Abstract
Background
Plant circadian clocks regulate many photoperiodic and diurnal responses that are conserved among plant species. The plant circadian clock system has been uncovered in the model plant, Arabidopsis thaliana, using genetics and systems biology approaches. However, it is still not clear how the clock system had been organized in the evolutionary history of plants. We recently revealed the molecular phylogeny of LHY/CCA1 genes, one of the essential components of the clock system. The aims of this study are to reconstruct the phylogenetic relationships of angiosperm clock-associated PRR genes, the partner of the LHY/CCA1 genes, and to clarify the evolutionary history of the plant clock system in angiosperm lineages.
Results
In the present study, to investigate the molecular phylogeny of PRR genes, we performed two approaches: reconstruction of phylogenetic trees and examination of syntenic relationships. Phylogenetic analyses revealed that PRR genes had diverged into three clades prior to the speciation of monocots and eudicots. Furthermore, copy numbers of PRR genes have been independently increased in monocots and eudicots as a result of ancient chromosomal duplication events.
Conclusions
Based on the molecular phylogenies of both PRR genes and LHY/CCA1 genes, we inferred the evolutionary process of the plant clock system in angiosperms. This scenario provides evolutionary information that a common ancestor of monocots and eudicots had retained the basic components required for reconstructing a clock system and that the plant circadian clock may have become a more elaborate mechanism after the speciation of monocots and eudicots because of the gene expansion that resulted from polyploidy events.
Background
Many organisms such as cyanobacteria, fruit flies, mammals and plants have an endogenous time-keeping mechanism, a circadian clock, to gauge daily and seasonal environmental changes. Circadian clock systems in plants regulate various photoperiodic and diurnal responses, such as photomorphogenic processes, floral transition, leaf movements, stomatal conductance, photosynthetic capacity, and volatile emissions (reviewed in [1]). Among these, means to discriminate the length of the photoperiod are conserved among plant species, and it is commonly thought that circadian clock system of plants shares a basic mechanism that controls photoperiodic responses.
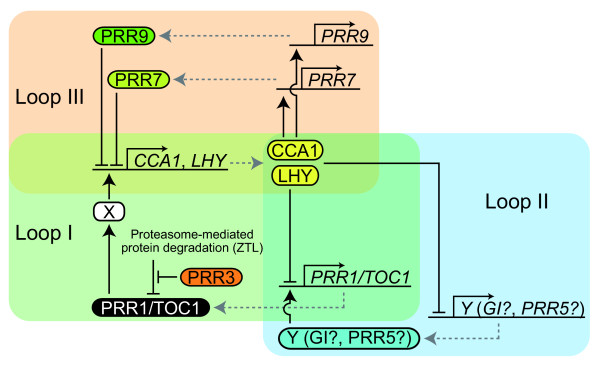
In the past decade, numerous molecular genetic analyses of the model plant Arabidopsis thaliana have uncovered the basic molecular network of the plant circadian clock (reviewed in [2,3]). Mathematical analyses have been used to develop a computational model of the plant clock system, which contains the main transcriptional feedback loop (Loop I) and two additional loops (Loops II and III) associated with the main loop (Figure 1) [4,5]. This multiple feedback loop system of the plant clock system is composed of two gene families, Pseudo-Response Regulators (PRRs) and Late Elongated Hypocotyl/Circadian Clock Associated 1 (LHY/CCA1), and two unknown factors ("X" and "Y"). The main feedback loop (Loop I) consists of two LHY/CCA1 genes, the Pseudo-Response Regulator 1/Timing of CAB2 Expression 1 (PRR1/TOC1) gene and the unknown factor "X". In this loop, the feedback regulatory network operates as follows: the evening-acting PRR1/TOC1 gene induces the morning-acting LHY and CCA1 genes via the unknown factor "X", and is in turn repressed by LHY/CCA1 [6]. The Loop I associates with Loop II via the PRR1/TOC1 gene and with Loop III via LHY/CCA1 genes [4,5]. Loop II is made up of PRR1/TOC1 and an unknown factor "Y". It has been proposed that the unknown factor "Y" is GIGANTEA and/or PRR5, although the true component has not yet been identified [7]. Loop III consists of LHY/CCA1 genes and two PRR genes, PRR7 and PRR9. Together, the gene families PRRs and LHY/CCA1s have key roles and form the complex regulatory network in the plant clock system.
Figure 1.
Model of the circadian clock system in Arabidopsis thaliana. Green, blue and orange shadings indicate loop I, II and III, respectively. This figure is modified from McClung [7] and Harmer [60].
Clock-associated PRR genes are conserved among angiosperm evolutionary lineages as are their partner LHY/CCA1 genes [8,9]. In eudicotyledonous plants, five copies of PRR genes have been identified in A. thaliana and Carica papaya and seven copies have been found in Populus trichocarpa [10-12]. In monocotyledonous plants, Oryza sativa has five PRR genes [13]. The expression patterns of PRR genes in A. thaliana and O. sativa share some common features. The five PRR genes in A. thaliana show diurnal and sequential-temporal expression patterns from dawn to dusk as follows; PRR9→PRR7→PRR5→PRR3→PRR1 [10]. The similar sequential expression pattern is found in homologous genes of O. sativa, which are expressed as follows; OsPRR73 (OsPRR37)→OsPRR95 (OsPRR59)→OsPRR1 [13]. In spite of these similarities in the copy numbers and the expression patterns of clock-associated PRR genes, it is still unclear how the PRR genes have evolved in monocots and eudicots and how they have been incorporated in the regulatory network of the clock system in the evolutionary history of plants.
Rapid accumulation of genomic sequence data offers new perspectives on the molecular phylogeny of genes in angiosperms [14]. Completion of genomic sequences for various plant species reveals that angiosperm genomes have undergone several ancient chromosomal or whole genome duplication events [11,15-17]. In monocot lineages, the ρ polyploidy event occurred before the speciation of O. sativa and Sorghum bicolor in commelinids [18,19]. On the other hand, four polyploidy events appear to have occurred in eudicot lineages. Among these polyploidy events, the γ triplication event took place near the base of the eudicot clade though the timing of this event is still being debated [11,14,16,17]. The draft genomic sequence analysis of C. papaya has revealed that the genome of A. thaliana underwent two polyploidy events (α and β) after the speciation of C. papaya and A. thaliana in eurosids II [11]. Furthermore, the β polyploidy event is thought to have occurred before the α event [14]. In eurosids I, the salicoid polyploidy event occurred within the Salicaceae lineages, which includes Populus [17]. The footprints of these chromosomal duplication events are the conserved order of the genes on the duplicated chromosomes in the present genomic sequences [20]. Thus, comparison of the order of genes surrounding duplicated genes provides molecular evolutionary information on their phylogenetic relationships [21,22].
In the present study, to clarify the phylogenetic relationships among angiosperm PRR genes, we (1) identified PRR genes using available genomic databases of eudicots (Vitis vinifera, P. trichocarpa, C. papaya, and A. thaliana) and monocots (O. sativa and S. bicolor) and (2) examined the evolutionary processes of angiosperm PRR genes by conventional phylogenetic reconstruction and examination of syntenic relationships. With these results, we reconstructed the molecular phylogeny of PRR genes in angiosperms and found that gene expansion of PRRs occurred via polyploidy events in monocots and eudicots. Taken together with the molecular phylogeny of the other major gene family of the plant clock system (LHY/CCA1s) [9], our data allow us to explore the evolutionary history of the multiple feedback loop system in angiosperm lineages.
Results
Identification of clock-associated PRR genes in angiosperms
There are five copies of the PRR genes in the genomes of O. sativa, S. bicolor, V. vinifera and C. papaya, six copies in A. thaliana, and eight copies in P. trichocarpa (see Additional files 1 and 2). The PRR1/TOC1 gene in C. papaya was not retrieved from the genomic sequence database because the nucleotide sequence of the C-terminal region of the gene has not yet been determined. The angiosperm PRR genes retained a highly conserved PR-domain at the N-terminus and a CCT-motif at the C-terminus (see Additional file 3). However, two PRR-like genes (PRR9b in A. thaliana, AtPRR9b, and PRR5c in P. trichocarpa, PtPRR5c) retained the CCT-motif but not the PR-domain (see Additional file 4). Thus, we examined the molecular phylogeny of PRR genes but excluded AtPRR9b and PtPRR5c in the present study.
Phylogenetic analysis of PRR gene family
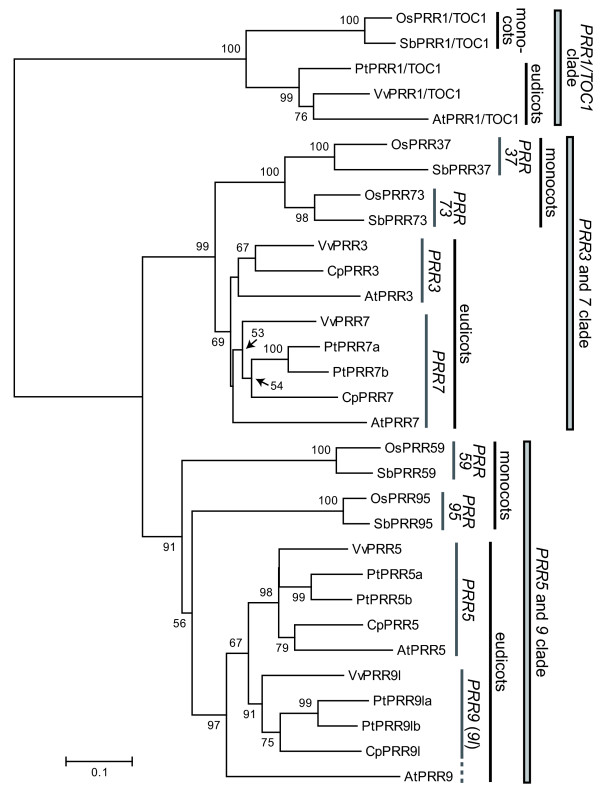
To deduce the evolutionary relationships among PRR genes in angiosperms, a phylogenetic tree was reconstructed using the minimum evolution (ME) method. Angiosperm PRR genes clearly separated into three clades (PRR1/TOC1 clade, PRR3 and 7 clade, and PRR5 and 9 clade) (Figure 2). This classification was consistent with the categorization of genomic structures of PRR genes that was apparent when exon-intron structures and insertions/deletions variation were examined (Figure 3, see Additional file 5). We found that each clade contains genes from both eudicots and monocots, suggesting that ancient PRR gene(s) diverged into three clades before the speciation of monocots and eudicots.
Figure 2.
Phylogenetic tree of angiosperm PRR genes. Amino acid sequences were aligned using TCoffee program [47]. The phylogenetic tree was reconstructed by the ME method from the numbers of amino acid substitutions estimated by the JTT model. PRR1/TOC1 genes were utilized as an outgroup in the phylogenetic trees. The numerals at the branch indicate bootstrap values calculated by the ME method with 1,000 replications. Bootstrap values >50% are shown.
Figure 3.
Comparison of the exon-intron structures of PRR genes around the region of PR-domain and CCT-motif. The amino acid sequences encoded by PRR genes were aligned using TCoffee program [47]. Accession numbers and gene IDs of the PRR genes are shown in Additional file 1. The numerals on the right side indicate the numbers of amino acid residues of each protein. Sequence similarity is indicated below the alignment using the symbols "asterisk," "colon," and "dot" for identical, highly similar, and weakly similar residues, respectively. Black and gray shadings on the alignments indicate a site of exon-intron boundary and one-amino acid deletion, respectively.
In all plant species examined, one copy of the PRR1/TOC1 gene was retained in the PRR1/TOC1 clade (Figure 2), whereas at least two copies were found in the PRR3 and 7 clade and the PRR5 and 9 clade. PRR1/TOC1 genes diverged into two clusters representing monocots and eudicots.
The PRR3 and 7 clade consisted of two different clusters, each of which exclusively consists of monocot or eudicot genes (Figure 2). Accordingly, the phylogenetic tree suggested that the gene duplication events producing monocotyledonous PRR37 and PRR73 or eudicotyledonous PRR3 and PRR7 occurred independently within monocot and eudicot lineages, respectively. After the gene duplication event in eudicots, orthologs of P. trichocarpa PRR3 appeared to be lost, whereas the P. trichocarpa PRR7 gene was duplicated into PRR7a and 7b.
In the PRR5 and 9 clade, the monocot PRR59 and PRR95 genes showed an earlier gene duplication that may have occurred in a common ancestor of monocots and eudicots (Figure 2). However, the bootstrap value supporting this branch was not very high, 56%. Eudicotyledonous PRR5 and PRR9/9-like (9l) genes formed a cluster in the phylogenetic tree. In this cluster, A. thaliana PRR9 was distantly related to other PRR5 and PRR9l genes, which was also observed in the phylogenetic trees reconstructed by the neighbor-joining, maximum likelihood and Bayesian methods (see Additional files 6 and 7). This topology within PRR5 and 9 clade might be the artefact caused by faster substitution rate of the AtPRR9 gene. Otherwise, sparse taxonomic sampling obscures the additional gene duplication and loss events occurred in eudicots. PRR5a in P. trichocarpa was more closely related to PRR5b than other PRR5, and similar close relationship was found between PRR9la and PRR9lb in P. trichocarpa (Figure 2). These findings indicated that the gene duplication events that produced PRR5a and 5b and PRR9la and 9lb occurred within rosids. Collectively, although the PRR3 and 7 clade and the PRR5 and 9 clade contained at least two copies of PRR genes in both monocots and eudicots, PRRs in the two clades are assumed to have independently duplicated in monocot and eudicot lineages.
Functional divergence among PRR gene clusters
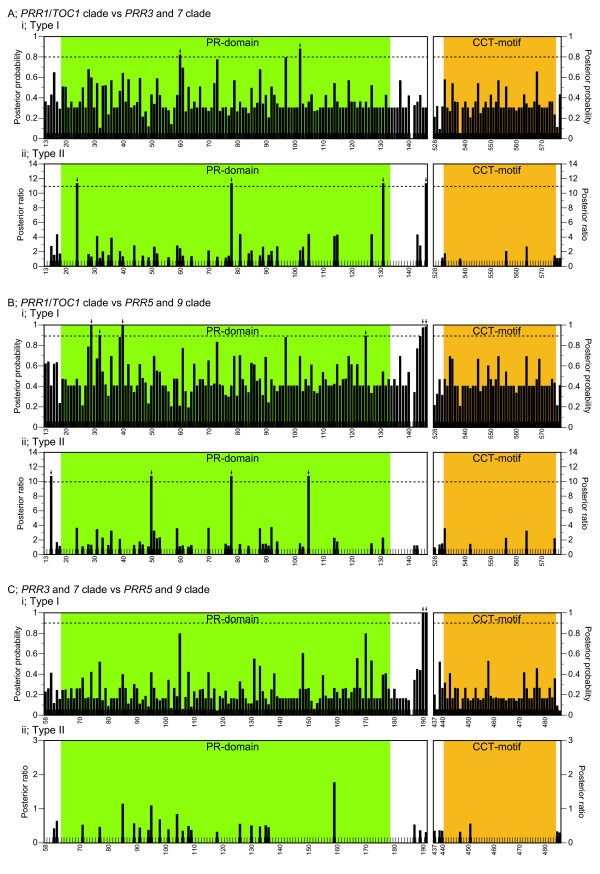
Clock associated-PRR genes were divided into the three gene clusters (PRR1/TOC1 clade, PRR3 and 7 clade, and PRR5 and 9 clade) that had been formed prior to the speciation of monocots and eudicots. Although the amino acid sequences of the genes were highly conserved in the PR-domain and CCT-motif, there were several amino acid changes that were distinctive among the three clades, which potentially contribute to functional differences (see Additional file 3). To detect amino acid substitutions that are potentially involved in functional divergence among the clades in PR-domain, CCT-motif and their flanking regions, we performed statistical analysis to estimate the coefficient of type I and type II functional divergences (θI and θII). In the type I functional divergence, sites are conserved in one gene cluster but variable in the sister clusters [23]. On the other hand, type II sites are fixed in both clusters but the amino acid residues are different between the clusters [24]. In the comparison among the PRR gene clusters, all of the coefficients for the type I functional divergence (θI) were significantly larger than zero (Table 1). In addition, the values of the type II functional divergence (θII) between PRR1/TOC1 clade and PRR3 and 7 clade and between PRR1/TOC1 clade and PRR5 and 9 clade were significantly different from zero while the value between PRR3 and 7 clade and PRR5 and 9 clade was not significantly greater than zero. Six sites that were above the empirical cutoff values were identified in the comparison between PRR1/TOC1 clade and PRR3 and 7 clade, ten sites in PRR1/TOC1 clade and PRR5 and 9 clade and two sites in PRR3 and 7 clade and PRR5 and 9 clade (Figures 4 and 5). Intriguingly, these sites were predominantly detected in the PR-domain and its flanking region rather than the CCT-motif (Figure 4). These results imply that the amino acid substitutions in the PR-domain, through which PRR proteins interact with other proteins (ZEITLUPE and PRRs) [25,26], may partially contribute to the functional divergence among the three gene clusters.
Table 1.
Coefficients of type I and type II functional divergences among the PRR gene clades.
| Comparison | Type I | Type II | ||
|---|---|---|---|---|
| θI | SE | θII | SE | |
| PRR1/TOC1 vs PRR3 and 7 | 0.366 | 0.109* | 0.196 | 0.093** |
| PRR1/TOC1 vs PRR5 and 9 | 0.473 | 0.075* | 0.175 | 0.088** |
| PRR3 and 7 vs PRR5 and 9 | 0.234 | 0.048* | -0.003 | 0.106 |
*P < 0.01, **P < 0.05
Figure 4.
Type I and type II functional divergences among the PRR gene clades. Coefficients of type I (i) and type II (ii) functional divergences between PRR1/TOC1 clade and PRR3 and 7 clade (A), between PRR1/TOC1 clade and PRR5 and 9 clade (B) and between PRR3 and 7 clade and PRR5 and 9 clade (C) were calculated by DIVERGE 2.0 [55,56] using the TCoffee alignment (see Additional file 3) and the ME tree (Figure 2). Right and left panels indicate the PR-domain and its flanking region, and the CCT-motif and its flanking region, respectively. Positions of amino acid residues correspond to AtPRR1/TOC1 in PRR1/TOC1 clade vs PRR3 and 7 clade, AtPRR1/TOC1 in PRR1/TOC1 clade vs PRR5 and 9 clade and to AtPRR3 in PRR3 and 7 clade vs PRR5 and 9 clade. Cutoff values of the posterior probability and posterior ratio were established empirically by sequentially removing the highest scoring sites from the alignment until θ = 0. The cutoff values are shown by broken lines. The value of θII between PRR3 and 7 clade and PRR5 and 9 clade was not set because the coefficient of the θII was not significantly greater than zero. Thus, there is no broken line shown in the bottom panel (C, ii). The regions represented in this figure are surrounded with blue boxes in Additional file 3. Arrows indicate sites above the empirical cutoff values. Green and yellow shadings on the panels indicate PR-domain and CCT-motif, respectively.
Figure 5.
Functionally divergent sites among the PRR gene clades. Sequence logos indicate amino acid variation of type I (θI) and type II (θII) sites that were above the empirical cutoff values in the comparison between PRR1/TOC1 clade and PRR3 and 7 clade (A), between PRR1/TOC1 clade and PRR5 and 9 clade (B) and between PRR3 and 7 clade and PRR5 and 9 clade (C). Amino acids are color-coded by physicochemical property. Positions of amino acid residues (bottom) correspond to AtPRR1/TOC1 in PRR1/TOC1 clade vs PRR3 and 7 clade, AtPRR1/TOC1 in PRR1/TOC1 clade vs PRR5 and 9 clade and to AtPRR3 in PRR3 and 7 clade vs PRR5 and 9 clade. The regions represented in this figure are surrounded with blue boxes in Additional file 3. Sequence logos were generated with WebLogo version 2.8.2 [57,58].
Phylogenetic relationships of PRR gene family inferred from chromosome syntenies
To clarify evolutionary events such as gene duplication and gene deletion among angiosperm PRR genes, we investigated chromosomal syntenies among the genomes of monocots or eudicots. Because ancient chromosome duplication events result in conserved gene order on the duplicated chromosomes [20], comparisons of gene organization and detection of chromosomal syntenies can provide molecular evolutionary information to understand the phylogenetic relationships of the genes [21,22].
In eudicots, the flanking region of PRR1/TOC1, PRR3, 5, 7 and 9 in A. thaliana showed a syntenic relationship with PRR1/TOC1, PRR3, 5, 7 and 9l in V. vinifera, P. trichocarpa, and C. papaya, respectively (Figure 6). In addition, the syntenies were also found in the flanking regions between AtPRR3 and VvPRR7, AtPRR7 and VvPRR3, AtPRR5 and VvPRR9l and AtPRR9 and VvPRR5 (data not shown). The former syntenic relationships between AtPRR3 and VvPRR3, AtPRR7 and VvPRR7, AtPRR5 and VvPRR5 and AtPRR9 and VvPRR9l were more conserved than the latter relationships between AtPRR3 and VvPRR7, AtPRR7 and VvPRR3, AtPRR5 and VvPRR9l and AtPRR9 and VvPRR5, respectively. Syntenic relationships were not found between the neighbouring regions of P. trichocarpa PRR1/TOC1 and those of other PRR1/TOC1s, or between the neighbouring region of C. papaya PRR9l and those of other PRR9/9ls. It is not clear whether these observations can be ascribed to chromosomal rearrangements or fragmentations of genomic sequences in small contigs.
Figure 6.
Chromosomal syntenies of flanking regions of eudicotyledonous PRR genes. Syntenic relationships of flanking regions of PRR1/TOC1 (A), PRR5 (B), PRR3 (C), PRR7 (D) and PRR9/9l (E) were examined using the comparative genomic tool, CoGe [29]. Syntenic relationships within A. thaliana or P. trichocarpa were analyzed by a comparative genomic tool, CoGe, and according to previous studies [17,27,28,59]. Diamonds colored with black (A. thaliana), green (C. papaya), blue (P. trichocarpa) and red (V. vinifera) indicate individual genes. White diamonds marked with arrows indicate PRR genes. Genes with no syntenic matches on the selected regions are not plotted. Orthologous genes are connected by broken lines. Diamonds with characters on the right side of strands indicate angiosperm polyploidy event (α, β and salicoid). The salicoid polyploidy event is shown as the diamond with the initial letter of salicoid (S). The lengths of the genomic regions are shown on the left.
In the genome of A. thaliana, the flanking regions of PRR genes showed syntenies with one or three partial regions of its genome (Figure 6). These syntenic relationships originated from the chromosomal duplications that arose from the β and α polyploidy events [27,28]. In the P. trichocarpa genome, two copies of each PRR5, PRR7, and PRR9l gene were located at the syntenic regions of chromosomes 12 and 15 (Figure 6B), those of chromosomes 8 and 10 (Figure 6D), and those of chromosomes 2 and 14 (Figure 6E), respectively. Tuskan et al., [17] showed that these syntenic regions were produced via the salicoid polyploidy event. Although flanking region of A. thaliana PRR3 shares syntenic relationships with partial regions of P. trichocarpa chromosomes 1 and 9, these two partial regions did not retain a PRR gene (Figure 6C).
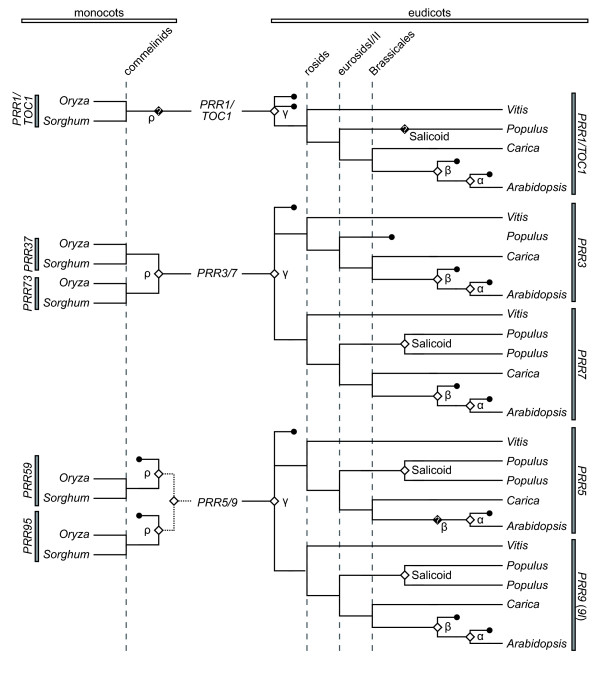
Next, we investigated the chromosomal syntenic relationships derived from the γ triplication event using the genomic information of V. vinifera [16]. There were chromosomal syntenies conserved between the flanking regions of VvPRR3 and VvPRR7 and between the flanking regions of VvPRR5 and VvPRR9l, which were originated from the γ triplication event ([16], see also the comparative genomic tool, CoGe [29]). The synteny of PRR3, 5, 7 and 9l in V. vinifera and homologous genes in other eudicotyledonous plants (A. thaliana, P. trichocarpa, and C. papaya), respectively, suggest that the ancestral PRR3/7 gene in eudicots was duplicated into PRR3 and PRR7, and PRR5/9 into PRR5 and PRR9/9l in the γ polyploidy event (Figure 7). After the γ polyploidy event, one copy of each PRR gene (PRR1/TOC1, PRR3, 5, 7 and 9/9l) has been conserved in the present genomes of V. vinifera and C. papaya, which apparently have not undergone additional polyploidy events. Although A. thaliana PRR genes were repeatedly duplicated by the β and/or α polyploidy events, one copy of each gene remains in the present A. thaliana genome, which is similar to V. vinifera and C. papaya genomes (Figures 6 and 7). In the genome of P. trichocarpa, PRR5, 7, and 9l were duplicated in the subsequent salicoid polyploidy event, but the PRR3 was lost prior to the salicoid polyploidy event or duplicated PRR3s were lost following the polyploidy event.
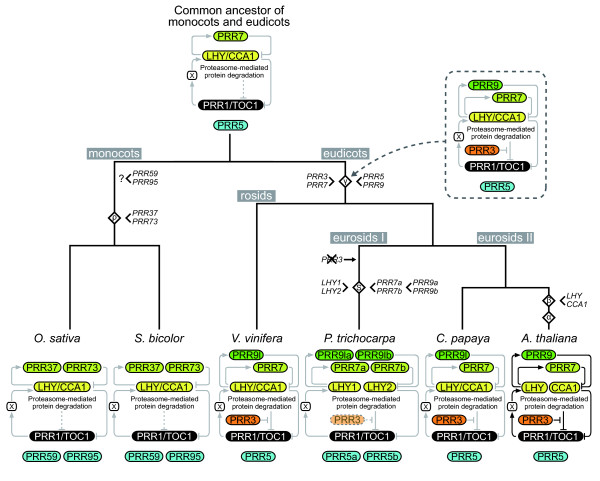
Figure 7.
Evolutionary processes of PRR genes in angiosperms reconstructed by phylogenetic analysis and syntenic relationships. Chromosomal syntenies among eudicots or monocots were analyzed by the comparative genomic tool, CoGe [29] or VISTA Browser [31], and according to previous studies [11,16-18,27,28,30,59]. Diamonds and circles indicate gene duplication and gene loss event, respectively. The timing of a gene duplication event that is not clear in the previous studies is shown by dotted line. Black diamonds with a question mark indicate that a gene duplication event derived from a polyploidy event is not resolved. Timings of plant speciation in commelinids, rosids, eurosids I/II and Brassicales are described by broken lines.
The flanking region of the PRR gene in O. sativa (OsPRR1/TOC1, OsPRR73, OsPRR59 and OsPRR95) showed conserved synteny with that of orthologous genes in S. bicolor [30]. On the other hand, only a few syntenic regions were identified between PRR37 of O. sativa and the orthologous gene of S. bicolor. In the genome of O. sativa, the neighbouring region of OsPRR37 showed synteny with that of OsPRR73. This syntenic relationship resulted from the chromosomal duplication that occurred in the ρ polyploidy event [18]. The ρ polyploidy event also resulted in conserved chromosomal synteny between the flanking region of OsPRR59 and a partial region of chromosome 8, and between the flanking region of OsPRR95 and a different partial region of chromosome 8. However, these partial regions of chromosome 8 have lost PRR genes ([18], see also the comparative genomic tool, VISTA Browser [31]). These results indicated that the gene duplication event resulting in PRR37 and PRR73 was the monocotyledonous ρ polyploidy event, and that PRR59 and PRR95 were duplicated via the ρ polyploidy event but one of the duplicated genes was lost from genomes of O. sativa and S. bicolor (Figure 7).
Discussion
The plant clock system consists of multiple interlocked feedback loops, which are comprised predominantly of two gene families, LHY/CCA1s and PRRs [4,5]. These gene families are conserved among both monocots and eudicots [8]. To clarify the evolutionary process of the plant clock system, we recently reported the molecular phylogeny of LHY/CCA1 genes in angiosperms [9]. Furthermore, in the present study, we reconstructed phylogenetic relationships among clock-associated PRR genes in monocots and eudicots using two approaches: reconstruction of phylogenetic trees and examination of syntenic relationships. Together, these phylogenetic analyses of the plant circadian clock related-genes, LHY/CCA1s and PRRs, are promising tools to unravel the evolutionary history of the plant clock system among angiosperm lineages.
Evolutionary processes of clock-associated PRR genes in angiosperms
PRR genes are conserved in angiosperms and at least five copies of PRR genes have been retained in their genomes (see Additional file 1). Angiosperm PRR genes are grouped into three clades (the PRR1/TOC1 clade, the PRR3 and 7 clade, and the PRR5 and 9 clade) that have already existed prior to the divergence of monocots and eudicots (Figures 2 and 3). After the speciation of monocots and eudicots, copy numbers of PRR genes independently increased in each lineage as a result of ancient chromosomal duplication events (Figure 7). In monocots, the ancestral PRR37/PRR73 was duplicated into PRR37 and PRR73 in the ρ polyploidy event that occurred before the speciation of O. sativa and S. bicolor [18,19]. In eudicots, the gene duplication events between PRR3 and PRR7 and between PRR5 and PRR9/9l are derived from the γ polyploidy event that took place before the speciation of Vitales (V. vinifera) and eurosid species (P. trichocarpa, C. papaya, and A. thaliana) [16]. In addition, our results show that PRR genes in P. trichocarpa have expanded more than those in other plant species (see Additional file 1). This expansion apparently resulted from the salicoid polyploidy event that occurred in the Populus lineage but not in other eudicots (V. vinifera, C. papaya, and A. thaliana) (Figure 7). Consequently, circadian clocks may have become more intricate networks after the speciation of monocots and eudicots if the additional genes have roles in the circadian networks.
In contrast to the increase in PRR genes via ancient chromosomal duplication events, the present genome of A. thaliana retains one copy of each PRR gene (PRR1/TOC1, PRR3, 5, 7, and 9) after β and α polyploidy events (Figure 7). Likewise, in Brassica rapa, PRR genes that were increased in a recent hexaploidization event have been reduced in its genome following the event, though the genome retains at least a set of PRR genes [32]. These results implied that dosages of the clock related-genes had been altered in the genomes during evolution. Since some reports showed that a gene dosage change of clock related genes influenced the clock regulatory network and downstream signals [33,34], plants may have flexibly modulated the complex network of the clock system after polyploidy events and subsequent gene deletion events. Indeed, allopolyploid Arabidopsis species can fine-tune a regulatory and stoichiometric balance of the circadian clock system to properly maintain a downstream homeostasis of the plants [35]. The history of gene duplications and deletions in the Arabidopsis clock system imply that the regulatory network of the clock system has maintained a degree of organization throughout the dynamic changes of copy numbers and functions of clock-related genes.
Phylogenetic footprint of the plant clock system in angiosperms
Loops I and III of the Arabidopsis clock system contain four PRR genes (PRR1/TOC1, PRR3, 7 and 9) and two LHY/CCA1 genes (LHY and CCA1) (Figure 1) [4,5,36]. Although the circadian clock-related genes in A. thaliana were duplicated via the β and α polyploidy events, the present genome of A. thaliana retains only one pair of LHY and CCA1 genes, which is derived from the β polyploidy event (Figure 8) [9]. As the β polyploidy event is assumed to have occurred in the Brassicaceae, LHY and CCA1 genes did not diverge before the speciation of A. thaliana and C. papaya, which is consistent with the fact that there is only one copy of the LHY/CCA1 gene in the genome of C. papaya [11]. Similar to the genome of A. thaliana, the C. papaya genome retained only one copy each of the PRR1/TOC1, PRR3, 7, and 9 genes (see Additional file 1). These results suggest that one copy of LHY/CCA1, PRR1/TOC1, PRR3, 7, and 9 was involved in the plant clock system in the common ancestor of A. thaliana and C. papaya.
Figure 8.
Proposed schematic diagram of the evolutionary process of the plant circadian clock system. For the plant clock system model, loop I and III are described in this diagram. Diamonds with characters indicate angiosperm polyploidy event (α, β, γ, salicoid and ρ). The salicoid polyploidy event is shown as the diamond with the initial letter of salicoid (S).
The evolutionary history of the plant clock system in two divergent members of Brassicales (A. thaliana and C. papaya) raises the question as to when the Arabidopsis-type clock apparatus arose in the evolutionary history of plants. Phylogenies of the circadian clock-related genes showed that a set of the genes, one copy of each LHY/CCA1, PRR1/TOC1, PRR3, 7 and 9l genes, is conserved in the genome of V. vinifera (Figures 7 and 8, see Additional file 1) [9,16]. Conservation of the set of clock-related genes suggests that the fundamental mechanism of the Arabidopsis-type clock apparatus was formed before the speciation of Vitales (V. vinifera) and eurosid species (P. trichocarpa, C. papaya, and A. thaliana), although it remains to be determined whether functional divergences between PRR3 and 7 and between PRR5 and 9/9l, which were duplicated in the γ triplication event, existed in the common ancestor of Vitales and eurosids (Figure 8).
Intriguingly, the clock system of P. trichocarpa might differ from the Arabidopsis clock system, because the P. trichocarpa PRR3 gene was lost and LHY/CCA1 and PRR7 and 9 were duplicated via the salicoid polyploidy event that occurred after the speciation of eurosids I and II (Figures 7 and 8). PRR3 protein in A. thaliana interacts with PRR1/TOC1 protein, which is a component of the main loop (Loop I), to inhibit its protein degradation [36]. The lack of PRR3 in Populus might influence the posttranslational regulation of PRR1/TOC1 protein or might be compensated by recruiting other PRR genes although conserved changes that would mark the PRR3 gene cluster were not identified in other Populus PRR proteins (data not shown). Furthermore, duplication of LHY/CCA1 and PRR7 and 9, but not PRR1/TOC1, could also affect the regulation mechanism of the Populus clock system, because a dosage balance in the plant clock system was ruined [33]. We recently revealed that Populus LHYs show typical diurnal expressions similar to LHY/CCA1 genes in other plant species [9,37-40], which is apparently contradictory to the speculation. The Populus clock system appears to retain a functional regulatory network in regard to the expression regulation of the LHY genes throughout the evolutionary changes of the circadian clock components.
The monocots O. sativa and S. bicolor retain one LHY/CCA1 gene and five PRR genes in their genomes (see Additional file 1) [9]. Phylogenetic analysis showed that the gene duplication events that produced PRR37 and 73, and PRR59 and 95 in monocots occurred separately and independently of the events that produced PRR3 and 7, and PRR5 and 9 in eudicots (Figure 7). This finding raises a complex question; what are the roles of these paralogous genes in the monocotyledonous clock system? The expression of PRR5 in A. thaliana is not regulated by light signals and reaches a peak of the diurnal rhythm around 8 h after dawn [10]. On the other hand, PRR9 in A. thaliana shows a light response expression, resulting in rhythmicity with peak expression just after dawn [10,41]. PRR59 and 95 in O. sativa have peak expressions around 9 h after dawn and are not induced by light signals [13], which appear to be more similar to the regulation of A. thaliana PRR5 expression than to that of A. thaliana PRR9 expression. In addition, peak expressions of O. sativa PRR37 and 73 are followed by expressions of PRP59 and 95, which may correspond to the sequential expression pattern of A. thaliana PRR7 and PRR5 [10,13]. These results collectively suggest that paralogous gene pairs PRR59/95 and PRR37/73 genes in monocots share functional roles with PRR5 and PRR7, respectively, in A. thaliana (Figure 8). Together, these data indicate that a common ancestor of monocots and eudicots may have had a main feedback loop (LHY/CCA1-PRR1/TOC1) that was not posttranslationally regulated by PRR3. Although the ancestral clock system appears to have been more simplified than that of the current Arabidopsis-type clock apparatus, it is assumed that the ancestral clock system have had the basic components reconstructing a primitive multiple feedback loop system.
Conclusions
The present study inferred the molecular phylogeny of angiosperm PRR genes that have key roles in the plant clock system. Clock-associated PRR genes diverged into three clades before the speciation of monocots and eudicots and, in addition, PRR3/7 and PRR5/9 underwent independent expansion in monocots and eudicots (Figure 7). Taken together with the molecular phylogeny of LHY/CCA1 genes [9], our studies suggest that the basic components of the Arabidopsis clock were established prior to the speciation of eudicots and monocots (Figure 8). Additional functional analyses and accumulation of genomic information from other plant species will clarify details of evolutionary and developmental processes of plant clock systems.
Methods
Retrieval of sequences of clock-associated PRR genes from draft genome sequences
PRR genes were retrieved from genomic databases for A. thaliana (TIGR Arabidopsis thaliana Database [42]) and O. sativa (Rice Annotation Project Database [43]). To identify PRR genes in S. bicolor, V. vinifera, P. trichocarpa, and C. papaya, TBLASTN searches were performed against the genomic databases using amino acid sequences encoded by PRR genes of A. thaliana or O. sativa as queries: JGI Sorghum bicolor v1.0 [44] for S. bicolor; Grape Genome Browser [45] for V. vinifera; JGI Populus trichocarpa v1.1 [46] for P. trichocarpa, and Papaya Genome Project v0.4 in CoGe [29] for C. papaya. Genes that retained the typical PR-domain at the N-terminal region and the CCT-motif at the C-terminal region were retrieved from these genomic databases. Genes that lacked the PR-domain or CCT-motif but showed significantly high similarity with typical PRR genes were also retrieved (those with E-values lower than 10-50 or >90% similarity). The genes retrieved from the genomic databases were aligned with PRR genes in A. thaliana and O. sativa using the TCoffee program [47]. Mispredicted genes, if found, were manually modified as follows. For predicted genes lacking a conserved portion of the PRR gene, we searched the database for expressed sequence tags (ESTs) of the gene (TIGR Plant Transcript Assemblies [48]) and re-annotated by assembling the predicted gene and relevant ESTs. In some cases, the open reading frame of the gene was repredicted by the Fgenesh+ program [49]. When the exon-intron boundary of a gene was mis-demarcated, we improved the boundary based on standard donor/acceptor splice sites without resulting in a frame shift.
Phylogenetic analysis
Amino acid sequences were deduced from nucleotide sequences of the predicted PRR genes and then aligned using the TCoffee program [47]. The alignment around the flanking region of the PR-domain was manually corrected based on the exon-intron structure. The number of amino acids substituted between each pair of PRR proteins was estimated by the Jones-Taylor-Thornton (JTT) model [50] with the complete- deletion option. From the number of estimated amino acid substitutions, a phylogenetic tree was reconstructed by the ME method [51]. Bootstrap values were calculated with 1,000 replications using the ME method [52]. PRR1/TOC1 genes were utilized as an outgroup in the phylogenetic trees. These procedures were performed using the MEGA4 software [53,54].
Detection of functional divergence
Type I and type II functional divergences among PRR gene clades was examined using the DIVERGE 2.0 software [24,55,56]. To calculate coefficient of type I and type II functional divergences (θI and θII), we used the protein sequence alignment constructed using the TCoffee program [47] and the topology and branch length of phylogenetic tree reconstructed by the ME method. Cutoff values of the posterior probability and posterior ratio were set empirically by sequentially removing the highest scoring sites from the alignment until θ = 0. Sequence logos were generated with WebLogo version 2.8.2 [57,58].
Identification of chromosomal synteny
Conservation of chromosomal synteny in V. vinifera, P. trichocarpa, C. papaya, and A. thaliana was determined as follows. First, we reconstructed the ancient gene organization of the flanking regions of A. thaliana PRR genes before the α and β polyploidy events using the chromosomal syntenies reported in previous studies [27,28]. Then, we compared the syntenic relationships between the ancient gene organization in A. thaliana and the flanking regions of PRR genes in V. vinifera, P. trichocarpa, and C. papaya using the comparative genomic tool, CoGe [11,29,59]. This process also reconstructed chromosomal syntenies in P. trichocarpa that were derived from the salicoid polyploidy event [17]. To clarify syntenic relationships derived from the γ polyploidy event, we used information on chromosomal syntenies within the genome of V. vinifera [16] and the comparative genomic tool, CoGe [29].
Syntenic relationships between the flanking regions of PRR genes in O. sativa and those in S. bicolor were reconfirmed using the chromosomal syntenies reported in previous studies [30] and a comparative genomic tool VISTA Browser [31]. To reconstruct chromosomal syntenies of the flanking regions of PRR genes derived from the monocotyledonous ρ polyploidy event, syntenic regions were identified according to methods reported previously using the O. sativa genomic sequence [18].
Authors' contributions
NT conceived of the study, performed all analyses and wrote the manuscript. MU, SS and CTS participated in coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
PRR genes in angiosperms used in the present study. aPlant classification refers to APGII [61]. bGene ID corresponds to the name obtained from Rice Annotation Project Database [43]. cGene ID corresponds to the name obtained from JGI Sorghum bicolor v1.0 [44]. dGene IDs correspond to the names obtained from TIGR Plant Transcript Assemblies [48]. eGene ID corresponds to the name obtained from Grape Genome Browser [45]. fGene IDs correspond to the names obtained from JGI Populus trichocarpa v1.1 [46]. gGene IDs correspond to the names obtained from TIGR Plant Genomics [62]. hGene IDs correspond to the names obtained from Comparative Genomics Homepage [29].*Genes, which appeared to be misprediced, were manually modified in this study. **Gene is misannotated.***Genes lack the PR-domain.
Predicted PRR genes in angiosperms. Angiosperm PRR genes were retrieved from genomic databases for A. thaliana (TIGR Arabidopsis thaliana Database, http://www.tigr.org/tdb/e2k1/ath1/), V. vinifera (Grape Genome Browser, http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/), P. trichocarpa (JGI Populus trichocarpa v1.1, http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html), C. papaya (Papaya Genome Project v0.4 in CoGe, http://synteny.cnr.berkeley.edu/CoGe/), O. sativa (Rice Annotation Project Database, http://rapdb.dna.affrc.go.jp/) and S. bicolor (JGI Sorghum bicolor v1.0, http://genome.jgi-psf.org/Sorbi1/Sorbi1.home.html).
Alignments of the amino acid sequences encoded by angiosperm PRR genes. Amino acid sequences were aligned using TCoffee program http://www.ebi.ac.uk/t-coffee/. Amino acid conservation was highlighted by the boxshade program http://www.ch.embnet.org/software/BOX_form.html. Identical and similar amino acid residues are highlighted with black and gray shading, respectively. Blue boxes indicate the regions that were represented in Figures 4 and 5.
Alignment of the amino acid sequences encoded by (A)AtPRR9s and (B)PtPRR5s. Sequence similarity is indicated below the alignment using the symbols "asterisk," "colon," and "dot" for identical, highly similar, and weakly similar residues, respectively. Black shadings indicate the PR-domain and the CCT-motif. Accession numbers and gene IDs of the PRR genes are shown in Additional file 1.
Nucleotide sequences around the region of the exon-intron boundaries of angiosperm PRR genes at the flanking region of PR-domain. Black and gray shadings on the alignments indicate a site of exon-intron boundary and one-amino acid deletion, respectively. Higher degree of conservation of nucleotide sequence is shown by the bigger size of letters.
Phylogenetic tree of PRR genes reconstructed by the Neighbor-Joining (NJ) method. Full-length amino acid sequences were aligned using TCoffee program. The phylogenetic tree was reconstructed by the NJ method from the numbers of amino acid substitutions estimated by applying the JTT model. The numerals at the branch indicate bootstrap values calculated by the NJ method with 1,000 replications. Bootstrap values >50% are shown.
Phylogenetic trees of PRR genes reconstructed by the Maximum likelihood (ML) and Bayesian methods. Full-length amino acid sequences were aligned using TCoffee program. The phylogenetic trees were reconstructed by the ML and Bayesian methods with applying the JTT model. The ML and Bayesian analyses were performed using the PhyML http://www.atgc-montpellier.fr/ and MrBayes http://mrbayes.csit.fsu.edu/ programs, respectively. PRR1/TOC1 genes were utilized as an outgroup in the phylogenetic trees. Support for the branches was calculated as percent of 100 bootstrap replications of the ML method (left) and Baysian posterior probabilities (right). The values >50% are shown.
Contributor Information
Naoki Takata, Email: naoki.takata@plantphys.umu.se.
Shigeru Saito, Email: sshigeru@nips.ac.jp.
Claire Tanaka Saito, Email: claire@iwate-u.ac.jp.
Matsuo Uemura, Email: uemura@iwate-u.ac.jp.
Acknowledgements
We thank Dr. Maria E. Eriksson (Umeå Plant Science Centre, Umeå University) for critical reading of the manuscript. This work was supported in part by a Grant-in-Aid for the 21st Century COE Program from the Ministry of Education, Culture, Sports, Science and Technology of Japan (K-3 to MU) and by a grant from the Japan Society for the Promotion of Science (19·9498 to NT).
References
- Yakir E, Hilman D, Harir Y, Green RM. Regulation of output from the plant circadian clock. FEBS J. 2007;274:335–345. doi: 10.1111/j.1742-4658.2006.05616.x. [DOI] [PubMed] [Google Scholar]
- McClung CR. Plant circadian rhythms. Plant Cell. 2006;18:792–803. doi: 10.1105/tpc.106.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P. Circadian clock signaling in Arabidopsis thaliana: from gene expression to physiology and development. Int J Dev Biol. 2005;49:491–500. doi: 10.1387/ijdb.041968pm. [DOI] [PubMed] [Google Scholar]
- Locke JC, Kozma-Bognár L, Gould PD, Fehér B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ. Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol. 2006;2:59. doi: 10.1038/msb4100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilinger MN, Farré EM, Taylor SR, Kay SA, Doyle FJ. A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol Syst Biol. 2006;2:58. doi: 10.1038/msb4100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- McClung CR. Comes a time. Curr Opin Plant Biol. 2008;11:514–520. doi: 10.1016/j.pbi.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Murakami M, Tago Y, Yamashino T, Mizuno T. Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 2007;48:110–121. doi: 10.1093/pcp/pcl043. [DOI] [PubMed] [Google Scholar]
- Takata N, Saito S, Saito CT, Nanjo T, Shinohara K, Uemura M. Molecular phylogeny and expression of poplar circadian clock genes, LHY1 and LHY2. New Phytol. 2009;181:808–819. doi: 10.1111/j.1469-8137.2008.02714.x. [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Mizuno T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol. 2000;41:1002–1012. doi: 10.1093/pcp/pcd043. [DOI] [PubMed] [Google Scholar]
- Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KL, Salzberg SL, Feng L, Jones MR, Skelton RL, Murray JE, Chen C, Qian W, Shen J, Du P, Eustice M, Tong E, Tang H, Lyons E, Paull RE, Michael TP, Wall K, Rice DW, Albert H, Wang ML, Zhu YJ, Schatz M, Nagarajan N, Acob RA, Guan P, Blas A, Wai CM, Ackerman CM, Ren Y, Liu C, Wang J, Wang J, Na JK, Shakirov EV, Haas B, Thimmapuram J, Nelson D, Wang X, Bowers JE, Gschwend AR, Delcher AL, Singh R, Suzuki JY, Tripathi S, Neupane K, Wei H, Irikura B, Paidi M, Jiang N, Zhang W, Presting G, Windsor A, Navajas-Pérez R, Torres MJ, Feltus FA, Porter B, Li Y, Burroughs AM, Luo MC, Liu L, Christopher DA, Mount SM, Moore PH, Sugimura T, Jiang J, Schuler MA, Friedman V, Mitchell-Olds T, Shippen DE, dePamphilis CW, Palmer JD, Freeling M, Paterson AH, Gonsalves D, Wang L, Alam M. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Carvajal GA, Morse AM, Davis JM. Transcript profiles of the cytokinin response regulator gene family in Populus imply diverse roles in plant development. New Phytol. 2008;177:77–89. doi: 10.1111/j.1469-8137.2007.02240.x. [DOI] [PubMed] [Google Scholar]
- Murakami M, Ashikari M, Miura K, Yamashino T, Mizuno T. The evolutionarily conserved OsPRR quintet: rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 2003;44:1229–1236. doi: 10.1093/pcp/pcg135. [DOI] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyère C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, Del Fabbro C, Alaux M, Di Gaspero G, Dumas V, Felice N, Paillard S, Juman I, Moroldo M, Scalabrin S, Canaguier A, Le Clainche I, Malacrida G, Durand E, Pesole G, Laucou V, Chatelet P, Merdinoglu D, Delledonne M, Pezzotti M, Lecharny A, Scarpelli C, Artiguenave F, Pè ME, Valle G, Morgante M, Caboche M, Adam-Blondon AF, Weissenbach J, Quétier F, Wincker P. French-Italian Public Consortium for Grapevine Genome Characterization. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, Sterck L, Aerts A, Bhalerao RR, Bhalerao RP, Blaudez D, Boerjan W, Brun A, Brunner A, Busov V, Campbell M, Carlson J, Chalot M, Chapman J, Chen GL, Cooper D, Coutinho PM, Couturier J, Covert S, Cronk Q, Cunningham R, Davis J, Degroeve S, Déjardin A, Depamphilis C, Detter J, Dirks B, Dubchak I, Duplessis S, Ehlting J, Ellis B, Gendler K, Goodstein D, Gribskov M, Grimwood J, Groover A, Gunter L, Hamberger B, Heinze B, Helariutta Y, Henrissat B, Holligan D, Holt R, Huang W, Islam-Faridi N, Jones S, Jones-Rhoades M, Jorgensen R, Joshi C, Kangasjärvi J, Karlsson J, Kelleher C, Kirkpatrick R, Kirst M, Kohler A, Kalluri U, Larimer F, Leebens-Mack J, Leplé JC, Locascio P, Lou Y, Lucas S, Martin F, Montanini B, Napoli C, Nelson DR, Nelson C, Nieminen K, Nilsson O, Pereda V, Peter G, Philippe R, Pilate G, Poliakov A, Razumovskaya J, Richardson P, Rinaldi C, Ritland K, Rouzé P, Ryaboy D, Schmutz J, Schrader J, Segerman B, Shin H, Siddiqui A, Sterky F, Terry A, Tsai CJ, Uberbacher E, Unneberg P, Vahala J, Wall K, Wessler S, Yang G, Yin T, Douglas C, Marra M, Sandberg G, Peer Y Van de, Rokhsar D. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Salse J, Bolot S, Throude M, Jouffe V, Piegu B, Quraishi UM, Calcagno T, Cooke R, Delseny M, Feuillet C. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell. 2008;20:11–24. doi: 10.1105/tpc.107.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, Zhang J, Zhang Y, Li R, Xu Z, Li S, Li X, Zheng H, Cong L, Lin L, Yin J, Geng J, Li G, Shi J, Liu J, Lv H, Li J, Wang J, Deng Y, Ran L, Shi X, Wang X, Wu Q, Li C, Ren X, Wang J, Wang X, Li D, Liu D, Zhang X, Ji Z, Zhao W, Sun Y, Zhang Z, Bao J, Han Y, Dong L, Ji J, Chen P, Wu S, Liu J, Xiao Y, Bu D, Tan J, Yang L, Ye C, Zhang J, Xu J, Zhou Y, Yu Y, Zhang B, Zhuang S, Wei H, Liu B, Lei M, Yu H, Li Y, Xu H, Wei S, He X, Fang L, Zhang Z, Zhang Y, Huang X, Su Z, Tong W, Li J, Tong Z, Li S, Ye J, Wang L, Fang L, Lei T, Chen C, Chen H, Xu Z, Li H, Huang H, Zhang F, Xu H, Li N, Zhao C, Li S, Dong L, Huang Y, Li L, Xi Y, Qi Q, Li W, Zhang B, Hu W, Zhang Y, Tian X, Jiao Y, Liang X, Jin J, Gao L, Zheng W, Hao B, Liu S, Wang W, Yuan L, Cao M, McDermott J, Samudrala R, Wang J, Wong GK, Yang H. The Genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3:266–281. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bocock PN, Morse AM, Dervinis C, Davis JM. Evolution and diversity of invertase genes in Populus trichocarpa. Planta. 2008;227:565–576. doi: 10.1007/s00425-007-0639-3. [DOI] [PubMed] [Google Scholar]
- Sampedro J, Lee Y, Carey RE, dePamphilis C, Cosgrove DJ. Use of genomic history to improve phylogeny and understanding of births and deaths in a gene family. Plant J. 2005;44:409–419. doi: 10.1111/j.1365-313X.2005.02540.x. [DOI] [PubMed] [Google Scholar]
- Gu X. Maximum-likelihood approach for gene family evolution under functional divergence. Mol Biol Evol. 2001;18:453–464. doi: 10.1093/oxfordjournals.molbev.a003824. [DOI] [PubMed] [Google Scholar]
- Gu X. A simple statistical method for estimating type-II (cluster-specific) functional divergence of protein sequences. Mol Biol Evol. 2006;23:1937–1945. doi: 10.1093/molbev/msl056. [DOI] [PubMed] [Google Scholar]
- Ito S, Matsushika A, Yamada H, Sato S, Kato T, Tabata S, Yamashino T, Mizuno T. Characterization of the APRR9 pseudo-response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 2003;44:1237–1245. doi: 10.1093/pcp/pcg136. [DOI] [PubMed] [Google Scholar]
- Kiba T, Henriques R, Sakakibara H, Chua NH. Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell. 2007;19:2516–2530. doi: 10.1105/tpc.107.053033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Thomas BC, Pederson B, Freeling M. Following tetraploidy in an Arabidopsis ancestor, gene were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006;16:934–946. doi: 10.1101/gr.4708406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CoGe. http://synteny.cnr.berkeley.edu/CoGe/
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev IV, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, McCann MC, Ming R, Peterson DG, Mehboob-ur-Rahman, Ware D, Westhoff P, Mayer KF, Messing J, Rokhsar DS. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- VISTA Browser. http://genome.lbl.gov/vista/index.shtml
- Kim JA, Yang TJ, Kim JS, Park JY, Kwon SJ, Lim MH, Jin M, Lee SC, Lee SI, Choi BS, Um SH, Kim HI, Chun C, Park BS. Isolation of circadian-associated genes in Brassica rapa by comparative genomics with Arabidopsis thaliana. Mol Cells. 2007;23:145–153. [PubMed] [Google Scholar]
- Más P, Alabadí D, Yanovsky MJ, Oyama T, Kay SA. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell. 2003;15:223–236. doi: 10.1105/tpc.006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Kim WY, Geng R. The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell. 2004;16:769–782. doi: 10.1105/tpc.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–331. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Wang L, Han L, Suh SS, Salomé PA, McClung CR, Somers DE. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem. 2008;283:23073–23083. doi: 10.1074/jbc.M803471200. [DOI] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 2002;16:2006–2020. doi: 10.1101/gad.999202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldis AD, Kousidis P, Kesanopoulos K, Prombona A. Light and circadian regulation in the expression of LHY and Lhcb genes in Phaseolus vulgaris. Plant Mol Biol. 2003;52:981–997. doi: 10.1023/A:1025433529082. [DOI] [PubMed] [Google Scholar]
- Ramos A, Pérez-Solís E, Ibáñez C, Casado R, Collada C, Gómez L, Aragoncillo C, Allona I. Winter disruption of the circadian clock in chestnut. Proc Natl Acad Sci USA. 2005;102:7037–7042. doi: 10.1073/pnas.0408549102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/S0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- Ito S, Matsushika A, Yamada H, Sato S, Kato T, Tabata S, Yamashino T, Mizuno T. Characterization of the APRR9 pseudo-response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 2003;44:1237–1245. doi: 10.1093/pcp/pcg136. [DOI] [PubMed] [Google Scholar]
- TIGR Arabidopsis thaliana Database. http://www.tigr.org/tdb/e2k1/ath1/
- Rice Annotation Project Database. http://rapdb.dna.affrc.go.jp/
- JGI Sorghum bicolor v1.0. http://genome.jgi-psf.org/Sorbi1/Sorbi1.home.html
- Grape Genome Browser. http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/
- JGI Populus trichocarpa v1.1. http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html
- TCoffee. http://www.ebi.ac.uk/t-coffee/
- TIGR Plant Transcript Assemblies. http://plantta.tigr.org/
- Fgenesh+ http://linux1.softberry.com/berry.phtml
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Rzhetsky A, Nei M. A simple method for estimating and testing minimum evolution trees. Mol Biol Evol. 1992;9:945–967. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- MEGA4. http://www.megasoftware.net/index.html
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Gu X, Velden K Vander. DIVERGE: phylogeny-based analysis for functional-structural divergence of a protein family. Bioinformatics. 2002;18:500–501. doi: 10.1093/bioinformatics/18.3.500. [DOI] [PubMed] [Google Scholar]
- DIVERGE 2.0. http://xgu.zool.iastate.edu/
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WebLogo 2.8.2. http://weblogo.berkeley.edu/
- Lyons E, Freeling M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008;53:661–673. doi: 10.1111/j.1365-313X.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- APGII. http://www.mobot.org/MOBOT/Research/APweb/welcome.html
- TIGR Plant Genomics. http://plantgenomics.tigr.org/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRR genes in angiosperms used in the present study. aPlant classification refers to APGII [61]. bGene ID corresponds to the name obtained from Rice Annotation Project Database [43]. cGene ID corresponds to the name obtained from JGI Sorghum bicolor v1.0 [44]. dGene IDs correspond to the names obtained from TIGR Plant Transcript Assemblies [48]. eGene ID corresponds to the name obtained from Grape Genome Browser [45]. fGene IDs correspond to the names obtained from JGI Populus trichocarpa v1.1 [46]. gGene IDs correspond to the names obtained from TIGR Plant Genomics [62]. hGene IDs correspond to the names obtained from Comparative Genomics Homepage [29].*Genes, which appeared to be misprediced, were manually modified in this study. **Gene is misannotated.***Genes lack the PR-domain.
Predicted PRR genes in angiosperms. Angiosperm PRR genes were retrieved from genomic databases for A. thaliana (TIGR Arabidopsis thaliana Database, http://www.tigr.org/tdb/e2k1/ath1/), V. vinifera (Grape Genome Browser, http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/), P. trichocarpa (JGI Populus trichocarpa v1.1, http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html), C. papaya (Papaya Genome Project v0.4 in CoGe, http://synteny.cnr.berkeley.edu/CoGe/), O. sativa (Rice Annotation Project Database, http://rapdb.dna.affrc.go.jp/) and S. bicolor (JGI Sorghum bicolor v1.0, http://genome.jgi-psf.org/Sorbi1/Sorbi1.home.html).
Alignments of the amino acid sequences encoded by angiosperm PRR genes. Amino acid sequences were aligned using TCoffee program http://www.ebi.ac.uk/t-coffee/. Amino acid conservation was highlighted by the boxshade program http://www.ch.embnet.org/software/BOX_form.html. Identical and similar amino acid residues are highlighted with black and gray shading, respectively. Blue boxes indicate the regions that were represented in Figures 4 and 5.
Alignment of the amino acid sequences encoded by (A)AtPRR9s and (B)PtPRR5s. Sequence similarity is indicated below the alignment using the symbols "asterisk," "colon," and "dot" for identical, highly similar, and weakly similar residues, respectively. Black shadings indicate the PR-domain and the CCT-motif. Accession numbers and gene IDs of the PRR genes are shown in Additional file 1.
Nucleotide sequences around the region of the exon-intron boundaries of angiosperm PRR genes at the flanking region of PR-domain. Black and gray shadings on the alignments indicate a site of exon-intron boundary and one-amino acid deletion, respectively. Higher degree of conservation of nucleotide sequence is shown by the bigger size of letters.
Phylogenetic tree of PRR genes reconstructed by the Neighbor-Joining (NJ) method. Full-length amino acid sequences were aligned using TCoffee program. The phylogenetic tree was reconstructed by the NJ method from the numbers of amino acid substitutions estimated by applying the JTT model. The numerals at the branch indicate bootstrap values calculated by the NJ method with 1,000 replications. Bootstrap values >50% are shown.
Phylogenetic trees of PRR genes reconstructed by the Maximum likelihood (ML) and Bayesian methods. Full-length amino acid sequences were aligned using TCoffee program. The phylogenetic trees were reconstructed by the ML and Bayesian methods with applying the JTT model. The ML and Bayesian analyses were performed using the PhyML http://www.atgc-montpellier.fr/ and MrBayes http://mrbayes.csit.fsu.edu/ programs, respectively. PRR1/TOC1 genes were utilized as an outgroup in the phylogenetic trees. Support for the branches was calculated as percent of 100 bootstrap replications of the ML method (left) and Baysian posterior probabilities (right). The values >50% are shown.