Abstract
Background
The phase II detoxification enzymes execute a major protective role against xenobiotics as well as endogenous toxicants. To understand how xenobiotics regulate phase II enzyme expression, acrylamide was selected as a model xenobiotic chemical, as it induces a large number and a variety of phase II enzymes, including numerous glutathione S-transferases (GSTs) in Caenorhabditis elegans.
Methodology/Principal Findings
To begin dissecting genetically xenobiotics response pathways (xrep), 24 independent mutants of C. elegans that exhibited abnormal GST expression or regulation against acrylamide were isolated by screening about 3.5×105 genomes of gst::gfp transgenic strains mutagenized with ethyl methanesulfonate (EMS). Complementation testing assigned the mutants to four different genes, named xrep-1, -2, -3, and -4. One of the genes, xrep-1, encodes WDR-23, a nematode homologue of WD repeat-containing protein WDR23. Loss-of-function mutations in xrep-1 mutants resulted in constitutive expression of many GSTs and other phase II enzymes in the absence of acrylamide, and the wild-type xrep-1 allele carried on a DNA construct successfully cured the mutant phenotype of the constitutive enzyme expression.
Conclusions/Significance
Genetic and cellular characterization of xrep-1 mutants suggest that a large number of GSTs and other phase II enzymes induced by acrylamide are under negative regulation by XREP-1 (WDR-23), which is likely to be a functional equivalent of mammalian Keap1 and a regulator of SKN-1, a C. elegans analogue of cap-n-collar Nrf2 (nuclear factor erythroid 2-related factor 2).
Introduction
Xenobiotics such as harmful food substances (e.g., acrylamide and 2-amino-1-methyl-6-phenylimidazol [4,5-b] pyridine), environmental pollutants (e.g., heavy metals like mercury and cadmium), or mycotoxins and exotoxins from contaminating microorganisms are direct or indirect threats that incur mutagenic, carcinogenic, teratogenic, endocrine disruptive, or other deleterious consequences [1]–[5]. Oxidative processes, such as oxidative phosphorylation indispensable for aerobic organisms to produce ATP via respiration, ironically produce highly active free radicals in the process: these radicals are thought to contribute to cancer, atherosclerosis, inflammation, hypertension, and diabetes [6]. Against all such exogenous and endogenous toxicants, phase II enzymes, with glutathione S-transferases (GSTs) as most prominent, are considered to play a major protective role [7]–[11]. As for GSTs, we should not dismiss other critical biochemical roles some GSTs play in such processes as eicosanoid or steroid hormone biosynthesis and amino acid metabolism [9], [12], [13].
Acrylamide, now recognized as a prevalent food substance, has been long known as a neurotoxin for many animals and a potential carcinogen for humans [1], [14]. Previously we reported in the nematode Caenorhabditis elegans [11] that acrylamide up-regulates a large number of phase II enzymes such as GSTs and UDP-glucuronosyl/glucosyl transferases (UGTs) and some phase I enzymes such as short-chain type dehydrogenases (SDRs). C. elegans offers many experimental advantages as a model for understanding diverse aspects of biology, including responses to xenobiotics [15]. In mammals, acrylamide might be detoxified mainly by GSTs and excreted in urine [16], [17]. Among GSTs that acrylamide up-regulated more than two-fold, those we studied displayed spatially varied expression patterns. For all these reasons we have selected acrylamide as a representative chemical for xenobiotics exposure.
To dissect genetically a xenobiotics response pathway (xrep) from a target of xenobiotics to the final destination of phase II enzyme expression, we isolated xrep gene mutants with abnormal GST expression or response to acrylamide. Here we report on one of four genes defined by these mutants, xrep-1, that encodes a WD repeat-containing protein, a nematode homologue of mammalian WDR23, and provide genetic and cellular evidence that the gene xrep-1 negatively regulates GSTs and some other phase II enzymes.
Results
Isolation and mapping of mutants showing abnormal GST regulation
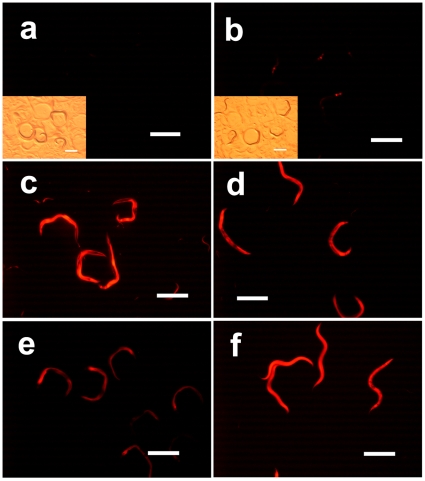
We used two gst::gfp transgenic animals, MJCU017 and MJCU047, to screen for mutants abnormally expressing GFP. MJCU017 and MJCU047, which have the chromosomally-integrated gst-4::gfp and gst-30::gfp fusion genes, respectively, emitted no detectable GFP signal in the absence of acrylamide, but emitted a very strong GFP signal from the whole body when treated with acrylamide (Fig. 1) [11], [18]. Mapping with a set of conventional marker mutants located both of these fusion genes as being integrated in linkage group X (data not shown). These animals were outcrossed at least three times with the wild-type strain N2.
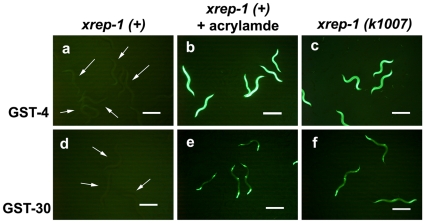
Figure 1. The xrep-1(k1007) mutants constitutively express GST-4 and GST-30 in the absence of acrylamide.
(a) MJCU017 wild-type xrep-1(+) animals. Without acrylamide, no GST-4::GFP expression is detected (arrows). (b) MJCU017 wild-type xrep-1(+) animals treated with 500 mg/L acrylamide for 24 hours at 20°C. GST-4::GFP expression is induced. (c) The GST-4::GFP expression pattern in the xrep-1(k1007) mutants. Without acrylamide, a GST signal is detected from the whole body. (d) MJCU047 wild-type xrep-1(+) animals. Without acrylamide, no GST-30::GFP expression is detected (arrows). (e) MJCU047 wild-type xrep-1(+) animals treated with 500 mg/L acrylamide for 24 hours at 20°C. GST-30::GFP expression is observed in the pharynx, hypodermis, and intestine. (f) The GST-30::GFP expression pattern in the xrep-1(k1007) mutants. GST-30::GFP expression is detected from the pharynx, hypodermis, and intestine. Scale bars, 500 µm.
We mutagenized these transgenic strains with ethyl methanesulfonate (EMS) and screened about 3.5×105 genomes to obtain 24 independent mutants. The mutants were then outcrossed more than three times with N2 wild type to reduce unwanted mutations. Complementation testing and linkage analysis assigned 16 of the mutations defined by these 24 mutants to the same gene on linkage group (LG) I (chromosome I): all of the 16 mutations were recessive, and the affected strains constitutively expressed GST in the whole body without acrylamide. Six, assigned to the same gene on LG II, were also recessive, and their mutants constitutively expressed GST in the body-wall muscle and pharynx only after they had reached the adult stage. Of the remaining two strains, one mutation on LG IV was dominant, with constitutively expressed GST throughout the whole body, whereas the other one, on LG I, was recessive with its mutant expressing no GST even when treated with acrylamide. We called these genes defined by the four complementation groups xrep-1, -2, -3, and -4, in the respective order described above.
The xrep-1 gene is wdr-23
In SNP mapping involving the Hawaiian wild-type strain CB4856 crossed with the xrep-1(k1007) mutation, one of the 16 xrep-1 alleles, we successfully assigned the gene xrep-1 to the middle of LG I between the SNP markers F21C3 and T23G11. Of the total 704 recombinants analyzed, no Hawaiian polymorphism was identified within the genomic region consisting of the cosmid clone D2030, indicating that the mutation site was in or near D2030 (Fig. 2a). We amplified 12 genes within the D2030 cosmid with T7 promoter-added primers and synthesized dsRNA for soaking RNAi. RNAi of the D2030.9 gene performed in MJCU017 resulted in the induction of GST-4 expression without acrylamide (Fig. S1). Furthermore, the DNA fragment of D2030.9 amplified from N2 genomic DNA rescued the xrep-1(k1007) mutant phenotype (data not shown).
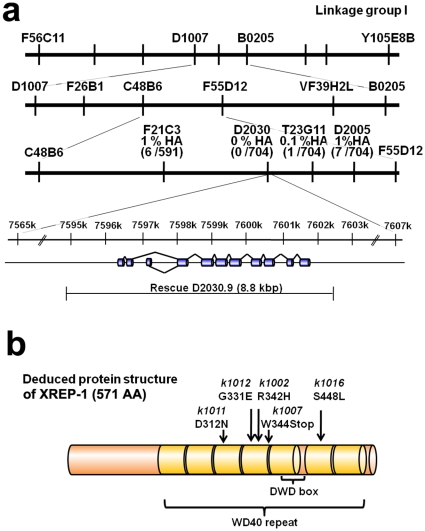
Figure 2. Genetic mapping and cloning of the xrep-1 gene.
(a) SNP mapping. The frequencies of Hawaiian polymorphism within F21C3, D2030, and T23G11 were 6 /591 (also designated as 1 % HA), 0 /704 (0 % HA), and 1 /704 (0.1 % HA), respectively. RNAi of D2030.9 was performed with MJCU017 by assaying the inducibility of GST-4 expression. Also, the genomic DNA fragment of D2030.9 was used to rescue the xrep-1(k1007) mutant phenotype. (b) Deduced protein structure of XREP-1A (see also Figure 4a-2) and locations of the five mutations identified in the present study. Amber-colored boxes represent seven WD40 domain repeats. DWD box indicates a DDB-1 (damaged DNA binding protein) WD40 binding domain. The k1002 mutation changes CGT to CAT resulting in R to H substitution at position 342. The k1007 mutation changes TGG to TGA resulting in W to protein chain termination at position 344. The k1011 mutation changes GAT to AAT resulting in D to N at position 312. The k1012 mutation changes GGA to GAA resulting in G to E at position 331. The k1016 mutation changes TCA to TTA, resulting in S to L at position 448.
D2030.9 encodes WDR-23, a nematode homologue of WDR23, a member of WD40 repeat-containing proteins, which are known to exist in yeast through plants and mammals [19]; and WDR-23 was recently reported to be involved in C. elegans GST-4 expression [20]. Five xrep-1 mutations sequenced so far have revealed four missense mutations and one nonsense mutation in the WD repeat domain, suggesting that this domain is important in the regulation of GST expression by keeping it from being induced in the absence of acrylamide (Fig. 2b). To avoid unnecessary confusion, we continue to use the gene name xrep-1 and its corresponding protein name XREP-1 instead of wdr-23 and WDR-23 unless necessary for clarification.
GST-4 expression is not totally but partially regulated by SKN-1
We reported previously that acrylamide-induced GST-4 expression was partially regulated by the transcription factor SKN-1 [11]. To examine whether the constitutive GST expression caused by the xrep-1 mutation was under SKN-1 control, we performed a series of knockdown experiments by feeding C. elegans RNAi constructs. As should be expected, the xrep-1(k1007) mutant constitutively emits a strong GFP signal from the whole body without acrylamide, and treating it with acrylamide did not further enhance this signal (Fig. 3). Knockdown of gfp resulted in the shutdown of both the constitutive and acrylamide-induced GFP signals except for that in the pharynx as expected (Fig. 3). The skn-1 knockdown also prevented the constitutive and acrylamide-induced GST-4 expression except for that in the pharynx and in body wall muscle (Figs. 3, S2). These results suggest that not all acrylamide-induced GST-4 expression is under SKN-1 regulation.
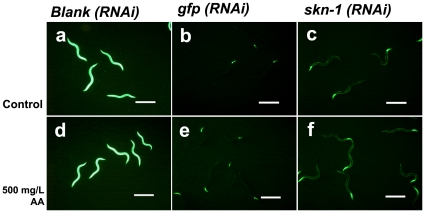
Figure 3. The xrep-1-induced GST-4 expression is partially regulated by the transcription factor SKN-1.
The GST-4 expression pattern of each RNAi in the xrep-1(k1007) mutant, without acrylamide (control) (a-c) or treated with 500 mg/L acrylamide (d-f). (a) Strong GFP signal is detected from the whole body. (b) GST-4 expression is suppressed by gfp(RNAi) except for that in the pharynx. (c) GST-4 expression is suppressed by skn-1(RNAi) except for that in the pharynx and body-wall muscle. (d) GST-4 expression is not further induced by acrylamide over that in the animals of a. (e) GST-4 expression pattern suppressed by gfp(RNAi) is unaffected by acrylamide. (f) GST-4 expression pattern suppressed by skn-1(RNAi) is not changed by acrylamide. Scale bars, 500 µm.
XREP-1 negatively regulates GST expression
Because the xrep-1(k1007) mutation constitutively expresses GST without acrylamide, we hypothesized that xrep-1 negatively regulated GST-4 expression and that acrylamide triggered GST induction by preventing normal XREP-1 function. To test this hypothesis, we first constructed the xrep-1(+)::rfp fusion gene and transferred it into the xrep-1(k1007) mutant. The xrep-1(+)::rfp completely rescued the xrep-1(k1007) mutant phenotype, but XREP-1::RFP fluorescence was too weak to be useful for the present experiment (data not shown). Therefore, we used a strain, MJCU058 {unc-119(ed3) III, kIs15[gst-4::rfp, gst-2::gfp, pDP#MM016B] IV}. This strain emitted no detectable GST-4::RFP fluorescence signal, with a hardly detectable weak constitutive GST-2::GFP signal from the mouth region (Fig. S3a). Treatment of MJCU058 with acrylamide increased the RFP signal, but did not change the GFP fluorescence signal (Fig. S3b). We introduced the xrep-1(k1007) mutation into the strain MJCU058 to construct MJCU059 {xrep-1(k1007) I; unc-119(ed3) III, kIs15 IV}, which, similarly to the fully induced GST-4::GFP expression pattern of MJCU017, constitutively expressed a strong GST-4::RFP signal from the whole body, without changing the level of the GST-2::GFP signal (Fig. S3c).
We then constructed an xrep-1(+)::gfp fusion gene (Fig. 4a) and introduced it into MJCU059 to obtain MJCU080 {kEx80[xrep-1(+)::gfp, pRF4]; xrep-1(k1007) I; unc-119(ed3) III, kIs15 IV}. The extrachromosomal array kEx80[xrep-1(+)::gfp, pRF4] of MJCU080 rescued the xrep-1(k1007) mutant phenotype and expressed GFP in the cytoplasm and nuclei of neurons, somatic gonads, intestine, and hypodermis over the whole body (Fig. 5). Because the extrachromosomal array of MJCU080 was mitotically unstable, this strain often produced animals mosaic for the xrep-1(+)::gfp transgene. Interestingly, although expectedly, those cells or tissues that retained the xrep-1(+)::gfp transgene, thus keeping the xrep-1(k1007) under rescue, emitted GFP signals. In contrast, cells or tissues without the xrep-1(+)::gfp transgene emitted no GFP signal, thus displaying the xrep-1 mutant phenotype as expected; that is, they emitted the RFP fluorescence signal (Figs. 5a–f). After a 48-hour acrylamide treatment, however, even those cells or tissues possessing xrep-1(+)::gfp that emitted the GFP signal were also induced to emit RFP fluorescence (Figs. 5g–l). This mosaic analysis confirmed our earlier hypothesis that XREP-1 repressed GST from being expressed in the absence of acrylamide and de-repressed GST expression in its presence.
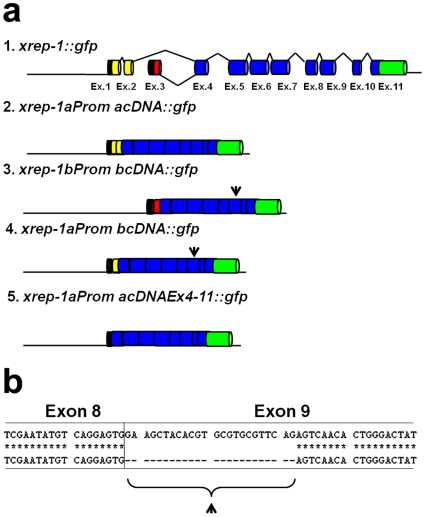
Figure 4. Structures of xrep-1 fusion constructs and partial DNA sequences of xrep-1 transcripts.
(a) Structures of various xrep-1::gfp fusion constructs. Two yellow boxes indicate exons 1 and 2 unique to xrep-1a, and a red box indicates exon 3 unique to xrep-1b. Black boxes indicate 5’ untranslated sequences, and a green box indicates gfp cDNA. The arrowhead indicates a shorter exon 9, whose sequence is shown in b. (b) Partial DNA sequence alignment of xrep-1a cDNA (upper lines) and xrep-1b cDNA (lower lines). Exon 9 in xrep-1b cDNA was 24 bp shorter than that in xrep-1a (arrowhead).
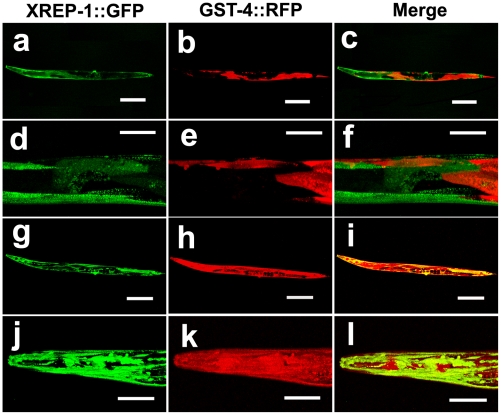
Figure 5. Mosaic analysis for xrep-1 functions.
(a, d) XREP-1::GFP mosaic expression pattern in MJCU080 {kEx80[xrep-1(+)::gfp, pRF4]; xrep-1(k1007) I; unc-119(ed3) III; kIs15 IV} without acrylamide. (b, e) GST-4::RFP expression pattern in MJCU080 without acrylamide. (c, f) Merged image. The xrep-1(k1007) phenotype is rescued in XREP-1::GFP-expressing cells or tissues. (g, j) XREP-1::GFP mosaic expression pattern in MJCU080, treated with 500 mg/L acrylamide. (h, k) GST-4::RFP expression pattern in MJCU080, treated with 500 mg/L acrylamide. (i, l) Merged image. GST-4::RFP expression is also detected in XREP-1::GFP-expressing cells or tissues. Scale bars, a, b, c, g, h, i, 200 µm; d, e, f, j, k, l, 50 µm. The images d, e, f, j, k, l are the respective enlargements of a, b, c, g, h, i. The images c, f, i, l are the respective merged images of a/b, d/e, g/h, j/k.
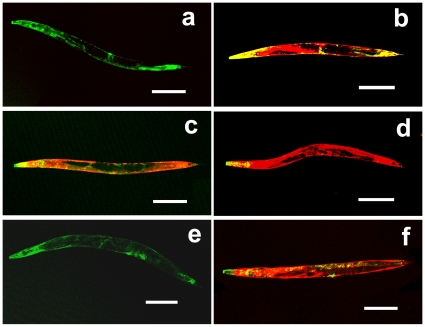
Notably, when MJCU080 animals were treated with acrylamide for 24 hours, the induction of GST expression was not so strong as would be expected from strains such as MJCU017 (Figs. 6a–b). Following 72 hours of acrylamide treatment, however, GST expression was well induced (Fig. 6c). We interpret the result to mean that this “super-repression” of acrylamide-induced GST expression was caused by over-expression from extra copies of the xrep-1(+)::gfp transgene. Its repression was eliminated when the animals were treated with xrep-1(RNAi) (Figs. 6d–f). This result further augments our hypothesis that XREP-1 controls GST expression through negative regulation, which is responsive to and inactivated or released by acrylamide. The XREP-1-mediated regulation of GST expression agrees with some functional evidence of this regulation: for instance, xrep-1(k1007) and xrep-1(RNAi) animals show more resistance to aldicarb than do their wild-type counterparts (manuscript in preparation).
Figure 6. Extra copies of the xrep-1(+)::gfp transgene suppress acrylamide-induced GST-4::RFP expression.
(a) Fluorescence plus bright field images of MJCU080 {kEx80[xrep-1(+)::gfp, pRF4]; xrep-1(k1007) I; unc-119(ed3) III; kIs15 IV}. Without acrylamide, no RFP signal is detected. (b) Fluorescence plus bright field images of MJCU080, treated with 500 mg/L of acrylamide for 24 hours at 20°C. Weak RFP signal is detected. (c) Fluorescence image of MJCU080, treated with 500 mg/L acrylamide for 72 hour at 20°C. RFP fluorescence signal is detected. (d) Fluorescence image of MJCU080, treated with xrep-1(RNAi) and 500 mg/L of acrylamide for 24 hours at 20°C. RFP fluorescence signal is detected. (e) Fluorescence image of MJCU080, treated with xrep-1(RNAi) for 24 hours at 20°C. RFP fluorescence signal is detected. (f) Fluorescence image of MJCU058 {unc-119(ed3) III; kIs15 IV}, treated with xrep-1(RNAi) and 500 mg/L of acrylamide for 24 hours at 20°C. RFP fluorescence signal is detected. Scale bars, 500 µm.
We constructed the chromosomally-integrated stable Is line MJCU085 {unc-119(ed3) III, kIs84[xrep-1(+)::gfp, pDP#MM016B]} and confirmed that its expression pattern did not differ from that for the Ex line MJCU080 (Fig. S4).
Functionality of complementary DNAs for two separate xrep-1 gene transcripts
The xrep-1 gene is predicted to encode a few protein isoforms according to WormBase (http://www.wormbase.org/). We obtained two transcripts from N2 animals, which were then used to synthesize xrep-1a and xrep-1b cDNAs. By sequencing them, we found that xrep-1a cDNA comprises 10 exons (exons 1, 2, and 4 to 11 with exclusion of exon 3) and xrep-1b cDNA consists of 9 exons (exons 3 to 11 with the exclusion of exons 1 and 2). Exons 4 to 11 were identical in both cDNAs except for the 9th exon; the 9th exon in xrep-1b cDNA was 24 bp shorter than that in xrep-1a (Fig. 4b). We then constructed xrep-1aProm acDNA::gfp and xrep-1bProm bcDNA::gfp fusion genes (Fig. 4a), which were used to transform MJCU059 {xrep-1(k1007) I; unc-119(ed3); kIs15 IV}, and obtained at least two lines of transgenic animals for each fusion gene. As observed with the xrep-1(+)::gfp transgene, xrep-1aProm acDNA::gfp expressed GST-4 in a variety of organs, tissues, or cell types in the body, such as pharynx, hypodermis, intestine, and neurons (Fig. S4), and completely rescued the xrep-1(k1007) mutant phenotype. GST-4 expression in the xrep-1aProm acDNA::gfp transgenic animal was not detected without acrylamide, but was induced with acrylamide (Figs. 7a–b). In contrast, the xrep-1bProm bcDNA::gfp transgene did not rescue the xrep-1(k1007) phenotype (Fig. 7c), as GST-4 was expressed in the absence of acrylamide (Fig. S4).
Figure 7. Expression patterns of xrep-1 cDNA::gfp fusion genes in the xrep-1(k1007) background.
GFP represents XREP-1 expression and RFP indicates GST-4 expression, without (a, c, d, e) or with 500 mg/L of acrylamide (b, f). (a) Fluorescence image of MJCU086. xrep-1aProm acDNA::gfp completely rescues xrep-1(k1007) GST-4::RFP expression. (b) Fluorescence image of MJCU086. After acrylamide treatment xrep-1aProm acDNA::gfp induces GST-4::RFP expression normally. (c) Fluorescence image of MJCU088. xrep-1bProm bcDNA::gfp does not rescue k1007 GST-4::RFP expression. (d) Fluorescence image of MJCU097. xrep-1aProm bcDNA::gfp does not rescue k1007 GST-4::RFP expression. (e) Fluorescence image of MJCU091. xrep-1aProm acDNAEx4-11::gfp completely rescues GST-4::RFP expression. (f) Fluorescence image of MJCU091. After acrylamide treatment xrep-1aProm acDNAEx4-11::gfp induces GST-4::RFP expression normally. Anterior is left, scale bars 200 µm.
We then constructed other gfp fusion genes, xrep-1aProm bcDNA::gfp and xrep-1aProm acDNAEx4-11::gfp (Fig. 4a); the former was an xrep-1b cDNA::gfp fusion gene transcribed from the xrep-1a start codon, whereas the latter was an xrep-1a cDNA::gfp fusion gene without exons 1 through 3. Both fusion genes were then introduced into MJCU059. These fusion genes exhibited the same expression pattern as did the xrep-1aProm acDNA::gfp gene (Fig. S4). Also the xrep-1aProm acDNAEx4-11::gfp fusion gene completely rescued the xrep-1(k1007) phenotype (Figs. 7e–f); however, the xrep-1aProm bcDNA::gfp did not (Fig. 7d).
Discussion
Of the phase II enzymes, GSTs are universally found in every organism from bacteria to humans and constitute a large family of enzymes that function as detoxifiers of both endogenous oxidative stress products and exogenous electrophilic chemical compounds [7]–[11]. Also very importantly, some GSTs are not just detoxifiers but multifunctional performers, as they participate in steroid and eicosanoid biosynthesis as well as in amino acid metabolism [9], [12], [13]. The C. elegans genome contains 52 gst-coding genes in the Alpha, Sigma, Omega, Zeta, and Pi classes (WormBase, http://www.wormbase.org/), and 18 of the GSTs were prominently up-regulated when animals were treated with acrylamide [11]. Because acrylamide is (a) found widely and abundantly in various foods as a hazardous food contaminant, (b) known as a potential carcinogen for humans, and (c) induces a large number and variety of phase II enzymes, we have selected this chemical as a model xenobiotic chemical probe that should represent a substantial number of xenobiotic compounds [11], [18], [21], [22], [23] (Fig. S5). To understand the genetic, molecular, and cellular processes of xenobiotics, we have attempted to dissect genetically a xenobiotics response pathway (xrep) in C. elegans by isolating mutants that respond abnormally to acrylamide. We have so far found four xrep genes: three genes (xrep-1 I, -2 II, and -3 IV) negatively and one gene (xrep-4 I) positively regulate GST expression.
The negatively regulating gene xrep-1 encodes XREP-1, a nematode homologue of the WD-repeat containing protein 23 or WDR23. WDR exists in a broad range of organisms from yeasts to mammals as well as plants. It participates in a variety of biochemical, cellular, and organismal processes, such as signal transduction, cytoskeletal dynamics, and RNA processing [19]. A family of WDR proteins functions as a substrate adapter for ubiquitin E3 ligase, and WDR domains are predicted to form a β-propeller structure, which acts as a dock for interaction with other proteins [19]. Another family of proteins predicted to form the β-propeller structure is a group of proteins containing Kelch-repeat domains, which are also considered to serve as substrate adaptors for ubiquitin E3 ligase [19]. One such protein called Keap1 represses the bZIP transcription factor Nrf2 via the Kelch-repeat domain for degradation through the ubiquitin-proteasome pathway in the absence of oxidative or electrophilic stresses. In the presence of such stresses, Nrf2 is freed from Keap1 into the nucleus where it induces the expression of phase II enzymes [8], [24]. In C. elegans, the bZIP transcription factor SKN-1, which is necessary for mesendodermal differentiation during early embryogenesis [25], is found to function similarly to Nrf2 by inducing phase II enzyme expression in response to oxidative stresses [26]–[30], sodium arsenite [30], and acrylamide [11]. According to our transcriptome analysis, a large number of genes coding for such detoxifying enzymes as GSTs, UGTs, and SDRs are negatively regulated by xrep-1 [23]. Thus, in C. elegans, XREP-1 might control SKN-1, similarly to the mammalian Keap1 for Nfr2 operative in the so-called antioxidant response element (ARE) pathway [9], [31]. An idea similar to this was recently reported [20].
Indeed, RNAi knockdown of skn-1 prevented the xrep-1(k1007) mutants and acrylamide-treated animals from expressing GST-4 in most parts of the C. elegans body, except for the pharynx and body-wall muscle (Fig. 3). Because the RNAi knockdown of gfp did not prevent GST-4 expression in the pharynx, however, we assume that RNAi itself did not work in the pharynx. GST-4 expression was not further induced in the xrep-1(k1007) mutants when they were treated with acrylamide (Fig. 3), thus suggesting that all observed acrylamide-induced GST-4 expression was under control through XREP-1. SKN-1 seemed to control GST-4 expression downstream of XREP-1 in the xenobiotics response pathway, albeit in a tissue-specific fashion, as GST-4 was expressed in the body-wall muscle of skn-1(RNAi) animals. All five xrep-1 mutation sites so far identified are located in the WDR domain of XREP-1 (Fig. 2). This result is consistent with the idea that this domain may be important for interaction with SKN-1.
Here we have an emerging picture of a functional similarity between the two pairs of proteins XREP-1/SKN-1 in worms and Keap1/Nrf2 in mammals, as both play a key role in the oxidative and electrophilic stress pathway. At the same time, however, this leaves us with fascinating evolutionary puzzles: (a) how the two counterparts XREP-1 and Keap1, considered as phylogenetically distant relatives (19), have converged to assume essentially an identical role in the pathway critically important in defending organisms against oxidative, xenobiotic, or other life-threatening stresses, (b) why or if really the nematode C. elegans lacks a homologue of the mammalian Keap1, which exists also in the insect Drosophila (32), and (c) why no or if any mammalian WD40 repeat proteins play a role similar to that for the C. elegans XREP-1.
We obtained two cDNAs, xrep-1a and xrep-1b, which were PCR products of transcripts from the N2 wild type, and analyzed their function. The xrep-1a cDNA consisting of the exons 4 to 11, which corresponds to a C-terminus of 775 amino acid residues, was sufficient to rescue the mutation xrep-1(k1007) and regulate acrylamide-induced GST expression just as does the entire xrep-1 gene (Fig. 7). Contrarily, the xrep-1b cDNA showed none of these functions. In the present experiment we could not detect any other transcripts, as implicated in WormBase (http://www.wormbase.org/). Thus, we have yet to know what the xrep-1b or any other transcripts of the xrep-1 gene are doing.
In summary, with the aid of C. elegans genetics we have so far identified four xrep genes that regulate the GST expression in the xenobiotics response pathway, and introduced here one of them, xrep-1. The gene xrep-1 that encodes a nematode homologue of WDR23 negatively regulates a large number of phase II enzymes [23]. Currently, we are studying our three remaining xrep genes while continuing isolation of new Xrep mutants to understand the xenobiotics response pathways and more generally the exogenous/endogenous stress response pathways and their regulation.
Materials and Methods
Nematode culturing and strains
Nematode culturing and handling were carried out at 20°C as described by Brenner [33]. Strains used in this experiments were N2 (Bristol strain), CB4856 (Hawaiian polymorphic strain), CB61 dpy-5(e61) I, CB1091 unc-13(e1091) I, CB120 unc-4(e120) II, CB364 dpy-18(e364) III, DP38 unc-119(ed3) III, CB138 unc-24(e138) IV, CB270 unc-42(e270) V, CB678 lon-2(e678) X, MJCU017 {unc-119(ed3) III, kIs17[gst-4::gfp, pDPMM#016B] X} [11], MJCU047 {unc-119(ed3) III, kIs41[gst-30::gfp, pDPMM#016B] X}, and MJCU058 {unc-119(ed3) III, kIs15[gst-4::rfp, gst-2::gfp, pDP#MM016B] IV}. NGM plates containing 500 mg/L (about 7 mM) acrylamide were prepared as described by Hasegawa et al. [11].
Strain construction
To make reporter constructs, all PCRs were performed with KOD Plus DNA polymerase (TOYOBO, Osaka, Japan) on N2 genomic DNA. PCR primer sequences are listed in Table S1. A fragment of C. elegans unc-54 3′UTR region was obtained by cutting the region with EcoRI and SpeI from the gfp vector pPD95.77 (kindly provided by A. Fire, Stanford University) and integrated into the equivalent restriction site of pDsRed-Monomer (Clontech, CA, USA) for rfp (red fluorescence protein) vector pMH06.12-Red. PCR primers for gst-2, gst-4, and gst-30 were designed to amplify predicted promoters for each gene, about 1.2–0.8 kbp upstream from the predicted start sites spanning over full coding regions without the stop codons. PCR-amplified DNA fragments were digested with the appropriate restriction enzymes (Table S1) and ligated into the gfp vector pPD95.77 (for gst-2::gfp and gst-30::gfp) or the rfp vector pMH06.12-Red (for gst-4::rfp). Each reporter construct (100 µg/mL) so obtained was co-injected with an equal concentration of pDP#MM016B into the gonadal arms of unc-119(ed3) adult hermaphrodites [34], [35] to obtain MJCU013 {unc-119(ed3) III; kEx13[gst-4::rfp, gst-2::gfp, pDP#MM016B]} and MJCU024 {unc-119(ed3) III; kEx24[gst-30::gfp, pDP#MM016B]}. Each transgene's extrachromosomal array was chromosomally integrated by the method of Mitani [36], and transgenic animals thus obtained were outcrossed at least three times with N2 to obtain MJCU058 {unc-119(ed3) III; kIs15[gst-4::rfp, gst-2::gfp, pDP#MM016B] IV} and MJCU047 {unc-119(ed3) III; kIs41[gst-30::gfp, pDPMM#016B] X}. Linkage mapping with the conventional markers (listed in the sub-section Nematode culturing and strains) located the fusion gene gst-4::rfp, gst-2::gfp of MJCU058 integrated in the linkage group IV and, similarly, the fusion gene gst-30::gfp of MJCU047 in X (data not shown). By the standard genetic methods, xrep-1(k1007) was transferred into MJCU058 {unc-119(ed3) III; kIs15 IV} to construct MJCU059 {xrep-1(k1007) I; unc-119(ed3) III; kIs15 IV}. Fluorescence expression patterns were observed with a Nikon SMZ800 dissection microscope equipped with a fluorescence filter.
Mutant isolation and mapping
We mutagenized the transgenic strains MJCU017 and MJCU047 with 50 mM ethyl methanesulfonate (EMS), following essentially the method described by Brenner [33] to obtain mutants expressing GST abnormally. Twenty-four mutants obtained independently were outcrossed at least three times with N2 wild type to reduce extraneous mutations. By complementation testing, 16 of 24 mutants were grouped as the same gene xrep-1. The allele xrep-1(k1007) was first mapped to linkage group I by using conventional markers, dpy-5(e61) I, unc-4(e120) II, dpy-18(e364) III, unc-24(e138) IV, unc-42(e270) V, lon-2(e678) X, and their positions were then narrowed by single-nucleotide polymorphism mapping [37]. We defined a genomic region between the SNP markers F21C3 and T23G11. No Hawaiian polymorphism was found within D2030 (total 704 recombinants inquired) indicating that the mutation site was nearby or within the cosmid clone D2030. We amplified 12 genes within D2030 with T7 promoter-added primers and synthesized dsRNA for soaking RNAi [38]. From the soaking RNAi results, the candidate gene D2030.9 was selected, PCR-amplified from the N2 wild-type genomic DNA, and co-injected with the pRF4 marker DNA into mutant animals following the method by Mello et al. [34].
xrep-1::gfp fusion gene construction
Five different gfp fusion genes, xrep-1(+)::gfp, xrep-1aProm acDNA::gfp, xrep-1bProm bcDNA::gfp, xrep-1aProm bcDNA::gfp, and xrep-1aProm acDNAEx4-11::gfp, were constructed. PCR primer sequences are listed in Table S2. The predicted xrep-1 promoter (2,110 bp upstream from the predicted start site) plus the deduced coding region was amplified with the primers XREP-1abp_HindIII_For and XREP-1ab_BamHI_Rev (Table S2) by PCR from the N2 genomic DNA and ligated into the pPD95.77 gfp expression vector to obtain the xrep-1(+)::gfp construct. Two isoforms of xrep-1 cDNA, xrep-1a and xrep-1b, were each amplified with the primers XREP-1ap_cDNA_For and XREP-1ab_BamHI_Rev (xrep-1a cDNA) or XREP-1bp_cDNA_For and XREP-1ab_BamHI_Rev (xrep-1b cDNA) (Table S2) by PCR from the two isoformic N2 cDNAs as templates originally derived from two respective transcripts. The xrep-1a or xrep-1b promoter was amplified with the primers XREP-1abp_HindIII_For and XREP-1ap_cDNA_Rev (xrep-1a promoter) or XREP-1abp_HindIII_For and XREP-1bp_cDNA_Rev (xrep-1b promoter) (Table S2) by PCR from the N2 genomic DNA. Either xrep-1a promoter and xrep-1a cDNA or xrep-1b promoter and xrep-1b cDNA were connected and ligated into the pPD95.77 gfp expression vector to obtain the xrep-1aProm acDNA::gfp or xrep-1bProm bcDNA::gfp construct (Figure 4a). Next, xrep-1b cDNA, connected to the xrep-1a promoter, was ligated into the pPD95.77 to obtain xrep-1aProm bcDNA::gfp (Figure 4a).
Furthermore, xrep-1a cDNA containing only exons 4 to 11 was amplified with the primers XREP-1ap_Exon4-11_For and XREP-1ab_BamHI_Rev (Table S2) from the N2 cDNA derived from the xrep-1a transcript. Again the xrep-1a promoter was amplified with primers XREP-1abp_HindIII_For and XREP-1ap_Exon4-11_Rev (Table S2) from the N2 genomic DNA and connected with the xrep-1a cDNA exon 4 to 11 and ligated into the pPD95.77 gfp expression vector to obtain xrep-1aProm acDNAEx4-11::gfp (Figure 4a). One hundred µg/mL of each reporter construct so obtained was co-injected with an equal concentration of pDP#MM016B or pRF4 into the gonadal arms of unc-119(ed3) or MJCU059 {xrep-1(k1007) I; unc-119(ed3) III; kIs15 IV} adult hermaphrodites as described [34], [35]. Integration of transgenic extrachromosomal arrays into chromosomes was performed as described by Mitani [36], and integrated lines were outcrossed two times with N2 wild type. Transgenic animals so obtained were MJCU080 {xrep-1(k1007) I; unc-119(ed3) III; kIs15 IV; kEx80[xrep-1(+)::gfp, pRF4]}, MJCU081 {xrep-1(k1007) I; unc-119(ed3) III; kIs15 IV; kEx81[xrep-1(+)::gfp, pRF4]}, MJCU085 {unc-119(ed3) III; kIs84[xrep-1(+)::gfp, pDP#MM016B]}, MJCU086 {xrep-1(k1007) I; unc-119(ed3) III; kIs15 IV; kEx86[xrep-1aProm acDNA::gfp, pRF4]}, MJCU087 {xrep-1(k1007) I; unc-119(ed3) III; kIs15 IV; kEx87[xrep-1aProm acDNA::gfp, pRF4]}, MJCU088 {xrep-1(k1007) I; unc-119(ed3) III; kIs15 IV; kEx88[xrep-1bProm bcDNA::gfp, pRF4]}, MJCU089 {xrep-1(k1007) I; unc-119(ed3) III; kIs15 IV; kEx89[xrep-1bProm bcDNA::gfp, pRF4]}, MJCU091 {xrep-1(k1007) I; unc-119(ed3) III; kIs15 IV; kEx91[xrep-1aProm acDNAEx4-11::gfp, pRF4]}, MJCU094 {xrep-1(k1007) I; unc-119(ed3) III; kIs15 IV; kEx94[xrep-1aProm acDNAEx4-11::gfp, pRF4]}, MJCU097 {xrep-1(k1007) I; unc-119(ed3) III; kIs15 IV; kEx97[xrep-1aPromb cDNA::gfp, pRF4]}, and MJCU098 {xrep-1(k1007) I; unc-119(ed3) III; kIs15 IV; kEx98[xrep-1a Prom bcDNA::gfp, pRF4]}.
RNAi
Gene fragments of skn-1 cDNA, xrep-1 cDNAEx4-11, or gfp were prepared by PCR amplification of C. elegans N2 cDNA, genomic DNA, or plasmid vector pPD95.77, respectively, with primers (Table S3). Each PCR fragment was digested with EcoRI and cloned into the EcoRI restriction site of the RNAi vector pPD129.36 (kindly provided by A. Fire, Stanford University). The PCR fragment-ligated plasmid or the blank vector pPD129.36 was used to transform E. coli HT115 [39].
For RNAi experiments, synchronized L1-stage animals were first cultured for 48 hours at 20°C on NGM (containing 50 µg/mL ampicillin and 12.5 µg/mL tetracycline) plates seeded with E. coli HT115 transformed with each different RNAi plasmid. The animals were then collected and transferred onto NGM plates with or without 500 mg/L acrylamide, seeded with each different E. coli HT115 RNAi bacteria. After 24 to 48 hours of incubation, the animals were observed for GFP expression with both a Nikon SMZ800 dissection microscope equipped with a fluorescence filter and a ZEISS Axiovert 200 microscope equipped with a confocal laser-scanning module.
Supporting Information
GST-4 expression is induced when the transgenic MJCU017 animals are treated with soaking RNAi of D2020.9 (xrep-1). (a) MJCU017 animals treated with blank RNAi. No GST-4::GFP expression is detected (arrows point to three animals). (b) MJCU017 animals treated with xrep-1(RNAi). GST-4::GFP expression is induced. Scale bar, 500 µm.
(1.29 MB TIF)
GST-4 expression induced by acrylamide and xrep-1(k1007) mutation is prevented by skn-1(RNAi) except for that in the pharynx and body-wall muscle. (a-c) GST-4 expression patterns in MJCU017 treated with 500 mg/L acrylamide. (d-f) GST-4 expression patterns in xrep-1(k1007) without acrylamide. (g-i) GST-4 expression patterns of skn-1(RNAi) in xrep-1(k1007) without acrylamide. Scale bars, 50 µm.
(1.33 MB TIF)
(a) Fluorescence plus bright field images of MJCU058 {unc-119(ed3) III; kIs15[gst-4::rfp, gst-2::gfp, pDP#MM016B] IV}. Without acrylamide, no fluorescence signal is detected. (b) Fluorescence image of MJCU058, treated with 500 mg/L of acrylamide at 20°C for 24 hours. RFP fluorescence signal is detected. (c) Fluorescence image of MJCU059 {xrep-1(k1007) I; unc-119(ed3) III; kIs15 IV}. RFP fluorescence signal is detected. (d-f) RFP fluorescence signal in the MJCU058 {unc-119(ed3) III; kIs15 IV} animal, treated with 500 mg/L of acrylamide at 20°C for 24 hours. (d) Head. (e) Vulva. (f) Tail. Scale bars, a-c, 500 µm; d-f, 50 µm.
(2.14 MB TIF)
Expression patterns of various xrep-1::gfp fusion genes. (a-c) XREP-1::GFP expression. (d-f) XREP-1aProm acDNA::GFP expression. (g-i) XREP-1bProm bcDNA::GFP expression patterns. (j-l) XREP-1aProm bcDNA::GFP expression. (m-o) XREP-1aProm acDNAEx4-11::GFP expression. Scale bars, 50 µm.
(1.79 MB TIF)
GST and UGT responses in transgenic animals against several xenobiotics. Young adult transgenics were transferred into NGM plates containing each xenobiotic and incubated at 25°C for 24 hours. Transgenic animals used in these experiments were MJCU017 {unc-119(ed3) III; kIs17[gst-4::gfp, pDP#MM016B] X}, MJCU028 {unc-119(ed3) III; kEx28[gsto-2b::cfp, pDP#MM016B]} (23), MJCU003 {kEx3[gst-7::gfp, pRF4]} (11), MJCU047 {unc-119(ed3) III; kIs41[gst-30::gfp, pDP#MM016B] X}, and MJCU050 {unc-119(ed3) III; kIs20[ugt-13p::gfp, pDP#MM016B] III} (21). Control, without xenobiotics; MeHg (500 nM Methylmercury); Paraquat (20 mM Paraquat); tBOOH (1 mM tert-Butyl hydroperoxide); AA (7 mM Acrylamide). (a) GST-4::GFP expression was not detected. Arrows indicate animals. (b) GST-4::GFP expression was induced when animals were treated with MeHg. (c) GST-4::GFP expression was slightly induced (arrows) when animals were treated with paraquat. (d) GST-4::GFP expression was induced when animals were treated with tBOOH. (f) Weak GSTO-2B::CFP expression was detected. (g-i) GSTO-2B::CFP expression was induced when animals were treated with MeHg, paraquat, and tBOOH. (k) Weak GST-7::GFP expression was detected. (l-n) GST-7::GFP expression was induced when animals were treated with MeHg, paraquat, and tBOOH. (p) GST-30::GFP expression was not detected. Arrows indicate animals. (q) GST-30::GFP expression was induced when animals were treated with MeHg (arrows). (r) GST-30::GFP expression was not induced when animals (arrows) were treated with paraquat. (s) GST-30::GFP expression was induced when animals were treated with tBOOH (arrows). (u) Weak UGT-13::GFP expression was detected. (v-x) UGT-13::GFP expression was induced when animals were treated with MeHg, paraquat, and tBOOH. (e, j, o, t, y) All of the GST- and UGT-fused GFP expressions were strongly induced when animals were treated with AA (11). Scale bars, 500 µm.
(1.64 MB TIF)
Primers for gst::reporter fusion genes.
(0.02 MB DOC)
Primers for xrep-1::reporter fusion genes.
(0.02 MB DOC)
Primers for RNAi.
(0.02 MB DOC)
Acknowledgments
We are grateful to the Caenorhabditis elegans Genetics Center for providing nematode strains, Ms. Satsuki Miwa, Viva Informatica, for useful comments and careful reading of this manuscript, and Mr. Takashi Komatsu and Hiroki Kawazoe for technical support.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a Grant-in-Aid for Scientific Research to JM and a JSPS research fund to KH of the Japanese Ministry of Education, Culture, Sports, Science and Technology and by the Chubu University special research fund to JM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Friedman M. Chemistry, biochemistry, and safety of acrylamide. A review. J Agric Food Chem. 2003;51:4504–4526. doi: 10.1021/jf030204+. [DOI] [PubMed] [Google Scholar]
- 2.Dybing E, O'Brien J, Renwick AG, Sanner T. Risk assessment of dietary exposures to compounds that are genotoxic and carcinogenic –An overview. Toxicol Lett. 2008;180:110–117. doi: 10.1016/j.toxlet.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Joseph P. Mechanisms of cadmium carcinogenesis. Toxicol Appl Pharmacol. 2009;238:272–279. doi: 10.1016/j.taap.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 4.Tan SW, Meiller JC, Mahaffey KR. The endocrine effects of mercury in humans and wildlife. Crit Rev Toxicol. 2009;39:228–269. doi: 10.1080/10408440802233259. [DOI] [PubMed] [Google Scholar]
- 5.Amzal B, Julin B, Vahter M, Wolk A, Johanson G, et al. Population toxicokinetic modeling of cadmium for health risk assessment. Environ Health Perspect. 2009;117:1293–1301. doi: 10.1289/ehp.0800317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benz CC, Yau C. Ageing, oxidative stress and cancer: paradigms in parallax. Nat Rev Cancer. 2008;8:875–879. doi: 10.1038/nrc2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao X, Dinkova-Kostova AT, Talalay P. Powerful and prolonged protection of human retinal pigment epithelial cells, keratinocytes, and mouse leukemia cells against oxidative damage: the indirect antioxidant effects of sulforaphane. Proc Natl Acad Sci USA. 2001;98:15221–15226. doi: 10.1073/pnas.261572998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 9.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 10.Lindblom TH, Dodd AK. Xenobiotic detoxification in the nematode Caenorhabditis elegans. J Exp Zool A Comp Exp Biol. 2006;305:720–730. doi: 10.1002/jez.a.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasegawa K, Miwa S, Isomura K, Tsutsumiuchi K, Taniguchi H, et al. Acrylamide-responsive genes in the nematode Caenorhabditis elegans. Toxicol Sci. 2008;101:215–225. doi: 10.1093/toxsci/kfm276. [DOI] [PubMed] [Google Scholar]
- 12.Johansson A-S, Mannervik B. Human glutathione transferase A3-3, a highly efficient catalyst of double-bond isomerization in the biosynthetic pathway of steroid hormones. J Biol Chem. 2001;276:33061–33065. doi: 10.1074/jbc.M104539200. [DOI] [PubMed] [Google Scholar]
- 13.Jowsey IR, Thomson AM, Flanagan JU, Murdock PR, Moore GBT, et al. Mammalian class Sigma glutathione S-transferases: catalytic properties and tissue-specific expression of human and rat GSH-dependent prostaglandin D2 synthases. Biochem J. 2001;359:507–516. doi: 10.1042/0264-6021:3590507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tareke E, Rydberg P, Karlsson P, Eriksson S, Törnqvist M. Analysis of acrylamide, a carcinogen formed in heated foodstuffs. J Agric Food Chem. 2002;50:4998–5006. doi: 10.1021/jf020302f. [DOI] [PubMed] [Google Scholar]
- 15.Leung MCK, Williams PL, Benedetto A, Au C, Helmcke KJ, et al. Caenorhabditis elegans: an emerging model in biomedical and environmental toxicology. Toxicol Sci. 2008;106:5–28. doi: 10.1093/toxsci/kfn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odland L, Romert L, Clemedson C, Walum E. Glutathione content, glutathione transferase activity and lipid peroxidation in acrylamide-treated neuroblastoma N1E 115 cells. Toxicol In Vitro. 1994;8:263–267. doi: 10.1016/0887-2333(94)90192-9. [DOI] [PubMed] [Google Scholar]
- 17.Sumner SCJ, Williams CC, Snyder RW, Krol WL, Asgharian B, et al. Acrylamide: a comparison of metabolism and hemoglobin adducts in rodents following dermal, intraperitoneal, oral, or inhalation exposure. Toxicol Sci. 2003;75:260–270. doi: 10.1093/toxsci/kfg191. [DOI] [PubMed] [Google Scholar]
- 18.Hasegawa K, Miwa S, Tajima T, Tsutsumiuchi K, Taniguchi H, et al. A rapid and inexpensive method to screen for common foods that reduce the action of acrylamide, a harmful substance in food. Toxicol Lett. 2007;175:82–88. doi: 10.1016/j.toxlet.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Hudson AM, Cooley L. Phylogenetic, structural and functional relationships between WD- and Kelch-repeat proteins. Subcell Biochem. 2008;48:6–19. doi: 10.1007/978-0-387-09595-0_2. [DOI] [PubMed] [Google Scholar]
- 20.Choe KP, Przybysz AJ, Strange K. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol Cell Biol. 2009;29:2704–2715. doi: 10.1128/MCB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasegawa K, Miwa S, Tsutsumiuchi K, Miwa J. Allyl isothiocyanate that induces GST and UGT expression confers oxidative stress resistance on C. elegans, as demonstrated by nematode biosensor. PloS ONE. 2010;5(2):e9267. doi: 10.1371/journal.pone.0009267. doi: 10.1371/journal.pone.0009267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasegawa K, Miwa S, Tsutsumiuchi K, Taniguchi H, Miwa J. Extremely low dose of acrylamide decreases lifespan in Caenorhabditis elegans. Toxicol Lett. 2004;152:183–189. doi: 10.1016/j.toxlet.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 23.Hasegawa K, Miwa J. Transcriptome analysis of the xrep-1(RNAi) phenocopy in Caenorhabditis elegans. Annu Rep Inst Biol Funct Chubu Univ, in press. 2010.
- 24.Kobayashi A, Kang MI, Watai Y, Tong KI, Shibata T, et al. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol. 2006;26:221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kahn NW, Rea SL, Moyle S, Kell A, Johnson, TE Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem J. 2008;409:205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- 27.An JH, Vranas K, Lucke M, Inoue H, Hisamoto N, et al. Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc Natl Acad Sci USA. 2005;102:16275–16280. doi: 10.1073/pnas.0508105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tullet JMA, Hertweck M, An JH, Baker J, Hwang JY, et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliveira RP, Abate JP, Dilks K, Landis J, Ashraf J, et al. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell. 2009;8:524–541. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park SK, Tedesco PM, Johnson TE. Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell. 2009;8:258–269. doi: 10.1111/j.1474-9726.2009.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 32.Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brenner S. The genetics of Caenorhabditis elegans. . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maduro M, Pilgrim D. Conservation of function and expression of unc-119 from two Caenorhabditis species despite divergence of non-coding DNA. Gene. 1996;183:77–85. doi: 10.1016/s0378-1119(96)00491-x. [DOI] [PubMed] [Google Scholar]
- 36.Mitani S. Genetic regulation of mec-3 gene expression implicated in the specification of the mechanosensory neuron cell types in Caenorhabditis elegans. Dev Growth Differ. 1995;37:551–557. doi: 10.1046/j.1440-169X.1995.t01-4-00010.x. [DOI] [PubMed] [Google Scholar]
- 37.Davis MW, Hammarlund M, Harrach T, Hullett P, Olsen S, et al. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics. 2005;6:118. doi: 10.1186/1471-2164-6-118. doi: 10.1186/1471-2164-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- 39.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2000;2:research0002.1–0002.10. doi: 10.1186/gb-2000-2-1-research0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GST-4 expression is induced when the transgenic MJCU017 animals are treated with soaking RNAi of D2020.9 (xrep-1). (a) MJCU017 animals treated with blank RNAi. No GST-4::GFP expression is detected (arrows point to three animals). (b) MJCU017 animals treated with xrep-1(RNAi). GST-4::GFP expression is induced. Scale bar, 500 µm.
(1.29 MB TIF)
GST-4 expression induced by acrylamide and xrep-1(k1007) mutation is prevented by skn-1(RNAi) except for that in the pharynx and body-wall muscle. (a-c) GST-4 expression patterns in MJCU017 treated with 500 mg/L acrylamide. (d-f) GST-4 expression patterns in xrep-1(k1007) without acrylamide. (g-i) GST-4 expression patterns of skn-1(RNAi) in xrep-1(k1007) without acrylamide. Scale bars, 50 µm.
(1.33 MB TIF)
(a) Fluorescence plus bright field images of MJCU058 {unc-119(ed3) III; kIs15[gst-4::rfp, gst-2::gfp, pDP#MM016B] IV}. Without acrylamide, no fluorescence signal is detected. (b) Fluorescence image of MJCU058, treated with 500 mg/L of acrylamide at 20°C for 24 hours. RFP fluorescence signal is detected. (c) Fluorescence image of MJCU059 {xrep-1(k1007) I; unc-119(ed3) III; kIs15 IV}. RFP fluorescence signal is detected. (d-f) RFP fluorescence signal in the MJCU058 {unc-119(ed3) III; kIs15 IV} animal, treated with 500 mg/L of acrylamide at 20°C for 24 hours. (d) Head. (e) Vulva. (f) Tail. Scale bars, a-c, 500 µm; d-f, 50 µm.
(2.14 MB TIF)
Expression patterns of various xrep-1::gfp fusion genes. (a-c) XREP-1::GFP expression. (d-f) XREP-1aProm acDNA::GFP expression. (g-i) XREP-1bProm bcDNA::GFP expression patterns. (j-l) XREP-1aProm bcDNA::GFP expression. (m-o) XREP-1aProm acDNAEx4-11::GFP expression. Scale bars, 50 µm.
(1.79 MB TIF)
GST and UGT responses in transgenic animals against several xenobiotics. Young adult transgenics were transferred into NGM plates containing each xenobiotic and incubated at 25°C for 24 hours. Transgenic animals used in these experiments were MJCU017 {unc-119(ed3) III; kIs17[gst-4::gfp, pDP#MM016B] X}, MJCU028 {unc-119(ed3) III; kEx28[gsto-2b::cfp, pDP#MM016B]} (23), MJCU003 {kEx3[gst-7::gfp, pRF4]} (11), MJCU047 {unc-119(ed3) III; kIs41[gst-30::gfp, pDP#MM016B] X}, and MJCU050 {unc-119(ed3) III; kIs20[ugt-13p::gfp, pDP#MM016B] III} (21). Control, without xenobiotics; MeHg (500 nM Methylmercury); Paraquat (20 mM Paraquat); tBOOH (1 mM tert-Butyl hydroperoxide); AA (7 mM Acrylamide). (a) GST-4::GFP expression was not detected. Arrows indicate animals. (b) GST-4::GFP expression was induced when animals were treated with MeHg. (c) GST-4::GFP expression was slightly induced (arrows) when animals were treated with paraquat. (d) GST-4::GFP expression was induced when animals were treated with tBOOH. (f) Weak GSTO-2B::CFP expression was detected. (g-i) GSTO-2B::CFP expression was induced when animals were treated with MeHg, paraquat, and tBOOH. (k) Weak GST-7::GFP expression was detected. (l-n) GST-7::GFP expression was induced when animals were treated with MeHg, paraquat, and tBOOH. (p) GST-30::GFP expression was not detected. Arrows indicate animals. (q) GST-30::GFP expression was induced when animals were treated with MeHg (arrows). (r) GST-30::GFP expression was not induced when animals (arrows) were treated with paraquat. (s) GST-30::GFP expression was induced when animals were treated with tBOOH (arrows). (u) Weak UGT-13::GFP expression was detected. (v-x) UGT-13::GFP expression was induced when animals were treated with MeHg, paraquat, and tBOOH. (e, j, o, t, y) All of the GST- and UGT-fused GFP expressions were strongly induced when animals were treated with AA (11). Scale bars, 500 µm.
(1.64 MB TIF)
Primers for gst::reporter fusion genes.
(0.02 MB DOC)
Primers for xrep-1::reporter fusion genes.
(0.02 MB DOC)
Primers for RNAi.
(0.02 MB DOC)