Abstract
Background
Teneurins are transmembrane proteins that assist morphogenetic processes in many organisms. ten-1 is the C. elegans teneurin homolog with two transcripts, ten-1a and ten-1b, that respectively encode a long (TEN-1L) and short (TEN-1S) form of the protein. We previously isolated a C. elegans mutant where one pharyngeal neuron was frequently misplaced, and now show that it corresponds to a novel allele of ten-1.
Results
The novel ten-1(et5) allele is a hypomorph since its post-embryonic phenotype is weaker than the null alleles ten-1(ok641) and ten-1(tm651). ten-1 mutants have defects in all pharyngeal neurons that we examined, and in vivo reporters show that only the long form of the ten-1 gene is expressed in the pharynx, specifically in six marginal cells and the M2 neurons. Defects in the pharyngeal M2 neurons were enhanced when the ten-1(ok641) mutation was combined with mutations in the following genes: mig-14, unc-5, unc-51, unc-52 and unc-129. None of the body neurons examined show any defects in the ten-1(ok641) mutant, but genetic interaction studies reveal that ten-1(ok641) is synthetic lethal with sax-3, unc-34 and unc-73, and examination of the hypodermal cells in embryos of the ten-1(ok641) mutant point to a role of ten-1 during hypodermal cell morphogenesis.
Conclusions
Our results are consistent with ten-1 normally providing a function complementary to the cytoskeletal remodeling processes that occur in migrating cells or cells undergoing morphogenesis. It is possible that ten-1 influences the composition/distribution of extracellular matrix.
Background
Teneurins are transmembrane proteins that participate in morphogenetic processes in many organisms [1,2]. Teneurins have a single transmembrane domain, a very large and cleavable extracellular domain containing eight EGF repeats, four NHL domains and more than 20 YD repeats, as well as a cleavable intracellular domain (ICD) that can be translocated to the nucleus. The Drosophila homologs, Ten-m and Ten-a, are the only pair-rule genes that do not encode traditional transcription factors [3-5]. Instead, they act at the cellular blastoderm stage, and cleavage of the ICD may allow it to directly regulate the transcription of target genes in alternate parasegments. Drosophila Ten-m is also important for several other developmental processes, including retina development [6], and peripheral nervous system development in imaginal disc-derived organs [7]. In vertebrates, the teneurin genes are expressed most prominently in developing neuronal tissues and are important for neuronal patterning and axon guidance [1,2,8]. The distinct expression profiles of various teneurins or teneurin isoforms in vertebrates, together with the neuronal defects observed in mutants, strongly suggest that teneurins act during cell communication to influence neurite outgrowth and guide axons [1,8,9]. Biochemical studies on the four mouse teneurins (Ten-m1 to Ten-m4) have shown that the EGF domains are important for teneurin homo- or heterodimerization via covalent disulfide links between the second and fifth EGF repeats [10,11], while the NHL and YD repeats form large glycosylated globular domains that may mediate homotypic or heterotypic interactions between cells that express the same or different forms of teneurin, as well as interactions with the extracellular matrix [1,2].
In C. elegans, the ten-1 gene can be transcribed from two distinct promoters to produce the transcripts ten-1a and ten-1b, which respectively encode two isoforms of the protein: TEN-1L, which contains all the teneurin domains described above (see Fig. 1F), and TEN-1S, which lacks the ICD but contains the rest of the protein including the transmembrane domain. Two null alleles of C. elegans ten-1 have previously been isolated using a reverse genetics approach. These ten-1 mutants are reported to have defects in the growth of several axons and in the development of the gonads and epidermis, and these defects correlated with disruptions in the extracellular matrix (ECM). These observations, together with the fact that ten-1 acts redundantly with genes encoding ECM components or adhesion molecules, such as integrin alpha (ina-1), dystroglycan (dgn-1), laminin alpha beta (epi-1) and nidogen (nid-1), have led to the conclusion that teneurin in C. elegans may have its primary function as an ECM organizer [2,12,13].
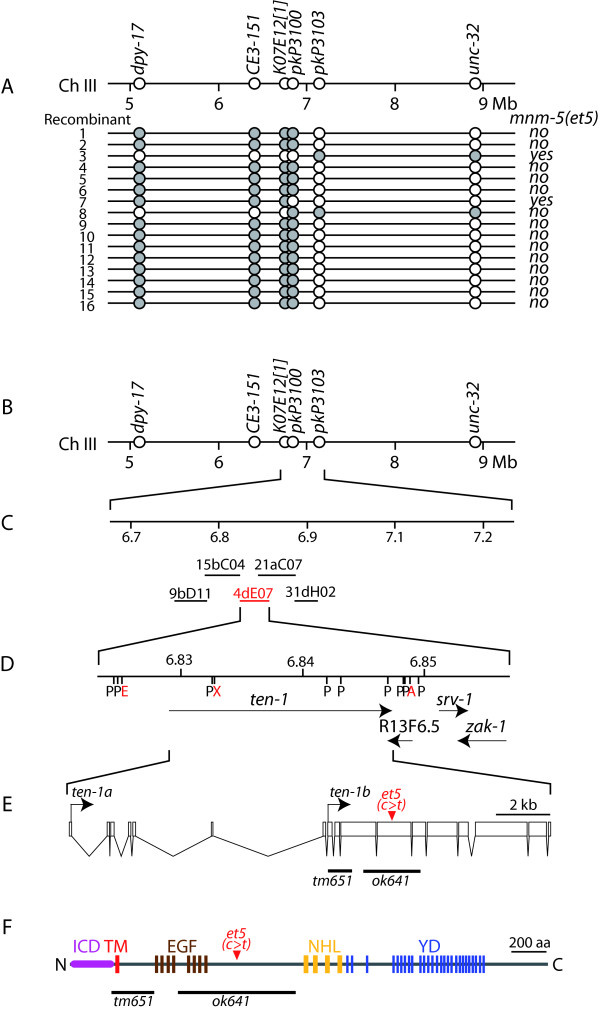
Figure 1.
Mapping of ten-1(et5). (A) Shows part of the map for chromosome III, including the positions of the genetic markers used to map mnm-5/ten-1(et5). Also shown are the structures of 16 informative recombinants among the F2 progeny of dpy-17 ten-1(et5) unc-32 mutants crossed with the Hawaiian strain CB4860. Open circles refer to the N2 allele at each locus. (B) and (C) Show an enlarged region of chromosome III and the locations of five fosmids that were tested for their ability to rescue the ten-1(et5) mutant. The full name for each fosmid includes the prefix "WRM06", and the rescuing fosmid WRM064dE07 is highlighted in red. (D) Partial restriction map of the WRM064dE07 fosmid, which contains the genes ten-1, srv-1, zak-1 and R13F6.5. The restriction enzyme sites AhdI, EcoNI, PstI and XhoI sites are respectively indicated as A, E, P and XhoI. (E) Structure of the ten-1 gene showing the two transcription starts, the regions deleted in the tm651 and ok641 alleles and the position of the et5 c>t nucleotide substitution. (F) Domain structure of the TEN-1 protein.
In the present study we provide novel insights into the functions of ten-1. Specifically we provide a detailed characterization of its embryonic expression, identify the expressing cells within the pharynx, show that ten-1 contributes to the development of several pharyngeal axons, and identify several genes with which ten-1 interacts. We also take advantage of the novel nature of the ten-1(et5) mutant, i.e. truncation just after the EGF domains, to draw structure-function conclusions.
Results
mnm-5(et5) is a novel allele of ten-1, and is therefore renamed ten-1(et5)
We previously isolated the mnm-5(et5) mutant in a forward genetics screen for mutants with abnormal M2 neurons [14], and later also found that it causes defects in the pharyngeal neurons NSMR and NSML [15]. By genetic mapping using recombinants between visible genetic markers, then between single nucleotide polymorphisms that distinguish the parental N2 strain from the Hawaiian strain CB4856, we defined the position of the mnm-5 gene to within a narrow region of chromosome III (Fig. 1A). We then tested five fosmids covering the genetic area of interest for their ability to rescue the mnm-5(et5) mutant (Fig. 1B-C). One fosmid, WRM064dE07, scored positive in this assay. From that fosmid we cut and purified an AhdI-EcoNI restriction fragment that contains the ten-1 gene but no other complete gene (Fig. 1D). This fragment also rescued the mnm-5(et5) mutant, showing that the mutation corresponds to a novel allele of the ten-1 gene. This was confirmed by sequencing: the et5 allele corresponds to a c>t point mutation that introduces a stop codon just downstream of the 8 EGF repeats in the extracellular domain of the TEN-1 protein (Fig. 1E-F). mnm-5(et5), which will henceforth be referred to as ten-1(et5).
ten-1(et5) is not a null allele
Two deletion mutant alleles of ten-1, i.e. alleles ok641 and tm651, have been characterized previously (see Fig. 1F). The ok641 allele is in frame and supports expression of a transcript [12] and the tm651 allele has an internal deletion of 890 base pairs that introduces a frameshift early in the coding sequence [13]. Both the ok641 and tm651 mutations are considered to be functional null alleles [13]. In their homozygous states these mutant alleles cause severe phenotypes including embryonic (~6%) and larval (~30%) lethality, and sterile adults or adults with vulva defects (17%), with less than 45% of L1s growing into fertile adults (Table 1). By comparison, we found that the novel ten-1(et5) allele exhibits the same rate of embryonic lethality as the null alleles (~6%), but reduced incidence of post-embryonic phenotypes, such that over 70% of homozygous et5 progeny grow into fertile adults (Table 1). ten-1(et5) is therefore not a null allele, which suggests an important function for the four EGF repeats present in the ten-1(et5) allele but absent from the tm651 and ok641 alleles (see Fig. 1F).
Table 1.
Characterization of visible phenotypes in ten-1 mutants.
| Genotype | Emb* (%) | Lvl* (%) | Vul/Ste* (%) | Fer* (%) | n |
|---|---|---|---|---|---|
| N2 | 0 | 0 | 0 | 100 | 207 |
| mnm-5(et5) | 5.8 | 18.6 | 2.5 | 73.1 | 191 |
| ten-1(ok641) | 6.1 | 30.9 | 18.3 | 44.7 | 211 |
| ten-1(tm651) | 6.5 | 33.8 | 16.1 | 43.5 | 186 |
*Emb: embryonic lethal; Lvl: larval lethal; Vul/Ste: vulva defective or sterile adult; Fer: fertile adult.
Expression of ten-1 transcriptional reporters
Others have reported on the expression profile of the two ten-1 isoforms in C. elegans [12,13]. We independently generated ten-1a::gfp and ten-1b::gfp transcriptional reporters and analyzed their expression during development and in adults. Our observations are generally consistent with the published ones. However, we also paid careful attention to embryonic and pharyngeal expression and made the following novel observations.
In wild-type embryos, ten-1a::gfp is first expressed in a cluster of cells in the anterior half at approximately 150 minutes after fertilization (Fig. 2A). These cells are precursors to the hypodermal cells, which are evident at 300 minutes post-fertilization, when the cells intercalate and begin the process of ventral closure (Fig. 2B), and to pharyngeal and intestinal cells, which are evident beginning at the bean stage (Fig. 2C). In later stages, strong expression of ten-1a::gfp persists in pharyngeal and intestinal cells, and appears in several head neurons (Fig. 2D-E). Examination of L1 larvae and adults allowed us to identify 8 pharyngeal cells that express ten-1a::gfp: the three marginal cells mc1, the three marginal cells mc3, and the neurons M2L and M2R (Fig. 2F-G). As reported previously, adults also express ten-1a::gfp in vulva muscles, the gonad distal tip cells, the intestine, several tail neurons including DVB and some other cells ([12]; data not shown).
Figure 2.

Expression of ten-1a/b transcriptional reporters. (A-E) Embryonic expression of ten-1a::gfp. The asterisk in (A) indicates the anterior cluster of GFP-positive cells in a ~150 min embryo. Arrowheads in (B) indicate intercalating hypodermal cells, while the asterisk indicates a cluster of pharyngeal precursor cells in a ~300 min embryo. (C-E) Shows that pharyngeal cells (p), intestinal (i) and some neurons (n) express ten-1a::gfp in ~350 min (dorsal view), 1.5-fold, and 2-fold stage embryos, respectively. (F-G) Show a L1 larva and the head of an adult, respectively. Note expression in the marginal cells (mc1 and mc3), head ganglion neurons (hg), the intestine (i) and the M2 pharyngeal neuron (M2). (H-J) Show a L1 ten-1(ok641) mutant larva expressing ten-1a::gfp. Note the deformed posterior end (indicated by arrowhead in H), and the poor connection between the posterior intestinal cells (indicated by arrowheads in J). (I) is an overlay of (H) and (J). (K-O) Embryonic expression of ten-1b::gfp. The asterisk in (K) indicates the anterior cluster of GFP-positive cells in a ~150 min embryo. Arrowheads in (L-M) indicate anterior and posterior clusters of hypodermal cells in ~300 min embryos shown from a dorsal or lateral perspective, respectively. (N-O) ten-1b::gfp expression in several neurons of the head and tail in 1.5 and 2-fold embryos, respectively. (P-T) Show expression of ten-1b::gfp ten-1(ok641) mutant embryos. Note abnormal development of the embryos and the persistence of strong GFP expression in dorsal hypodermal cells (asterisks in R; arrowheads in S and T) at stages where the expression is declining or lost from wild-type embryos. (U) Shows a L1 larva, with several head ganglion neurons (hg) and tail neurons (n) expressing GFP. (V) Shows an adult pharynx expressing ten-1b::gfp and stained with DiI, which labels the taste sensory amphid neurons (red). Note the absence of overlap between the DiI and GFP signals. (W-X) Show the head of an adult ten-1(ok641) mutant expressing ten-1a::gfp. Note the presence of GFP-positive axons forming an ectopic nerve ring anterior to the metacorpus (arrowheads), which is never seen in wild-type controls. The scale bar in (A) represents 10 μm and applies to all panels except (H-J), which have a separate scale bar.
The expression of the ten-1b reporter differs from that of ten-1a. Initial expression is detected in fewer anterior cells at 150 minutes post-fertilization (Fig. 2K), and becomes restricted to anterior neuronal cells and posterior hypodermal cells by 300 minutes (Fig. 2L-M). By the 1.5-fold stage (~460 minutes post-fertilization), hypodermal cell expression gradually fades away, and strong expression is found only in neurons of the head (Fig. 2N). This pattern persists to the end of embryogenesis (Fig. 2O). As previously reported, post-embryonic expression is found in head and tail neurons and some other cells (Fig. 2U; [12]). We used DiI to show that the ten-1b::gfp positive neurons are not amphid or phasmid neurons since they did not pick up the dye (Fig. 2V). Expression of the ten-1b reporter is never observed within the pharynx.
ten-1 reporters help visualize mutant phenotypes
The expression of ten-1a::gfp and ten-1b::gfp was examined in the ten-1(ok641) null mutant background to try and detect expression changes or morphogenesis defects in the ten-1 expressing cells. Mutant embryos usually did not differ from wild-type until the bean stage. After this stage, ~6% of the ten-1(ok641) mutant embryos failed to complete hypodermal cell intercalation and also failed to fold properly their posterior end, such that the 1.5- and 2-fold stages were delayed and poorly formed. The observed defects in hypodermal cell intercalation in mutant embryos coincides with persistent strong expression of ten-1b::gfp in dorsal hypodermal cells well beyond the developmental stages at which it becomes barely detectable in control embryos (Fig. 2P-T).
Mutant animals that survived through embryogenesis exhibit phenotypes easily visualized using the ten-1 transcriptional reporters. Firstly, many L1 larvae have deformations in their posterior half, and the ten-1a::gfp reporter reveals that the intestinal cells in these larvae are misshapen and poorly bound to one another; this phenotype may account for the 30% embryonic lethality observed in this mutant (Fig. 2H-J). Secondly, and as previously reported [12], many of the escapers that grow into healthy-looking adults actually have gross defects in the anatomy of their vulva muscles and often fail to open a vulva slit (data not shown). These defects in vulva morphogenesis likely contribute to the sterility/exploded vulva phenotypes observed at a frequency of 15-20% in the ten-1(ok641) mutant background. Expression of the ten-1b::gfp reporter was generally normal in the ten-1(ok641) mutant escapers that matured into L1s without posterior defects. However, we noted a low incidence of animals exhibiting misplaced axon trajectories within the head (~2%; N = 400), such that they appeared as a separate nerve ring just anterior to the pharyngeal metacorpus (Fig. 2W-X).
In summary, cells that express the ten-1 gene embryonically or post-embryonically often exhibit developmental defects in the ten-1(ok641) mutant background. This result suggests that ten-1 may generally act cell-autonomously.
Pharyngeal neuron defects in ten-1 mutants
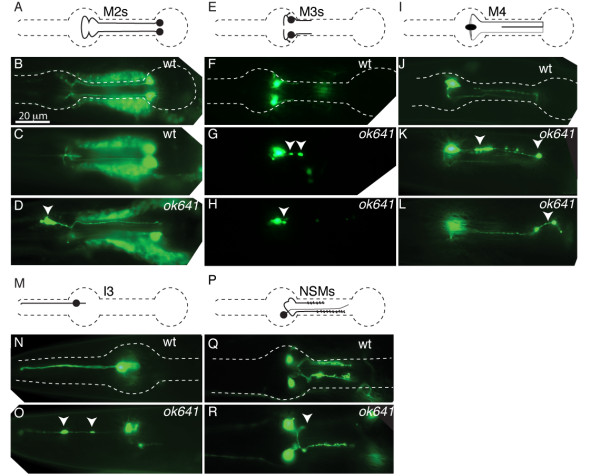
As mentioned earlier, the ten-1(et5) mutant was isolated by virtue of its M2 pharyngeal neurons defects [14], and later also found to have defects in the neurons NSMR and NSML [15]. To determine if other pharyngeal neurons depend on functional ten-1 for their development, we examined the trajectories of the pharyngeal neurons for which we could obtain specific GFP reporters in both the ten-1(et5) and the more severe ten-1(ok641) mutants. Specifically, we examined the M1, M2, M3, M4, I3 and NSM neurons, and found that ten-1 contributes to the guidance of all these neurons, but that its role varies in importance (Fig. 3 and Table 2). The most typical defects are the presence of large varicosities and truncations of the axons, which are both indicative of growth cones stalling during axon elongation. Misguided axon trajectories are also observed with variable frequencies in the motor neurons M2, M3 and M4. Of the neurons examined, M4 has the most complex trajectory and showed the strongest requirement for functional ten-1, with more than 60% of neurons being defective in the ten-1(et5) and ten-1(ok641) mutant backgrounds. Only the M2 neuron exhibited misplaced cell bodies, with a frequency of roughly 10%, as previously reported. The two alleles examined, et5 and ok641, produced a very similar range of phenotypes with similar severities and penetrance.
Figure 3.
Pharyngeal neuron defects in ten-1 mutants. Each series shows the following: a cartoon representation of the normal morphology of the examined neuron, photographs of the wild-type neurons visualized with a specific GFP reporter (N2; see materials and methods), and mutant animals (ok641) with typical defects indicated by arrowheads. (C) Shows a different focal plane of the worm shown in (B) to make the ends of the distal M2s visible. The arrowheads indicate a misplaced M2 neuron cell body (D), conspicuous varicosities (G, K, O), ectopic branches (H), misguided axons (L) or truncated axons (R). The scale bar in (B) represents 20 μm and applies to all photographs.
Table 2.
Scoring of pharyngeal neurons in the ten-1(et5) and ten-1(ok641) mutants.
| Neuron | Genotype | Normal % | Varicosities % | Truncated % | Misguided % | Others * | n |
|---|---|---|---|---|---|---|---|
| M2 | wt | 99.8 | 0.2 | 419 | |||
| et5 | 87.7 | 0.2 | 3.9 | 8.3 | 432 | ||
| ok641 | 86.9 | 1.3 | 3.3 | 8.5 | 459 | ||
| M3 | wt | 100 | 130 | ||||
| et5 | 60.3 | 5.7 | 13.6 | 20.4 | 191 | ||
| ok641 | 60.8 | 15.2 | 24.0 | 216 | |||
| M4 | wt | 100 | 114 | ||||
| et5 | 58 | 30.4 | 5.8 | 5.8 | 102 | ||
| ok641 | 34.9 | 42.5 | 11.0 | 11.6 | 181 | ||
| I3 | wt | 99.1 | 0.9 | 222 | |||
| et5 | 89.6 | 10.3 | 203 | ||||
| ok641 | 92.0 | 8.0 | 210 | ||||
| NSM | wt | 98.9 | 1.1 | 271 | |||
| et5 | 82.5 | 6.6 | 10.9 | 241 | |||
| ok641 | 79.6 | 7.8 | 12.6 | 165 |
Notes
* M2 neuron: indicates the percentage of M2 neurons that were misplaced into the isthmus or metacorpus.
* M3 neuron: indicates cases of missing M3 neuron cell bodies.
* M4 neuron: indicates supernumerary ectopic outgrowths.
ten-1(ok641) mutants have no obvious body neuron defects
Chiquet-Ehrismann and co-workers previously reported neuronal defects outside the pharynx in animals treated with RNAi against ten-1 as well as in ten-1(ok641) mutant animals [12]. This surprised us in view of the fact that our initial description of the ten-1(et5) mutant had not revealed any such defects [14] and because the viable ten-1 mutant individuals are not uncoordinated. We used several neuronal reporters to examine carefully whether ten-1 is important for the development of neurons outside of the pharynx. First, we introduced a pan-neuronal GFP reporter, i.e. evIs111, in the ten-1(ok641) mutant. We detected no obvious abnormalities in the extrapharyngeal nervous system, including the positioning of lateral body neurons, and the trajectories of several lateral neurons that can be seen using evIs111 (Additional file 1: fig. S1 A-B). Similar results were obtained with the other presumed null mutant, ten-1(tm651) (data not shown). We also examined carefully the body neurons that express ten-1b::gfp and found that these too were normal in the ten-1(ok641) background, except for the ~2% of larvae that had mispositioned nerve ring axons to positions anterior of the metacorpus (Fig. 2X). The six microtubule-rich mechanosensory neurons were also specifically scored in ten-1(ok641) mutants, this time using a mec-7::gfp reporter. Four of these neurons (ALML, ALMR, PLML and PLMR) project on the lateral side of the body, while the two others (AVM and PVM) originate from the lateral side, navigate ventrally into the nerve cord then project anteriorly [16,17]. No defects were observed in the trajectories of these six neurons (Additional file 1: fig. S1 C-D). Finally, we obtained control and ten-1(ok641) strains carrying a ten-1b::gfp reporter (a kind gift from R. Chiquet-Ehrismann; [12]) and scored these for possible neuronal defects in the GFP-positive neurons. Again we observed no neuronal defects in randomly picked L4 or adult worms (Additional File 1: Fig S2 A-D), except in those rare instances (<3%; N > 100) where a viable worm also exhibited obvious morphological defects (Additional file 1: fig. S2E). From our analysis of various neurons in ten-1 mutants, we conclude that ten-1 is not generally important for neuronal development (e.g. no gross defects in extrapharyngeal neuroanatomy or in specific mechanosensory neurons), but rather is specifically required by some neurons (e.g. several pharyngeal neurons). It seems likely that any neuronal defects in body neurons are secondary to body morphogenesis defects.
Enhanced body muscle expression of unc-129 in ten-1(ok641) mutants
To further test the possibility that some C. elegans body neurons may rely on ten-1 during their development, we introduced an unc-129::gfp reporter in ten-1(ok641) mutant animals. This reporter in expressed in the DA/DB neurons that project circumferentially from the ventral cord to innervate the dorsal body muscles, and also weakly in the dorsal muscle cells [18]. Consistent with the lack of any obvious Unc phenotype, no defects were observed in the DA/DB neurons of ten-1(ok641) mutants. Unexpectedly however, we discovered a 20-fold increase in the expression levels of the reporter in the dorsal muscle cells of mutants, as judged by the intensity of the GFP signal (Fig. 4). It seemed possible that the muscle cells compensate for ten-1 loss by expressing and secreting more of the unc-129 guidance signal. This possibility prompted us to investigate the trajectories of the DA/DB neurons in a ten-1(ok641);unc-129(ev55) double mutant background. As shown in Fig. 4F, the double mutant shows the same frequency of DA/DB defects as in the unc-129 single mutant, suggesting that ten-1 does not contribute at all to the guidance of these axons, although it obviously plays a role in regulating their level of TGF-β expression from the unc-129 locus.
Figure 4.
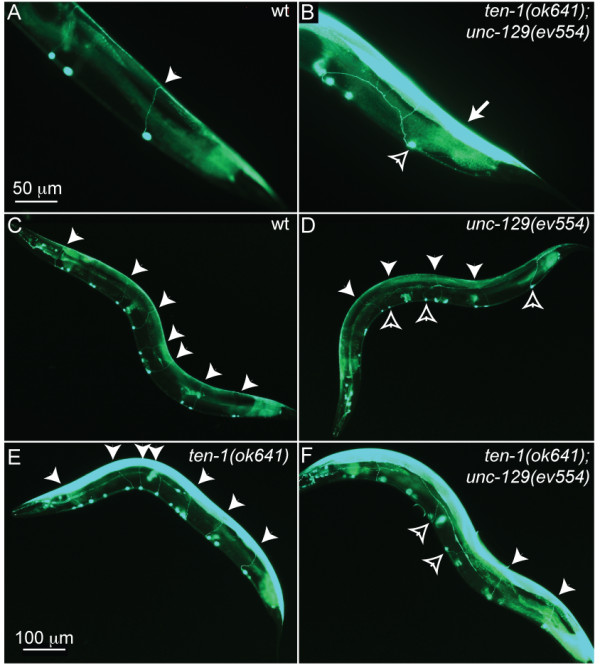
unc-129::gfp expression in the ten-1(ok641) and unc-129(ev554) mutants. All panels show worms transgenic for a unc-129::gfp reporter expressed in the DA/DB neurons and in the dorsal muscle cells. (A) Shows a control worm in which one clear circumferential axon is in indicated by the filled arrowhead. (B) Shows a ten-1(ok641);unc-129(ev554) double mutant where the same axon errs and branches abnormally (open arrowhead), and the dorsal muscle expression is dramatically enhanced compared to control (arrow). (C-D) Show lower magnification views of animals with the indicated genotypes. Filled arrowheads indicate axons that projected correctly circumferentially, while open arrowheads indicate axons that erred and branched abnormally, failing to reach the dorsal side. The scale bar in (A) applies also to (B), while the scale bar in (C) applies to panels (C-F).
ten-1 enhances the severity of mutations affecting growth cone and/or cytoskeleton regulators
The pharyngeal neuron defects in the ten-1 mutant are never 100% penetrant even in the most severely affected neurons, which indicate redundancy with other genes and pathways. In our previous study of the M2 neurons we discovered that mutations in the sax-3, unc-5 and unc-6 genes cause misplacement of the M2 neurons similar to those seen in ten-1(et5) but at a much lower penetrance [14]. To explore if ten-1 acts in or in parallel with netrin, ROBO or other pathways, we performed a genetic interaction study. The mutations tested are listed in Table 3. Single and double mutants were scored for viability and for M2 neuron trajectories, and the results are presented in Table 4.
Table 3.
Lists of genes tested for possible interaction with ten-1.
| Gene(allele) | Allele type | Function/pathway |
|---|---|---|
| mig-14(ga62) | hypomorph | Wnt-secretion factor |
| sax-3(ky123) | null | receptor/robo slit |
| slt-1(e15) | null | ligand/robo slit |
| unc-5(e53) | null | receptor/netrin |
| unc-6(ev400) | null | ligand/netrin |
| unc-34(e315) | nonsense | Enabled VASP homolog/netrin and robo |
| unc-40(e271) | null | receptor/netrin |
| unc-51(e369) | dominant negative | serine threonine kinase |
| unc-52(e1421) | possible null | perlecan homolog |
| unc-73(e396) | null | guanine nucleotide exchange factor |
| unc-129(ev554) | null | secreted TGF-β |
Table 4.
Pharyngeal M2 neuron defects in ten-1 mutants.
| Genotype | wt (%) | Misplac (%) | Ipsil (%) | Trunc (%) | Other (%) | N |
|---|---|---|---|---|---|---|
| etIs2 | 99.8 | 0.2 | 419 | |||
| mnm-5(et5); etIs2 | 87.7 | 8.3 | 3.9 | 0.2 | 432 | |
| ten-1(ok641); etIs2 | 86.9 | 8.5 | 3.3 | 1.3 | 459 | |
| ten-1(tm651); etIs1 | 83.3 | 14.6 | 2.0 | 246 | ||
| mig-14;etIs2 | 85.5 | 0.9 | 0.4 | 3.0 | 10.3 | 234 |
| mig-14;etIs2 ok641 | 36.4 | 26.6 | 8.9 | 12.1 | 15.9 | 214 |
| sax-3; etIs2 | 68.9 | 5.1 | 7.2 | 2.9 | 15.7 | 235 |
| sax-3; etIs2 ok641 | LETHAL | - | ||||
| slt-1; etIs2 | 97.3 | 1.9 | 0.8 | 262 | ||
| slt-1; etIs2 ok641 | 93.9 | 1.1 | 3.0 | 1.9 | 264 | |
| unc-5; etIs2 | 76.8 | 16.0 | 7.2 | 250 | ||
| unc-5; etIs2 ok641 | 35.6 | 6.2 | 42.9 | 15.3 | 177 | |
| unc-6; etIs2 | 23.7 | 0.4 | 37.9 | 37.9 | 253 | |
| unc-6; etIs2 ok641 | 42.2 | 6.6 | 42.2 | 8.3 | 0.6 | 469 |
| unc-34; etIs2 | 91.8 | 4.9 | 3.3 | 245 | ||
| unc-34; etIs2 ok641 | LETHAL | - | ||||
| unc-40; etIs2 | 78.3 | 6.5 | 14.8 | 0.4 | 230 | |
| unc-40; etIs2 ok641 | 78.6 | 2.5 | 10.7 | 8.2 | 280 | |
| unc-51; etIs2 | 83.7 | 5.9 | 10.4 | 270 | ||
| unc-51; etIs2 ok641 | 49.2 | 7.4 | 33.5 | 9.7 | 268 | |
| unc-52; etIs2 | 94.4 | 3.7 | 1.8 | 215 | ||
| unc-52; etIs2 ok641 | 40.8 | 7.1 | 42.9 | 9.2 | 184 | |
| unc-73; etIs2 | 50.4 | 19.9 | 29.7 | 236 | ||
| unc-73; etIs2 ok641 | LETHAL | - | ||||
| unc-129; etIs2 | 91.7 | 2.0 | 6.3 | 204 | ||
| unc-129; etIs2 ok641 | 74.1 | 8.6 | 8.2 | 9.1 | 220 |
Notes
wt: normal
Misplac: M2 cell body mispositioned into the metacorpus
Ipsil: ipsilateral outgrowth
Trunc: truncated distal end
Contra: contralateral outgrowth
Post: posterior outgrowth
Other: Contralateral outgrowths in the case of sax-3;etIs2, unc-6; etIs2 ok641 and unc-40; etIs2. Ectopic dorsal branch, posterior branch, misplaced cell into isthmus, growth posterior from metacorpus back into isthmus in the case of mig-14; etIs2 and mig-14;ok641 etIs2.
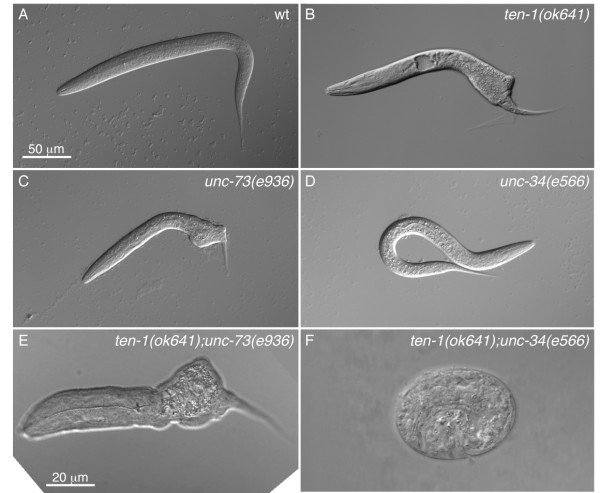
A striking observation concerns the synthetic embryonic or L1 larval lethality obtained when combining the ten-1(ok641) mutation with mutations in the sax-3(ky123), unc-34(e315) or unc-73(e396) genes (see Fig. 5). The synthetic lethality demonstrates important roles for these genes in early embryonic processes, and likely reflects the complementarity between the ECM environment to which ten-1 contributes [13], and the cytoskeletal changes regulated by sax-3, unc-34 and unc-73 in response to that environment as embryonic cells migrate, adhere and otherwise interact with each other.
Figure 5.
Emb and Lvl synthetic phenotypes involving ten-1 mutants. Larvaes of the indicated genotypes are shown. Note that the posterior defect seen in the ten-1(ok641) and unc-73(e936) single mutants have low penetrance (<5%) while the defects in the two shown double mutants (E-F) are 100% penetrant. Scale bar in (A) applies to all panels.
Another striking finding is that the hypomorphic mutation mig-14(ga62) and the null mutation ten-1(ok641) synergistically enhanced their effects on M2 cell position and axon trajectories (Table 4). In particular, the double mutant had over 25% misplaced M2 cell bodies, the highest incidence ever observed for any mutant or double mutant. mig-14 encodes the sole C. elegans homolog of the Wnt-secretion factor Wntless, and it plays an important role in anterior-posterior guidance during cell migration and axon elongation [19,20]. Our results suggest that ten-1 is also important for these processes, acting either in parallel with mig-14 or by increasing the activity of the mig-14 pathway.
With regards to the distal M2 ends, which are dependent on growth cones for their development [14], we found that most mutations tested increased the occurrence of defects when combined with the ten-1(ok641) mutation, especially in the ipsilateral outgrowth class that is characterized by the distal ends failing to migrate dorsally within the metacorpus, erring instead anteriorly in their original focal plane (Table 4). The most dramatic genetic interactions that affected ispilateral outgrowth involved ten-1(ok641) together with unc-51 (a serine/threonine kinase important for the localization of guidance receptors; [21]) or with unc-52 (which encodes the C. elegans perlecan; [22,23]): while the single mutants individually displayed about 5% ipsilateral outgrowth, over 30% of the M2 neurons showed this defect in the double mutants (Table 4).
ajm-1::GFP reveals a morphogenesis defect during embryogenesis in ten-1 mutants
The embryonic/larval lethality observed in several double mutants involving ten-1(ok641) strongly suggests an important role for ten-1 during early development (see Table 4 and Fig. 5). The expression of ten-1 reporters in the hypodermal cells of early embryos (Fig. 2 and 3) prompted us to examine the behavior of these cells in the ten-1(ok641) mutant using a marker of adherance junctions, i.e. ajm-1::GFP, to visualize the hypodermal cells as they intercalate and change shape during ventral closure and embryonic elongation. The parental strain SU93, which carries the ajm-1::GFP transgene, occasionally exhibits early embryonic arrest, mostly at pre-bean stages, at a frequency of 1.8% (N = 281 bean to 2-fold stage embryos scored), indicating that the transgene in itself may have some effect on embryogenesis. When the same transgene is introduced in the ten-1(ok641) background, embryonic phenotypes are evident at a frequency of 6.3% (N = 268 bean to 2-fold stage embryos scored), suggesting that ten-1(ok641) is responsible for about 4.5% embryonic defects, which is consistent with the study of the single mutant described earlier (Table 1). ten-1(ok641) embryos also exhibited phenotypes not seen in the parental strain SU93. In particular, posterior morphogenesis defects at the comma or later stages were specific for the ten-1(ok641) mutants (Fig. 6). The hypodermal cells are grossly disorganized in some embryos (Fig. 6B and 6D), while in others they failed to fuse or became mispositioned such as to cause bulges in the posterior half (Fig. 6F). These results are in general agreement with published work [12]. When larvae with deformed posterior halves are carefully examined by DIC microscopy, it was obvious that their muscle quadrants have developed successfully (Fig. 6G), but that their intestinal cells are grossly malformed (Fig. 6H).
Figure 6.

Hypodermal cell defects in ten-1(ok641) embryos. All animals shown are ten-1(ok641) mutants carrying the ajm-1::gfp transgene, which allows the visualization of the adherence junctions that surround the hypodermal cells. (A) A 1.5-fold embryo that is developing normally. Note the seam cells indicated by asterisks, and the ongoing hypodermal cell fusions on the dorsal side (discontinuous lines of GFP expression). (B) An example of a 2-fold embryo in which the posterior half has developed abnormally. Note the poorly organized pattern of ajm-1::gfp expression. (C) An early 3-fold stage embryo that is developing normally. Note the evenly spaced seam cells (asterisks) and the symmetrical distribution of the ajm-1::gfp pattern. (D) An early 3-fold embryo in which the posterior half has developed abnormally. Note in particular the meandering and disorganized pattern of ajm-1::gfp distribution. (E) Ventral aspect of an L1 larva that has developed normally. Again, note the regular shapes of the hypodermal cells. (F) Ventral aspect of an L1 larva with a deformity in its posterior half, which corresponds to a misshaped hypodermal cell (arrow). (G) DIC image of a ten-1(ok641) L1 larva with focus on the two muscle quadrants with normal appearance (asterisks) in spite of the posterior deformity (arrow). (H) DIC image of a ten-1(ok641) L1 larva with focus on the deformed intestine (asterisks) and posterior deformity (arrow).
Discussion
The ten-1(et5) allele is a hypomorph
Our observations complement those of Chiquet-Ehrismann and co-workers [12,13]. In their studies, they described the expression profiles of the ten-1a and ten-1b forms of the gene, the phenotypes induced by RNAi against ten-1, and the phenotypes of two null mutant alleles (ok641 and tm651). They also documented defects in basement membranes in the mutants, such as abnormal or deficient laminin distribution around the pharynx and developing gonad, and demonstrated that ten-1 acts in part redundantly with genes encoding the extracellular matrix components dystroglycan (dgn-1) and laminin (epi-1), and one integrin adhesion molecule (ina-1).
Our study adds several novel observations that will each be discussed separately. Firstly, we isolated a novel allele of ten-1, namely the et5 allele. The novel et5 allele is hypomorphic since it causes a milder post-embryonic phenotype than either the ok641 or tm651 alleles, which are both considered functional null alleles [13]. The TEN-1 protein expressed from the ten-1 allele will be truncated after the eight extracellular EGF domains. The null ok641 allele is predicted to encode a protein truncated between the fourth and fifth EGF domain. Our observations suggest that important post-embryonic functions may reside within the four EGF domains present in the et5 allele but absent from the ok641 allele. The second and fifth EGF repeats are important for homo- and hetero-dimerization [10,11], and the et5 allele is therefore expected to be able to multimerize. Because the et5 allele exhibits embryonic phenotypes with the same penetrance and severity as the null alleles, it seems possible that some embryonic functions of the TEN-1 protein are dependent on motifs located on the C-terminal side of the EGF domains. The NHL and YD repeats present in that part of the protein are expected to mediate homotypic or heterotypic interactions between cells that express teneurin, as well as interactions with the extracellular matrix [1,2]. These functions are critical during morphogenetic events and may also account for the roles of ten-1 in regulating extracellular deposition and composition, to which ten-1 clearly contributes [13].
ten-1 may act cell-autonomously during the morphogenesis of hypodermal and vulva muscle cells
Both ten-1a and ten-1b are expressed in hypodermal cells during early embryonic development, and that expression ends by the time that these cells have completed their elongation, intercalation and ventral closure. An interesting observation is that the expression of ten-1b is enriched in the posterior hypodermal cells, the same cells that will show the most common morphogenesis defects in the ten-1 mutants. Similarly, ten-1a is expressed post-embryonically in the vulva muscles, and these will often develop abnormally in the mutant. These two observations suggest that ten-1 may act cell-autonomously during the morphogenesis of hypodermal and vulva muscle cells. The ten-1 homologs in Drosophila and vertebrates are also expressed in embryos at sites and times of morphogenetic cell movements, and this is possibly an ancestral function for the gene [7,8,24,25].
ten-1 participates in the development of all examined pharyngeal neurons
Only the ten-1a form is expressed in the pharynx, with expression in the marginal cells mc1 and mc3, and in the neurons M2L and M2R. However, all pharyngeal neurons examined exhibit defects in the ten-1 mutant. This suggests that the marginal cells mc1 and mc3 play an important role during pharyngeal neuron development. This is a reasonable hypothesis since the posterior part of mc1 occupies the metacorpus and mc3 occupies the posterior bulb, which are the two regions where most of the neuronal defects were observed. Another interesting possibility is that the two M2 neurons influence the development of the other pharyngeal neurons. The M2 neurons each send a long straight axon trajectory through the isthmus that develops without growth cones, and it is possible that it provides a pioneer axon function [26]. Other pharyngeal axons could grow through the isthmus by using growth cones that navigate along the M2 axon. In the absence of ten-1, these growth cones may stall, halt permanently or err in incorrect directions, which could explain the observed axon defects in and outside the isthmus for several of the pharyngeal axons studied (e.g. the M3, M4 and NSM neurons). Consistently, no varicosities or trajectory defects are ever observed within the isthmus for the M2 neurons.
ten-1 interacts with several morphogenesis/axon guidance genes
ten-1 is synthetic lethal with mutations in the genes sax-3, unc-34 and unc-73. These genes are important for several axon guidance decisions during C. elegans development: sax-3 is the receptor for the guidance molecule slt-1 [27-29], and unc-34 and unc-73 regulate cytoskeleton dynamics in growth cones [30-33]. All three genes are also important for morphogenetic processes in early development, as evidenced by the low penetrance body shape abnormalities seen in these mutants. However, ten-1 must usually provide a function that allows most embryos to develop successfully in the absence of any one of these three genes. Chiquet-Ehrismann and co-workers have shown that ten-1 is important for organizing the ECM [13], which leads us to interpret our results in the following way. On the one hand, the single mutants of sax-3, unc-34 or unc-73 have defects in the regulation of cytoskeletal dynamics within cells undergoing morphogenesis but retain enough activity to succeed with this process provided that the extracellular environment is not also compromised. Conversely, the ten-1 mutant has defects in the composition/distribution of extracellular matrix important for morphogenetic processes but retains enough of it to complete embryogenesis provided that the ability of the cells to regulate their cytoskeletal dynamics is not compromised. Double mutants fall below essential thresholds and therefore fail. The most obvious developmental failure in ten-1 mutants is their deformed posterior half (Figs. 5 and 6), which correlates with the hypodermal cells failing to intercalate properly and to drive the convergent extension and contraction-driven elongation of which they are responsible [34,35]. The enhancement of the embryonic morphogenesis phenotype observed in the ten-1 and sax-3, unc-34 or unc-73 double mutants implicates all four genes in posterior hypodermal morphogenesis. Both ten-1a and ten-1b are expressed in the hypodermal cells of the posterior half, and a cell-autonomous function for ten-1 may explain why the posterior half is more susceptible to developmental failure.
It is worth noting that mutations in several genes have previously been reported to be synthetic lethal with the ten-1(ok641): dgn-1 (dystroglycan), nid-1 (nidogen), epi-1 (laminin alpha-beta) and ina-1 (integrin) [13]. Pursuing with the above reasoning, it would appear again that two essential forces are in play: ECM integrity on the one hand (involving dystroglycan, nidogen and laminin) and cellular interaction/response to the ECM on the other (involving the integrin gene ina-1). When the ECM is too compromised, as in double mutants involving ten-1 and an ECM component gene, the embryo cannot undergo normal morphogenesis and is not viable. Conversely, when a partially compromised extracellular matrix mutant (ten-1) is combined with a mutation affecting interaction/response to the ECM (such as ina-1) then that too is not viable.
The observation that the ten-1 mutation also enhanced the penetrance of M2 neuron defects for the unc-5, unc-129, and mig-14 axon guidance pathways shows that ten-1 provides a function important for all these disparate pathways within the pharynx. Again, this function is likely to be related to providing a suitable ECM for the retention of guidance cues, and acting as a substrate for growth cone migration. The strong synergy observed between ten-1 and unc-51 is also interpreted in the same way: unc-51 is important for the transport of several guidance receptors on the plasma membrane of growth cones [21,36,37], and impairing both these receptors and the extracellular matrix that provides their cues and substrates has a severe impact on axon development.
ten-1 genetically interacts with unc-52 and other extracellular matrix genes
Another dramatic enhancement in the frequency of M2 distal end defects was observed when the ten-1 mutation was combined with unc-52, which encodes the worm homolog of perlecan, an extracellular matrix component [22,38,39]. This result suggests that ten-1 and unc-52 act contribute redundantly to the formation of extracellular matrix suitable for the guidance of growth cones within the pharynx, just as ten-1 complements the activities of dystroglycan (dgn-1), laminin alpha beta (epi-1), nidogen (nid-1), collagen (cle-1) and perlecan (unc-52) in the extracellular matrix outside the pharynx [13].
ten-1 is not generally essential for neuronal development
An apparent discrepancy between the present work and that of Chiquet-Ehrismann and co-workers regards extrapharyngeal neuron development. In contrast to their published observations [12], we were unable to document defects in neurons outside the pharynx, except for a low frequency (~2%) of L1 larvae having misplaced nerve ring axons and in a low frequency (<3%) of randomly picked L4 or adult worms that happened to have obvious hypodermal morphogenesis defects. It is important to note that the previously reported neuronal defects in ten-1(ok641) mutants were not quantified carefully, and in any case were most likely secondary to hypodermal morphogenesis defects.
Neuronal phenotypes in mouse and worm teneurin mutants
Are the roles that ten-1 plays during pharyngeal neuron development evolutionarily conserved? That is obviously a difficult question. In the C. elegans ten-1 mutant, the defects in the pharyngeal neurons show much variation indicative of general pathfinding defects. This is consistent with the fact that ten-1 mutations typically enhance whatever defects are present in other mutants (e.g. mig-14 or unc-5) when mutations are combined. This appears to be in contrast with mouse teneurin mutants that exhibit quite specific path decision errors. Specifically, and as an example, the Ten-m3 mutant exhibits characteristic defects in ipsilateral, but not contralateral, guidance decisions during the development of the visual circuitry [40]. Indeed, Ten-m3 is expressed in a graded fashion consistent with a role as a guidance molecule for retinal fibers, and may guide their growth by mediating homotypic adhesion [41]. It therefore appears that at least some teneurin genes may guide neuronal development in mouse in ways that are more sophisticated than in nematodes.
Conclusions
1. The novel allele ten-1(et5) is hypomorphic, which suggests that important post-embryonic function resides within the four EGF domains present in the et5 allele but absent from the null ok641 allele.
2. C. elegans ten-1 participates in the guidance of all tested pharyngeal neurons, and the ten-1(ok641) null mutation is synthetic lethal with mutations in cytoskeleton regulators (sax-3, unc-34, unc-73) and enhances the pharyngeal guidance defects of several mutations in axon guidance genes (e.g. mig-14, unc-5, unc-51, unc-52 and unc-129. ten-1 therefore complements these pathways during morphogenesis and axon guidance, perhaps by regulating the composition of the extracellular matrix.
3. ten-1 is not generally essential for neuronal development since neuronal defects outside of the pharynx were only rarely observed in ten-1 null mutants.
Methods
Strains
Worms were maintained at 20°C using standard methods [42]. The Bristol N2 strain was used as wild-type reference [43], and all strains were obtained from the C. elegans Genetics Center (St-Paul, Minnesota), unless stated otherwise. The following mutations were studied:
LG I: unc-40(e271), unc-73(e396),
LG II: mig-14(ga62), unc-52(e1421)
LG III: dpy-17(e164), mnm-5(et5), ten-1(ok641), ten-1(tm651), unc-32(e189)
LG IV: rac-2(ok326), unc-5(e53), unc-129(ev554)
LG V: unc-34(e315), unc-51(e369)
LG X: sax-3(ky123), slt-1(e15), unc-6(ev400)
Transgenes
In some cases, the plasmid pRF4, containing rol-6(su1006) which causes a Roller phenotype, was used as a transformation marker [44].
The following transgenes were used:
etIs1 and etIs2 which carry a ric-19::gfp translational reporter as well as pRF4, integrated into linkage groups IV and III, respectively [14].
evIs79, which carries unc-129::gfp [18]. This was a gift from Joe Culotti.
jcIs1, which consists of pJS191 (ajm-1::gfp), pRF4 and C45D3 (unc29(+)) DNAs [45].
zdIs13, which carries a tph-1::gfp transcriptional reporter expressed in the NSM pharyngeal neuron and HSN extrapharyngeal neuron [46]. This was a gift from S. Clark.
muIs32, which carries a mec-7::gfp transcriptional reporter expressed in the mechanosensory neurons. This was a gift from Cynthia Kenyon.
etEx106, which is an extrachromosomal array carrying a ser-7b::gfp transcriptional reporter [47] expressed in the M4 neurons as well as the plasmid pRF4.
Sequencing of the ten-1(et5) mutation
Six fragments that covered the entire ten-1 gene were PCR amplified with PFU Ultra (Stratagene) on single lysed mnm-5(et5) or wild type worms as templates with the following primers: ten-1_fragment1_for: 5'-cgccgtcgtctgtgttcgaaac-3'+ ten-1_fragment1_rev: 5'-caaagctcctcaagaactactac-3', ten-1_fragment2_for: 5'-ctagtaacagatgatgaggcggc-3'+ ten-1_fragment2_rev: 5'-cgattcaccttcgaagtcttaggc-3', ten-1_fragment3_for: 5'-catggaagcaataagagccatatc-3'+ ten-1_fragment3_rev: cgcaccgttttagaattggtgac-3', ten-1_fragment4_for: 5'-ctaatgcgaaaggaggcagaagcc-3'+ ten-1_fragment4_rev: 5'-cagtctaccgaatcccaacctgac-3', ten-1_fragment5_for: 5'-ctgtaatggaaggggacgatgtgac-3'+ ten-1_fragment5_rev: 5'-caaactgccatccgaatcatcacc-3, ten-1_fragment6_for: 5'-cgtgatagggaatattggagactc-3'+ ten-1_fragment6_rev: 5'-cgttcacgccaccgacaaaatgtc-3'. The products were cloned into the pCR-BluntII-TOPO vector (Invitrogen) and sequenced by MWG Biotech (Germany).
Generation of transgenic animals
Germline transformation was performed as described by Mello et al. 1991 and the dominant rol-6 (su1006) was used as a marker for transgenic worms, [44]. Plasmids were prepared with a Qiagen miniprep kit (Qiagen) and used with the following concentrations: pRF4 (rol-6) of 50 ng/μl, test plasmids of 25 ng/μl, and pBSKS (Stratagene) of 25 ng/μl.
Detection of the ten-1 mutant alleles
When generating double mutants or lines carrying transgenes, it was often necessary to rely on PCR to detect the ten-1 alleles. This was done as follows. The ten-1(ok641) deletion was detected by PCR with TAQ DNA polymerase (Roche), using the following primers: forward-16: 5'-caccgttactaagccttcacgg-3'and reverse-8: 5'-ccactggaaaacgattgaggttt-3'which produces a product of 3 kb in wild type and 1 kb in the deletion mutant. The absence of wild type sequence in the mutant was confirmed with the following primers: forward-18: 5'-cttcgagtcattgccaattcaag-3'and reverse-8 which produces no band in the deletion mutant and a band of 1 kb in wild type.
Similarly, the ten-1(tm651) deletion was detected by PCR with TAQ DNA polymerase (Roche), using the following primers: forward-15: 5'-cagacctcatacgtctggaggagc-3' and reverse-10: 5'-cgtccgaacctgttggagatcc-3' which produces a product of 2350 bp in wild type and 1463 bp in the deletion mutant. The mutant with absent wild type sequence was confirmed with following primers: forward-16: 5'-caccgttactaagccttcacgg-3' and reverse-11: 5'-caactcggcttcgttcgttgat-3' which produces no band in the deletion mutant and a band of 280 bp in wild type.
Construction of plasmids
Lysed wild type worms were used as template for PCR, amplifications were performed with PFU Ultra (Stratagene), the PCR products were gel purified with Qiagen Gel Extraction Kit (Qiagen), subcloned into the pCR-BluntII-TOPO vector (Invitrogen) and further transferred into pPD95.77 (Addgene).
pten-1a:gfp
This plasmid was created by amplification of 5520 bp 5'-UTR of the ten-1 long form, ten-1a, with the following primers: ten-1a_gfp_forward: 5'- cgcatgccgttcattttccgtgtcaac-3' (SphI site in bold) and ten-1a_gfp_reverse: 5'-cctgcagattaggcggtgggccttgc-3' (PstI site in bold). The PCR product was subcloned into the SphI and PstI sites of pPD95.77.
pten-1b:gfp
This plasmid was constructed by amplification of 3246 bp 5'-UTR of the ten-1 short form, ten-1b, with the following primers: ten-1b_gfp_forward: 5'-cgcatgcccatatgtctcttagtttagc-3'(SphI site in bold) and ten-1b_gfp_reverse: 5'-cctgcagggatcaccattgttcatagtgc-3' (PstI site in bold) and the PCR product was subcloned into the SphI and PstI sites of pPD95.77.
Scoring of Neurons
M2 neuron
These neurons were visualized using etIs2, which carries a ric-19::gfp translational reporter expressed in the M2 neurons [14], or by using microinjection to generate transgenic lines carrying extrachromosomal arrays bearing both the pRIC-19::GFP [48] and pRF4 plasmids, and maintaining the transgenic lines by picking rollers.
M3 neuron
Transgenic lines carrying extrachromosomal arrays bearing the plasmid pQC105 and pRF4 were used to monitor the M3 neurons. pQC105 carries a mnm-2::gfp transcriptional reporter that is strongly expressed in the M3 neurons [49].
M4 neuron
Transgenic lines carrying extrachromosomal arrays bearing the plasmid pser-7b::gfp (a transcriptional reporter for ser-7b; [47]) and pRF4 were used to monitor the M4 neurons.
I3 neuron
Transgenic lines carrying extrachromosomal arrays bearing the plasmids pten-1a:gfp and pRF4 were used to monitor the I3 neurons.
Ventral motor neurons
The DA/DB motor neurons were visualized using evIs79, which carries unc-129:gfp. The expression levels of this reporter were also quantified with the free software ImageJ (Rasband, WS. ImageJ, U.S. National Institute of Health, Bethesda, Maryland, USA http://rsb.info.nih.gov/ij/).
Mechanosensory neurons
These neurons were visualized using the muIs32 transgene, which contains the mec-7::gfp transcriptional reporter.
Viability assay
For each genotype tested, adults were allowed to lay eggs during a period of 2 hours. Ten eggs were then gently transferred to each of ten plates. The worms from these plates were monitored daily for viability and any visible phenotype, and were transferred daily to fresh plates beginning from day three.
Authors' contributions
CM and MP designed most of the experiments. CM mapped the et5 allele and identified the mutation. CM, VV and MP constructed most of the strains, characterized their phenotypes and scored the various pharyngeal and body neurons. GJ characterized the expression profiles of the GFP reporters. MP wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplementary Figures. Fig. S1. Extrapharyngeal neurons are normal in the ten-1(ok641) mutant. The pan-neuronal evIs111 transgene was used to visualize the entire nervous system in (A) wild-type or (B) ten-1(ok641) mutant adults. No trajectory defects or abnormal varicosities were observed in the mutant. Asterisks indicate cell bodies that are analogously positioned in wild-type and mutant: any differences are within the variation range found in wild-type. The mec-7::gfp transgene was also used to score the mechanosensory neurons in (C) wild-type and in (D) ten-1(ok641) mutant adults. Again, no differences were observed between wild-type and mutant animals (N>100 worms examined for each treatment). Meaning of lowercase annotations: vul (vulva), vnc (ventral nerve cord), tg (tail ganglion), nr nerve ring). Fig. S2. Body neurons expressing ten-1b::gfp only rarely exhibit defects in the ten-1(ok641) mutant. In young larvae (L2), there is evidence of incomplete fasciculation in the ventral cords of both wild-type and ten-1(ok641) worms (arrows in A-B). By the L4 stage wild-type and mutants have nicely fasciculated ventral nerve cords (arrows in C-D), and two well defined lateral axons on each side (arrowheads in C-D indicate one such pair). Only occasionally (<3%; N > 100) are abnormalities observed in the neurons of randomly picked L4s or adults, and this is fond only in those animals with deformed body shapes. The upper worm in (E) is a pregnant hermaphrodite with a deformed posterior half, and several errant axon trajectories, two of which are indicated by arrowheads. The worm just below it is also a mutant but has a perfectly normal nervous system. Scale bars represent 50 μm.
Contributor Information
Catarina Mörck, Email: catarina.morck@cmb.gu.se.
Vivekanand Vivekanand, Email: iitbiotech@gmail.com.
Gholamali Jafari, Email: genetics_human@yahoo.com.
Marc Pilon, Email: marc.pilon@cmb.gu.se.
Acknowledgements
We thank Claes Axäng and Henrik Gradstedt for help with the et5 mapping. This research was supported by the following agencies: Vetenskaprådet, Cancerfonden, Åhlén StiftelseStiftelse, Magnus Bergvalls Stiftelse, Carl Tryggers Stiftelse, and Erik Philip-Sörensens Stiftelse.
References
- Kenzelmann D, Chiquet-Ehrismann R, Tucker RP. Teneurins, a transmembrane protein family involved in cell communication during neuronal development. Cell Mol Life Sci. 2007;64:1452–6. doi: 10.1007/s00018-007-7108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RP, Chiquet-Ehrismann R. Teneurins: a conserved family of transmembrane proteins involved in intercellular signaling during development. Dev Biol. 2006;290:237–45. doi: 10.1016/j.ydbio.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Levine A, Bashan-Ahrend A, Budai-Hadrian O, Gartenberg D, Menasherow S, Wides R. Odd Oz: a novel Drosophila pair rule gene. Cell. 1994;77:587–98. doi: 10.1016/0092-8674(94)90220-8. [DOI] [PubMed] [Google Scholar]
- Levine A, Gartenberg D, Yakov R, Lieberman Y, Budai-Hadrian O, Bashan-Ahrend A, Wides R. The genetics and molecular structure of the Drosophila pair-rule gene odd Oz (odz) Gene. 1997;200:59–74. doi: 10.1016/S0378-1119(97)00375-2. [DOI] [PubMed] [Google Scholar]
- Rakovitsky N, Buganim Y, Swissa T, Kinel-Tahan Y, Brenner S, Cohen MA, Levine A, Wides R. Drosophila Ten-a is a maternal pair-rule and patterning gene. Mech Dev. 2007;124:911–24. doi: 10.1016/j.mod.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Kinel-Tahan Y, Weiss H, Dgany O, Levine A, Wides R. Drosophila odz gene is required for multiple cell types in the compound retina. Dev Dyn. 2007;236:2541–54. doi: 10.1002/dvdy.21284. [DOI] [PubMed] [Google Scholar]
- Levine A, Weiss C, Wides R. Expression of the pair-rule gene odd Oz (odz) in imaginal tissues. Dev Dyn. 1997;209:1–14. doi: 10.1002/(SICI)1097-0177(199705)209:1<1::AID-AJA1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Kenzelmann D, Chiquet-Ehrismann R, Leachman NT, Tucker RP. Teneurin-1 is expressed in interconnected regions of the developing brain and is processed in vivo. BMC Dev Biol. 2008;8:30. doi: 10.1186/1471-213X-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XH, Brandau O, Feng K, Oohashi T, Ninomiya Y, Rauch U, Fassler R. The murine Ten-m/Odz genes show distinct but overlapping expression patterns during development and in adult brain. Gene Expr Patterns. 2003;3:397–405. doi: 10.1016/S1567-133X(03)00087-5. [DOI] [PubMed] [Google Scholar]
- Feng K, Zhou XH, Oohashi T, Morgelin M, Lustig A, Hirakawa S, Ninomiya Y, Engel J, Rauch U, Fassler R. All four members of the Ten-m/Odz family of transmembrane proteins form dimers. J Biol Chem. 2002;277:26128–35. doi: 10.1074/jbc.M203722200. [DOI] [PubMed] [Google Scholar]
- Oohashi T, Zhou XH, Feng K, Richter B, Morgelin M, Perez MT, Su WD, Chiquet-Ehrismann R, Rauch U, Fassler R. Mouse ten-m/Odz is a new family of dimeric type II transmembrane proteins expressed in many tissues. J Cell Biol. 1999;145:563–77. doi: 10.1083/jcb.145.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabikowski K, Trzebiatowska A, Chiquet-Ehrismann R. ten-1, an essential gene for germ cell development, epidermal morphogenesis, gonad migration, and neuronal pathfinding in Caenorhabditis elegans. Dev Biol. 2005;282:27–38. doi: 10.1016/j.ydbio.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Trzebiatowska A, Topf U, Sauder U, Drabikowski K, Chiquet-Ehrismann R. Caenorhabditis elegans teneurin, ten-1, is required for gonadal and pharyngeal basement membrane integrity and acts redundantly with integrin ina-1 and dystroglycan dgn-1. Mol Biol Cell. 2008;19:3898–908. doi: 10.1091/mbc.E08-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörck C, Axäng C, Pilon M. A genetic analysis of axon guidance in the C. elegans pharynx. Dev. Biol. 2003;260:158–175. doi: 10.1016/S0012-1606(03)00238-0. [DOI] [PubMed] [Google Scholar]
- Axäng C, Rauthan M, Pilon M. Developmental genetics of the C. elegans pharyngeal neuron NSM. BMC Dev Biol. 2008;8:38. doi: 10.1186/1471-213X-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Au M. Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science. 1989;243:1027–33. doi: 10.1126/science.2646709. [DOI] [PubMed] [Google Scholar]
- O'Hagan R, Chalfie M. Mechanosensation in Caenorhabditis elegans. Int Rev Neurobiol. 2006;69:169–203. doi: 10.1016/S0074-7742(05)69006-X. [DOI] [PubMed] [Google Scholar]
- Colavita A, Krishna S, Zheng H, Padgett RW, Culotti JG. Pioneer axon guidance by UNC-129, a C. elegans TGF-beta. Science. 1998;281:706–9. doi: 10.1126/science.281.5377.706. [DOI] [PubMed] [Google Scholar]
- Pan CL, Baum PD, Gu M, Jorgensen EM, Clark SG, Garriga G. C. elegans AP-2 and retromer control Wnt signaling by regulating mig-14/Wntless. Dev Cell. 2008;14:132–9. doi: 10.1016/j.devcel.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PT, Lorenowicz MJ, Silhankova M, Coudreuse DY, Betist MC, Korswagen HC. Wnt signaling requires retromer-dependent recycling of MIG-14/Wntless in Wnt-producing cells. Dev Cell. 2008;14:140–7. doi: 10.1016/j.devcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Ogura K, Goshima Y. The autophagy-related kinase UNC-51 and its binding partner UNC-14 regulate the subcellular localization of the Netrin receptor UNC-5 in Caenorhabditis elegans. Development. 2006;133:3441–50. doi: 10.1242/dev.02503. [DOI] [PubMed] [Google Scholar]
- Merz DC, Alves G, Kawano T, Zheng H, Culotti JG. UNC-52/Perlecan affects gonadal leader cell migrations in C. elegans hermaphrodites through alterations in growth factor signaling. Dev. Biol. 2003;256:173–186. doi: 10.1016/S0012-1606(03)00014-9. [DOI] [PubMed] [Google Scholar]
- Rogalski TM, Williams BD, Mullen GP, Moerman DG. Products of the unc-52 gene in Caenorhabditis elegans are homologous to the core protein of the mammalian basement membrane heparan sulfate proteoglycan. Genes Dev. 1993;7:1471–84. doi: 10.1101/gad.7.8.1471. [DOI] [PubMed] [Google Scholar]
- Baumgartner S, Martin D, Hagios C, Chiquet-Ehrismann R. Tenm, a Drosophila gene related to tenascin, is a new pair-rule gene. EMBO J. 1994;13:3728–40. doi: 10.1002/j.1460-2075.1994.tb06682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RP, Chiquet-Ehrismann R, Chevron MP, Martin D, Hall RJ, Rubin BP. Teneurin-2 is expressed in tissues that regulate limb and somite pattern formation and is induced in vitro and in situ by FGF8. Dev Dyn. 2001;220:27–39. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1084>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Pilon M. Fishing lines, time-delayed guideposts, and other tricks used by developing pharyngeal neurons in Caenorhabditis elegans. Dev Dyn. 2008;237:2073–80. doi: 10.1002/dvdy.21636. [DOI] [PubMed] [Google Scholar]
- Fujisawa K, Wrana JL, Culotti JG. The slit receptor EVA-1 coactivates a SAX-3/Robo mediated guidance signal in C. elegans. Science. 2007;317:1934–8. doi: 10.1126/science.1144874. [DOI] [PubMed] [Google Scholar]
- Watari-Goshima N, Ogura K, Wolf FW, Goshima Y, Garriga G. C. elegans VAB-8 and UNC-73 regulate the SAX-3 receptor to direct cell and growth-cone migrations. Nat Neurosci. 2007;10:169–76. doi: 10.1038/nn1834. [DOI] [PubMed] [Google Scholar]
- Zallen JA, Yi BA, Bargmann CI. The conserved immunoglobulin superfamily member SAX-3/Robo directs multiple aspects of axon guidance in C. elegans. Cell. 1998;92:217–227. doi: 10.1016/S0092-8674(00)80916-2. [DOI] [PubMed] [Google Scholar]
- Kubiseski TJ, Culotti J, Pawson T. Functional analysis of the Caenorhabditis elegans UNC-73B PH domain demonstrates a role in activation of the Rac GTPase in vitro and axon guidance in vivo. Mol Cell Biol. 2003;23:6823–35. doi: 10.1128/MCB.23.19.6823-6835.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Strumpf N, Culotti JG. VAB-8, UNC-73 and MIG-2 regulate axon polarity and cell migration functions of UNC-40 in C. elegans. Nat Neurosci. 2007;10:161–8. doi: 10.1038/nn1835. [DOI] [PubMed] [Google Scholar]
- Shakir MA, Gill JS, Lundquist EA. Interactions of UNC-34 Enabled with Rac GTPases and the NIK kinase MIG-15 in Caenorhabditis elegans axon pathfinding and neuronal migration. Genetics. 2006;172:893–913. doi: 10.1534/genetics.105.046359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven R, Kubiseski TJ, Zheng H, Kulkarni S, Mancillas J, Morales A Ruiz, Hogue CW, Pawson T, Culotti J. UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell. 1998;92:785–95. doi: 10.1016/S0092-8674(00)81406-3. [DOI] [PubMed] [Google Scholar]
- Chin-Sang ID, Chisholm AD. Form of the worm: genetics of epidermal morphogenesis in C. elegans. Trends Genet. 2000;16:544–551. doi: 10.1016/S0168-9525(00)02143-0. [DOI] [PubMed] [Google Scholar]
- Simske JS, Hardin J. Getting into shape: epidermal morphogenesis in Caenorhabditis elegans embryos. BioEssays. 2001;22:12–23. doi: 10.1002/1521-1878(200101)23:1<12::AID-BIES1003>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Ogura K-I, Wicky C, Magnenat L, Tobler H, Mori I, Müller F, Ohshima Y. Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes Dev. 1994;8:2389–2400. doi: 10.1101/gad.8.20.2389. [DOI] [PubMed] [Google Scholar]
- Okazaki N, Yan J, Yuasa S, Ueno T, Kominami E, Masuho Y, Koga H, Muramatsu M. Interaction of the UNC-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain: possible role of vesicular transport in axonal elongation. Mol. Brain Res. 2000;85:1–12. doi: 10.1016/S0169-328X(00)00218-7. [DOI] [PubMed] [Google Scholar]
- Rogalski TM, Gilchrist EJ, Mullen GP, Moerman DG. Mutations in the unc-52 gene responsible for body wall muscle defects in adult Caenorhabditis elegans are located in alternatively spliced exons. Genetics. 1995;139:159–69. doi: 10.1093/genetics/139.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski TM, Mullen GP, Bush JA, Gilchrist EJ, Moerman DG. UNC-52/perlecan isoform diversity and function in Caenorhabditis elegans. Biochem Soc Trans. 2001;29:171–6. doi: 10.1042/BST0290171. [DOI] [PubMed] [Google Scholar]
- Leamey CA, Merlin S, Lattouf P, Sawatari A, Zhou X, Demel N, Glendining KA, Oohashi T, Sur M, Fassler R. Ten_m3 regulates eye-specific patterning in the mammalian visual pathway and is required for binocular vision. PLoS Biol. 2007;5:e241. doi: 10.1371/journal.pbio.0050241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamey CA, Glendining KA, Kreiman G, Kang ND, Wang KH, Fassler R, Sawatari A, Tonegawa S, Sur M. Differential gene expression between sensory neocortical areas: potential roles for Ten_m3 and Bcl6 in patterning visual and somatosensory pathways. Cereb Cortex. 2008;18:53–66. doi: 10.1093/cercor/bhm031. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Hodgkin JA. In: The Nematode Caernorhabditis elegans. Wood WB, editor. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. Methods; pp. 587–606. [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler WA, Simske JS, Williams-Masson EM, Hardin JD, White JG. Dynamics and ultrastructure of developmental cell fusions in the Caenorhabditis elegans hypodermis. Curr. Biol. 1998;8:1087–1090. doi: 10.1016/S0960-9822(98)70447-6. [DOI] [PubMed] [Google Scholar]
- Clark SG, Chiu C. C. elegans ZAG-1, a Zn-finger-homeodomain protein, regulates axonal development and neuronal differentiation. Development. 2003;130:3781–3794. doi: 10.1242/dev.00571. [DOI] [PubMed] [Google Scholar]
- Ray P, Schnabel R, Okkema PG. Behavioral and synaptic defects in C. elegans lacking the NK-2 homeobox gene ceh-28. Dev Neurobiol. 2008;68:421–33. doi: 10.1002/dneu.20599. [DOI] [PubMed] [Google Scholar]
- Pilon M, Peng X-R, Spence AM, Plasterk RHA, Dosch H-M. The diabetes autoantigen ICA69 and its Caenorhabditis elegans homologue are conserved regulators of neuroendocrine secretion. Mol. Biol. Cell. 2000;11:3277–3288. doi: 10.1091/mbc.11.10.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauthan M, Mörck C, Pilon M. The C. elegans M3 neuron guides the growth cone of its sister cell M2 via the Krüppel-like zinc finger protein MNM-2. Dev. Biol. 2007;311:185–199. doi: 10.1016/j.ydbio.2007.08.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures. Fig. S1. Extrapharyngeal neurons are normal in the ten-1(ok641) mutant. The pan-neuronal evIs111 transgene was used to visualize the entire nervous system in (A) wild-type or (B) ten-1(ok641) mutant adults. No trajectory defects or abnormal varicosities were observed in the mutant. Asterisks indicate cell bodies that are analogously positioned in wild-type and mutant: any differences are within the variation range found in wild-type. The mec-7::gfp transgene was also used to score the mechanosensory neurons in (C) wild-type and in (D) ten-1(ok641) mutant adults. Again, no differences were observed between wild-type and mutant animals (N>100 worms examined for each treatment). Meaning of lowercase annotations: vul (vulva), vnc (ventral nerve cord), tg (tail ganglion), nr nerve ring). Fig. S2. Body neurons expressing ten-1b::gfp only rarely exhibit defects in the ten-1(ok641) mutant. In young larvae (L2), there is evidence of incomplete fasciculation in the ventral cords of both wild-type and ten-1(ok641) worms (arrows in A-B). By the L4 stage wild-type and mutants have nicely fasciculated ventral nerve cords (arrows in C-D), and two well defined lateral axons on each side (arrowheads in C-D indicate one such pair). Only occasionally (<3%; N > 100) are abnormalities observed in the neurons of randomly picked L4s or adults, and this is fond only in those animals with deformed body shapes. The upper worm in (E) is a pregnant hermaphrodite with a deformed posterior half, and several errant axon trajectories, two of which are indicated by arrowheads. The worm just below it is also a mutant but has a perfectly normal nervous system. Scale bars represent 50 μm.