expression under the control of promoters for key markers (such as lysozyme) have been used to activate conditional reporter genes so that all downstream daughter cells express a stable marker (9). It has to be cautioned, however, that ‘key’ markers are seldom completely specific, progenitor cells can express low levels of the marker and marker positive cells may not uniformly turn on the reporter, all of which may complicate the interpretation of such lineage marking experiments (74, 75). Parabiotic mice and adoptive transfer into mice in which cell populations have been depleted by genetic means may provide complementary approaches.
The transcription factors controlling monocyte and macrophage subtype-specific programs in vivo need also to be identified. Models that faithfully reproduce defined differentiation stages in culture and the development of new genetic tools of inducible gene regulation that enable precise differentiation stage-specific manipulation of transcription factors in vivo could accelerate discovery in this area. The use of new model systems for the study of phagocytes, such as Drosophila melanogaster, which lack lymphocytes but possess a powerful innate immune system, is perfectly suited for genetic studies and in vivo imaging and has the potential to accelerate discovery of genetic pathways that control the development and functions of phagocytes in vivo (76-78).
Another challenge will be to translate knowledge obtained from mouse and other models into a better understanding of the mononuclear phagocyte system and its role in inflammation and associated diseases in humans. A large number of studies have attempted to recapitulate in vitro some of the heterogeneity of DCs and macrophages. On the basis of these studies, functional subsets of macrophages and DCs (e.g. tolerogenic DCs) inside tissues have been proposed to mediate important functions in vivo, such as tissue repair, angiogenesis, or tolerance to self. Laser capture dissection techniques are being developed to obtain gene expression data from macrophages isolated from tissues (79). Flow cytometry-based cell sorting approaches also hold promise for isolating pure macrophage and DC populations from various tissues and organs. These techniques are especially important in order to translate some of the findings made in mice into the human system. Global expression profiling of mouse DC, macrophages, and monocyte subsets might assist in the identification of corresponding human populations. Human hemato-lymphoid-system mouse models that will provide in vivo DC and macrophage read-outs from human hematopoietic stem cells and progenitor cells will also be important (80). Finally, studies of patients with deficiencies for genes involved in myeloid cell differentiation and functions will be critical to assess the relevance of models derived from studies in the mouse and in vitro experiments in human cells (81).
Acknowledgments
The diagram depicted in Fig. 2 is the result of a collective discussion at a recent workshop on « Monocytes, Macrophages and Dendritic Cell heterogeneity” organised March 2-7, 2009 by the Fondation des Treilles (France) (http://www.lestreilles.com/ssimages/index.html). We thank V. Bronte, I. Charo, J. Van Ginderachter, G. MacPherson, A. Mantovani, E. Pamer, R. Steinman and F. Swirski both for their input into Fig.1 and for their ideas that we have incorporated into this review. Any misrepresentation is the responsibility of the authors. Work by FG and KL on the development of monocytes, macrophages and dendritic cells is funded by MRC G0900867 to FG, and NIH R01 HL058108 to K.L.
References and Notes
- 1.van Furth R, Cohn ZA. J Exp Med. 1968 Sep 1;128:415. doi: 10.1084/jem.128.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinman RM, Lustig DS, Cohn ZA. J Exp Med. 1974 Jun 1;139:1431. doi: 10.1084/jem.139.6.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auffray C, Sieweke MH, Geissmann F. Annu Rev Immunol. 2009;27:669. doi: 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- 4.Swirski FK, et al. Science. 2009 Jul 31;325:612. doi: 10.1126/science.1175202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serbina NV, Jia T, Hohl TM, Pamer EG. Annu Rev Immunol. 2008;26:421. doi: 10.1146/annurev.immunol.26.021607.090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon S. Cell. 2002 Dec 27;111:927. doi: 10.1016/s0092-8674(02)01201-1. [DOI] [PubMed] [Google Scholar]
- 7.Banchereau J, Steinman RM. Nature. 1998 Mar 19;392:245. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 8.Mellman I, Steinman RM. Cell. 2001 Aug 10;106:255. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 9.Liu K, et al. Science. 2009 Apr 17;324:392. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waskow C, et al. Nat Immunol. 2008 Jun;9:676. doi: 10.1038/ni.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merad M, et al. Nat Immunol. 2002 Dec;3:1135. doi: 10.1038/ni852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corcoran L, et al. J Immunol. 2003 May 15;170:4926. doi: 10.4049/jimmunol.170.10.4926. [DOI] [PubMed] [Google Scholar]
- 13.Colonna M, Trinchieri G, Liu YJ. Nat Immunol. 2004 Dec;5:1219. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- 14.Sasmono RT, et al. Blood. 2003 Feb 1;101:1155. doi: 10.1182/blood-2002-02-0569. [DOI] [PubMed] [Google Scholar]
- 15.Dai XM, et al. Blood. 2002 Jan 1;99:111. doi: 10.1182/blood.v99.1.111. [DOI] [PubMed] [Google Scholar]
- 16.Cecchini MG, et al. Development. 1994 Jun;120:1357. doi: 10.1242/dev.120.6.1357. [DOI] [PubMed] [Google Scholar]
- 17.Wiktor-Jedrzejczak W, Gordon S. Physiol Rev. 1996 Oct;76:927. doi: 10.1152/physrev.1996.76.4.927. [DOI] [PubMed] [Google Scholar]
- 18.Kawasaki ES, et al. Science. 1985 Oct 18;230:291. doi: 10.1126/science.2996129. [DOI] [PubMed] [Google Scholar]
- 19.Lin H, et al. Science. 2008 May 9;320:807. [Google Scholar]
- 20.McKenna HJ. Blood. 2000;95:3489. [PubMed] [Google Scholar]
- 21.Onai N, Obata-Onai A, Tussiwand R, Lanzavecchia A, Manz MG. J Exp Med. 2006 Jan 23;203:227. doi: 10.1084/jem.20051645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D'Amico A, Wu L. J Exp Med. 2003;198:293. doi: 10.1084/jem.20030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onai N, et al. Nat Immunol. 2007 Nov;8:1207. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- 24.Fogg DK, et al. Science. 2006 Jan 6;311:83. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- 25.Varol C, et al. J Exp Med. 2007 Jan 22;204:171. doi: 10.1084/jem.20061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varol C, et al. Immunity. 2009 Sep 18;31:502. doi: 10.1016/j.immuni.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 27.Naik SH, et al. Nat Immunol. 2007 Nov;8:1217. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- 28.Naik SH, et al. Nat Immunol. 2006 Jun;7:663. doi: 10.1038/ni1340. [DOI] [PubMed] [Google Scholar]
- 29.Kabashima K, et al. Immunity. 2005 Apr;22:439. doi: 10.1016/j.immuni.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Bogunovic M, et al. Immunity. 2009 Sep 3; [Google Scholar]
- 31.Geissmann F, Jung S, Littman DR. Immunity. 2003 Jul;19:71. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 32.Auffray C, et al. J Exp Med. 2009 Mar 16;206:595. doi: 10.1084/jem.20081385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Nat Neurosci. 2007 Dec;10:1538. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- 34.Merad M, Manz MG. Blood. 2009 Apr 9;113:3418. doi: 10.1182/blood-2008-12-180646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chorro L, et al. J Exp Med. 2009 doi: 10.1084/jem.20091586. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright DE, Wagers AJ, Gulati AP, Johnson FL, Weissman IL. Science. 2001 Nov 30;294:1933. doi: 10.1126/science.1064081. [DOI] [PubMed] [Google Scholar]
- 37.Massberg S, et al. Cell. 2007 Nov 30;131:994. doi: 10.1016/j.cell.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu L, Shortman K. Semin Immunol. 2005 Aug;17:304. doi: 10.1016/j.smim.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Li J, Park J, Foss D, Goldschneider I. J Exp Med. 2009 Mar 16;206:607. doi: 10.1084/jem.20082232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaensson E, et al. J Exp Med. 2008 Sep 1;205:2139. doi: 10.1084/jem.20080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulz O, et al. J Exp Med. 2009 in press. [Google Scholar]
- 42.Yrlid U, Jenkins CD, MacPherson GG. J Immunol. 2006 Apr 1;176:4155. doi: 10.4049/jimmunol.176.7.4155. [DOI] [PubMed] [Google Scholar]
- 43.Feinberg MW, et al. Embo J. 2007 Sep 19;26:4138. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alder JK, et al. J Immunol. 2008 Apr 15;180:5645. doi: 10.4049/jimmunol.180.8.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scatizzi JC, et al. Am J Pathol. 2006 May;168:1531. doi: 10.2353/ajpath.2006.050555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mildner A, et al. Nat Neurosci. 2007 Dec;10:1544. doi: 10.1038/nn2015. [DOI] [PubMed] [Google Scholar]
- 47.Narni-Mancinelli E, et al. J Exp Med. 2007 Sep 3;204:2075. doi: 10.1084/jem.20070204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunay IR, et al. Immunity. 2008 Aug 15;29:306. doi: 10.1016/j.immuni.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robben PM, LaRegina M, Kuziel WA, Sibley LD. J Exp Med. 2005 Jun 6;201:1761. doi: 10.1084/jem.20050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Movahedi K, et al. Blood. 2008 Apr 15;111:4233. doi: 10.1182/blood-2007-07-099226. [DOI] [PubMed] [Google Scholar]
- 51.Shechter R, et al. PLoS Med. 2009 Jul;6:e1000113. doi: 10.1371/journal.pmed.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Auffray C, et al. Science. 2007 Aug 3;317:666. doi: 10.1126/science.1142883. [DOI] [PubMed] [Google Scholar]
- 53.Nahrendorf M, et al. J Exp Med. 2007 Nov 26;204:3037. doi: 10.1084/jem.20070885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martinez FO, Helming L, Gordon S. Annu Rev Immunol. 2009;27:451. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 55.Grunewald M, et al. Cell. 2006 Jan 13;124:175. doi: 10.1016/j.cell.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 56.De Palma M, et al. Cancer Cell. 2005 Sep;8:211. doi: 10.1016/j.ccr.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 57.Taylor PR, Gordon S. Immunity. 2003 Jul;19:2. doi: 10.1016/s1074-7613(03)00178-x. [DOI] [PubMed] [Google Scholar]
- 58.Sallusto F, Lanzavecchia A. J Exp Med. 1994 Apr 1;179:1109. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geissmann F, et al. Immunol Cell Biol. 2008 Jul;86:398. doi: 10.1038/icb.2008.19. [DOI] [PubMed] [Google Scholar]
- 60.Geissmann F, et al. J Exp Med. 1998 Mar 16;187:961. doi: 10.1084/jem.187.6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stein M, Keshav S, Harris N, Gordon S. J Exp Med. 1992 Jul 1;176:287. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martinez FO, Sica A, Mantovani A, Locati M. Front Biosci. 2008;13:453. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 63.Gleissner CA, et al. Circulation Research. 2009 in press. [Google Scholar]
- 64.Zenke M, Hieronymus T. Trends Immunol. 2006 Mar;27:140. doi: 10.1016/j.it.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 65.Cisse B, et al. Cell. 2008 Oct 3;135:37. [Google Scholar]
- 66.Hildner K, et al. Science. 2008 Nov 14;322:1097. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sarrazin S, et al. Cell. 2009 Jul 23;138:300. [Google Scholar]
- 68.Bakri Y, et al. Blood. 2005 Apr 1;105:2707. doi: 10.1182/blood-2004-04-1448. [DOI] [PubMed] [Google Scholar]
- 69.Laslo P, et al. Cell. 2006 Aug 25;126:755. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 70.Sieweke MH, Graf T. Curr Opin Genet Dev. 1998;8:545. doi: 10.1016/s0959-437x(98)80009-9. [DOI] [PubMed] [Google Scholar]
- 71.Aziz A, et al. Mol Cell Biol. 2006 Sep;26:6808. doi: 10.1128/MCB.00245-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.O'Connell RM, et al. J Exp Med. 2008 Mar 17;205:585. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. Proc Natl Acad Sci U S A. 2007 Jan 30;104:1604. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miyamoto T, et al. Dev Cell. 2002 Jul;3:137. doi: 10.1016/s1534-5807(02)00201-0. [DOI] [PubMed] [Google Scholar]
- 75.Shigematsu H, et al. Immunity. 2004 Jul;21:43. doi: 10.1016/j.immuni.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 76.Martinez-Agosto JA, Mikkola HK, Hartenstein V, Banerjee U. Genes Dev. 2007 Dec 1;21:3044. doi: 10.1101/gad.1602607. [DOI] [PubMed] [Google Scholar]
- 77.Babcock DT, et al. Proc Natl Acad Sci U S A. 2008 Jul 22;105:10017. doi: 10.1073/pnas.0709951105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cuttell L, et al. Cell. 2008 Oct 31;135:524. doi: 10.1016/j.cell.2008.08.033. [DOI] [PubMed] [Google Scholar]
- 79.Trogan E, et al. Proc Natl Acad Sci U S A. 2002 Feb 19;99:2234. doi: 10.1073/pnas.042683999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Manz MG. Immunity. 2007 May;26:537. doi: 10.1016/j.immuni.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 81.Casanova JL, Abel L. Annu Rev Immunol. 2002;20:581. doi: 10.1146/annurev.immunol.20.081501.125851. [DOI] [PubMed] [Google Scholar]