Abstract
Background
Expansins form a large multi-gene family found in wheat and other cereal genomes that are involved in the expansion of cell walls as a tissue grows. The expansin family can be divided up into two main groups, namely, alpha-expansin (EXPA) and beta-expansin proteins (EXPB), with the EXPB group being of particular interest as group 1-pollen allergens.
Results
In this study, three beta-expansin genes were identified and characterized from a newly sequenced region of the Triticum aestivum cv. Chinese Spring chromosome 3B physical map at the Sr2 locus (FPC contig ctg11). The analysis of a 357 kb sub-sequence of FPC contig ctg11 identified one beta-expansin genes to be TaEXPB11, originally identified as a cDNA from the wheat cv Wyuna. Through the analysis of intron sequences of the three wheat cv. Chinese Spring genes, we propose that two of these beta-expansin genes are duplications of the TaEXPB11 gene. Comparative sequence analysis with two other wheat cultivars (cv. Westonia and cv. Hope) and a Triticum aestivum var. spelta line validated the identification of the Chinese Spring variant of TaEXPB11. The expression in maternal and grain tissues was confirmed by examining EST databases and carrying out RT-PCR experiments. Detailed examination of the position of TaEXPB11 relative to the locus encoding Sr2 disease resistance ruled out the possibility of this gene directly contributing to the resistance phenotype.
Conclusions
Through 3-D structural protein comparisons with Zea mays EXPB1, we proposed that variations within the coding sequence of TaEXPB11 in wheats may produce a functional change within features such as domain 1 related to possible involvement in cell wall structure and domain 2 defining the pollen allergen domain and binding to IgE protein. The variation established in this gene suggests it is a clearly identifiable member of a gene family and reflects the dynamic features of the wheat genome as it adapted to a range of different environments and uses.
Accession Numbers: ctg11 =FN564426
Survey sequences of TaEXPB11ws and TsEXPB11 are provided request.
Background
Cereal plant crops are vital to the overall health of the world's population and genome sequencing is an important step in the genetic improvement of crops. While hexaploid wheat (Triticum aestivum L.) accounts for nearly one-fifth of the entire world's daily calories [1], the sequencing of its genome has been restricted by high sequencing costs associated with its large genome size (~16,000 Mb) and high (~80%) repetitive content [2]. The published physical map of the largest wheat chromosome 3B [3], which itself is twice the size of the entire rice genome, has allowed researchers to target specific regions that have been identified to contain agronomically important traits such as fungal resistance or grain quality. Projects co-coordinated within the International Wheat Genome Sequencing Consortium (IWGSC) on chromosome 3B aim to tackle the challenges associated with genome sequencing through collaboration, and facilitate the study of significant multi-gene families.
One such multi-gene family found extensively in the wheat and other cereal genomes are the expansins. It has been estimated that the hexaploid wheat genome contains more than 95 expressed members [4], much higher than the rice genome. Expansins belong to a large group of proteins found within the structure of plant cell walls and are considered to be involved in the expansion of cell walls as a tissue grows [5]. The proposed model of expansin action is that these proteins modify the cell-wall matrix to enable growth and development of plant cells [6-8] and, as a result, expansins have been implicated in providing resistance to certain diseases [9]. The latter was of particular interest because it was located in a region of the wheat genome being sequenced in order to define disease resistance genes in the region. Expansins were originally isolated from cucumber seedlings and have 'acid growth' characteristics, where they can stimulate cell enlargement in the response to acid pH [10]. Expansins have now been reported in many plants such as cotton [Gossypium hirsutum; [11,12]], tomato [Lycopersicon esculentum; [13]], Arabidopsis [Arabidopsis thaliana; [14,15]] and pea [Pisum sativum; [16]]. cDNA clones have also been isolated from wheat [4,17-19] and barley [Hordeum vulgare; [20]].
The multi-gene expansin family can be divided up into two main groups, namely, α-expansin (EXPA) and beta-expansin proteins (EXPB), which share very limited (~20%) amino acid similarity even though both are associated with cell-loosening activity [6]. The beta-expansin proteins were originally viewed exclusively as group 1 pollen allergens but are now considered to be important in cell wall changes during growth in vegetative tissues of grasses and dicotyledon plants [18], most notably in development and growth zones of tissues such as roots [20,21]. The group 1 pollen allergen domain-containing beta-expansin proteins are highly expressed in mature pollen of grass species and are thought to have a role in pollen tube penetration [14,22]. Pollen-triggered allergic reactions (e.g. hayfever and seasonal asthma) affect up to 25% of adults in industrialized nations [23]. Group 1 allergens bind to group 1 specific IgE antibodies [24] and a well studied example is the pollen allergen, Phl p 2 in timothy grass (Phleum pratense) where a specific protein domain has been identified as a binding site [25].
A previous study analysed the sera of patients that had undergone positive double blind, placebo-controlled food challenge to hexaploid wheat and identified the gene TaEXPB11 as one of 12 genes that encoded proteins binding to the IgE from wheat sensitive patients [19]. In the present study, we present the identification of three beta-expansin genes through a genome sequencing study of the short-arm of chromosome 3B in hexaploid wheat (T. aestivum cv Chinese Spring). Structural analyses of the three TaEXPB11-Chinese Spring variants indicate a localised gene duplication producing non-coding gene copies. Re-analysis of recombinant lines using specific markers from the expansin genes indicated that they were not linked to the Sr2 resistance phenotype of wheat cv. Hope, arguing against an involvement in the resistance. Comparative sequence analysis was also undertaken on selected cultivars and a T. spelta wheat accession demonstrating changes within the gene-coding sequence of TaEXPB11 produced protein structural changes. Expression of TaEXPB11, experimentally and within EST databases, was also assessed. Our study provides novel insights into the structure of beta-expansins and their variation across different wheat genomes.
Results
Ctg11 wheat genome sequencing
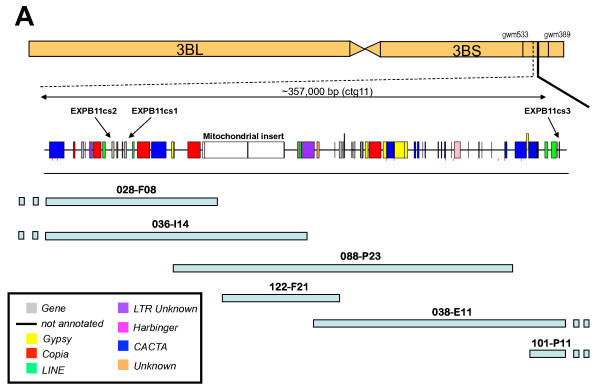
The physical map of chromosome 3BS of hexaploid wheat (cv. Chinese Spring and cv. Hope) has been compiled through DNA fingerprinting of the flow-sorted chromosome 3B BAC library and anchoring BAC contigs to genetic maps [4,26]. The shotgun sequencing of BACs located within a minimum tilling path of ca 1.3 Mb across the Sr2 locus on chromosome 3BS [26] was carried out to identify possible candidate genes for this disease resistance locus (R. Mago et al. 2010, in preparation). The BAC clones characterized in this study, and related work (Choulet et al. 2010; Wicker et al. in preparation), indicated the presence of an active expansin gene which was a candidate for the Sr2 locus because, as a cell wall component, the product of the gene has been associated with disease resistance [27]. A 357 kb region the ctg11 genome sequence containing three beta-expansin genes was annotated in detail for the present study (Figure 1)
Figure 1.
Genomic location, bacterial artificial chromosome (BAC) clone map and sequence annotation of 357 kb from the hexaploid wheat (T. aestivum cv Chinese Spring) ctg11 genomic sequence of chromosome 3B containing three beta-expansin genes.
The 357 kb ctg11 sequence contained nine complete or partial gene-coding regions (Table 1) giving it a gene density of one gene per 44.6 kb. The sequence contained 133,134 bp (33.2%) repetitive DNA in the form of transposable elements (TEs) and a 51,666 bp mitochondrial genome insert. The presence of the mitochondrial DNA insert in normal genomic DNA has been confirmed by assaying the genomic sequence for mtDNA-chromosomal DNA junctions (data not shown). Further genome sequencing was carried out using 454-technology sequencing of BAC clone 3B_036_I14 (Zhang, J. and Kong, XY., unpublished), which contained two full-length beta-expansin genes and the mitochondrial insert, in order to confirm the presence of the mitochondrial insert and improve the sequence assembly in some regions. Additional BAC sequencing was also carried out on the syntenic ctg11 region of wheat cv. Hope (R. Mago et al. 2010, in preparation).
Table 1.
Gene-coding annotation of the 357,000 bp sub-sequence of ctg11
| Rice Genome | Wheat EST Analysis | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Length (nt) | # Exons | Predicted protein length (aa) | Rice Chr. | Top Rice protein hit (MSU Rice annotation version 5) | Wheat EST | E-value | Wheat Unigene set |
| EXPB11cs2 | 1233 | 3 | 275 | 3 | LOC_Os03g01270 | CJ674809 | 1e-178 | Ta.31031 |
| EXPB11cs1 | 1342 | 3 | 290 | 3 | LOC_Os03g01270 | CJ674809 | 2e-105 | Ta.31031 |
| FMO1 | 1720 | 4 | 469 | 7 | LOC_Os07g02140 | CK207166 | 9e-124 | Ta.32721 |
| RGA1 | 3037 | 3 | 553 | 1 | LOC_Os01g36640 | CJ948865 | 2e-78 | Ta.41149 |
| WDL1 | 1841 | 7 | 394 | 11 | LOC_Os11g38010 | CJ661904 | 3e-104 | Ta.57217 |
| DPC1 | 2707 | 3 | 230 | 1 | LOC_Os01g25880 | CK212399 | 0.0 | Ta.55136 |
| UTG1 | 1470 | 1 | 490 | 11 | LOC_Os11g38650 | BU100894 | 6e-152 | Ta.51207 |
| ADP1 | 3805 | 6 | 308 | 5 | LOC_Os05g04860 | CK210567 | 2e-66 | Ta.4597 |
| EXPB11cs3 | 291 | 1 | 97 | 3 | LOC_Os03g01270 | BQ608206 | 6e-129 | Ta.31031 |
Structural characterisation and validation of three beta-expansin genes in ctg11
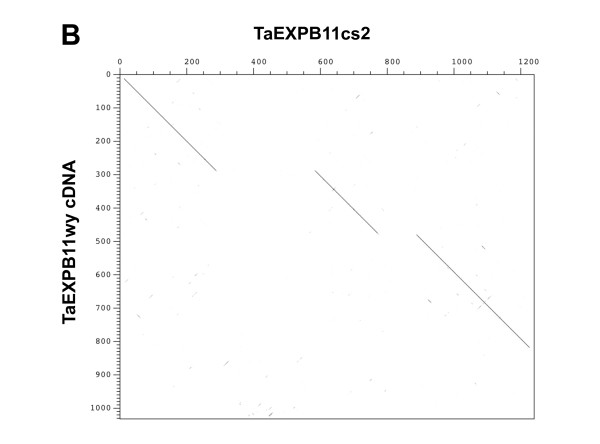
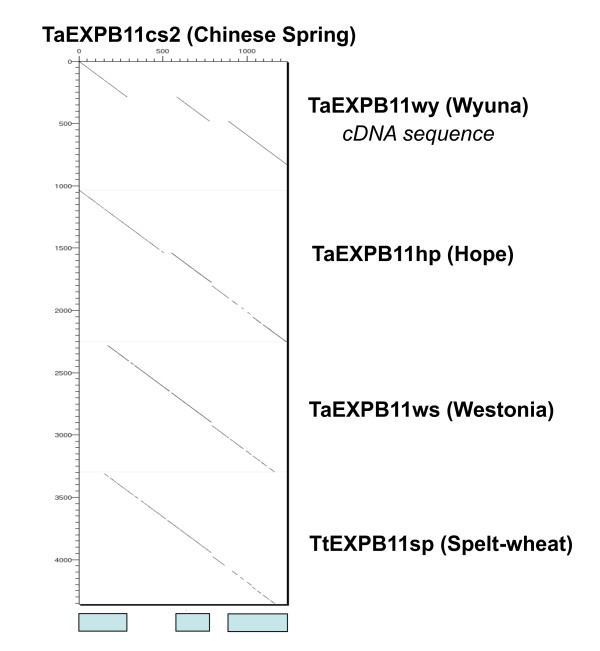
The 357 kb subsequence of ctg11 was masked for repetitive elements by running RepeatMasker (Smit and Green, http://www.repeatmasker.org/) using the Triticeae repeat sequence (TREP) database http://wheat.pw.usda.gov/ITMI/Repeats/. Gene predictions were then searched against a protein subsection of the TREP database using BLASTP to ensure no repetitive predicted proteins were included in the analysis. Two highly similar gene models were predicted and found within 7,418 bp of each other with a third similar truncated copy located over 320 kb away (Figure 1). Annotation of the genomic sequence identified that the three copies of sequences were related to the beta-expansin family [27], with the second copy of the three genes being very similar to a published TaEXPB11 cDNA (Figure 2) [19]. The published TaEXPB11 sequence was recovered from an endosperm cDNA library from wheat cv. Wyuna [28].
Figure 2.
Dot matrix to show the nucleotide sequence comparison between the TaEXPB11cs2 and TaEXPB11wy cDNA sequence (isolated from wheat cv. Wyuna [19]).
The coding regions of the beta-expansin that has the highest sequence similarity to the published TaEXPB11 cDNA has 6 amino acid differences (98.2% nucleotide sequence similarity) resulting from single nucleotide polymorphism (SNP) differences; these SNPs are not unexpected considering the published TaEXPB11 derived from wheat cv. Wyuna and the genomic sequence from wheat cv. Chinese Spring. We propose to name this gene TaEXPB11cs2 with the names of copies located upstream being TaEXPB11cs1 and TaEXPB11cs3 respectively. The TaEXPB11 from wheat cv. Wyuna [19] will be referred to as TaEXPB11wy in this study.
TaEXPB11cs1 had a lower sequence similarity (73.36%) to the TaEXPB11wy cDNA and a slightly lower similarity to the identified rice homolog OsEXPB7 (63% compared to 64.83% in TaEXPB11cs2). The TaEXPB11cs3 gene was very closely related to TaEXPB11wy except for the fact that the gene is truncated, containing only one exon (97 amino acids or 35.4% of the full-length TaEXPB11 gene). The coding regions of all the genes were found to have high sequence similarity (>80% at the nucleotide level) to a full-length wheat cDNA from a recently published database of 11,902 full-length wheat cDNA sequences (KOMUGI; http://www.shigen.nig.ac.jp/dnadb/index.jsp).
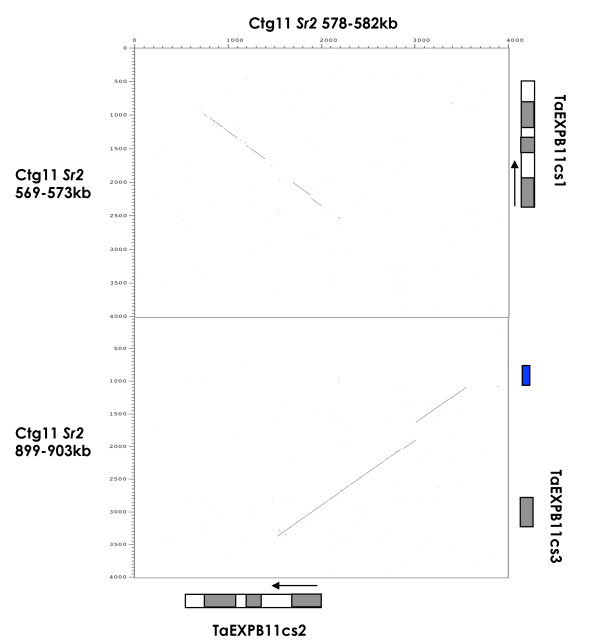
Comparing the nucleotide sequence to TaEXPB11wy identified the intron-exon structure of TaEXPB11cs2 (Figure 2). TaEXPB11cs1 and TaEXPB11cs2 (compared in Figure 3) both have three exons with intron 1 (348 bp in TaEXPB11cs2) being more than twice the size of intron 2 (122 bp), while the truncated gene TaEXPB11cs3 does not contain any introns. TaEXPB11cs1 and has a 42 bp insertion at position 90-132, within the first exon of the gene when compared to TaEXPB11cs2. This insertion in TaEXPB11cs1 contains a stop codon which suggests that it is probably a pseudo-gene.
Figure 3.
Dot-matrix plot of 4 kb of the genomic sequence surrounding TaEXPB11cs2 (horizontal axis) against 4 kb of the two other beta-expansin genes (TaEXPB11cs1 and TaEXPB11c3) found within the wheat cv. Chinese Spring assembled ctg11 genome sequence. The exon regions of each gene have been annotated. The blue box indicates a small 269 bp CACTA DNA transposon located near the TaEXPB11cs3 gene fragment.
The intron 1 sequences from TaEXPB11cs1 and TaEXPB11cs2 were compared to identify whether or not they represented recent duplications. Both genes sequences have a 92% sequence similarity over 1,514 bp. Outside the coding sequence of the genes, 28 bp of sequence 5' from the start codon and 26 bp 3' from the end codon, are conserved between TaEXPB11cs1 and TaEXPB11cs2. The comparison in Figure 3 between TaEXPB11cs2 and TaEXPB11cs3 demonstrates that the first intron is found to be well conserved between both sequences, as well as 880 bp of genomic sequence 5' to the start of the first exon. The beta-expansin genes amplified from the genomic DNA of a number of varieties of wheat (see later), showed that the respective TaEXPB11cs2-type genes were readily distinguished from TaEXPB11cs1 based on sequence divergence in intron 1. Intron 2 was not as diagnostic for distinguishing the TaEXPB11cs1 and TaEXPB11cs2 gene categories. Exon 3 in TaEXPB11cs1 showed a characteristic 35 bp insertion that was not present in any of the other TaEXP11cs type genes.
The ages of both TaEXPB11cs1 and TaEXPB11cs3 (proposed duplicates of TaEXPB11cs2) were estimated using methods used to date LTR retrotransposons [29]. The nucleotide sequence used for comparison between TaEXPB11cs2 and the two proposed gene duplications was from the start codon of each gene or gene fragment (in the case of TaEXPB11cs3), covering all of exon 1 and part of the intron 1 sequence (452 bp in total). The TaEXPB11cs1 duplication was identified to have duplicated 5.69 million years ago (MYA) with a standard deviation of 0.77 MYA, while the TaEXPB11cs3 duplication was much younger at only 1.59 MYA (standard deviation of 0.38 MYA.
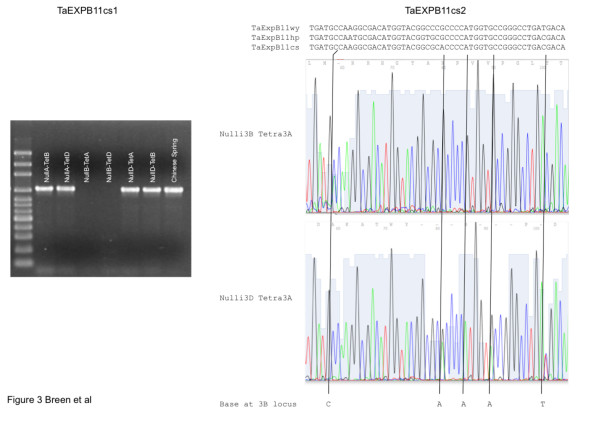
Validation of sequence structure for the expansin region assembled in Figure 1, in genomic DNA was carried out for TaEXPB11cs1 and TaEXPB11cs3 because their structure was unusual and it was important to ensure that changes had not occurred during the BAC cloning process. For TaEXPB11cs3, primers were designed to amplify DNA fragments from the borders of the regions that had a high similarity to TaEXPB11cs2 and these were predicted to generate fragments 686 bp and 856 bp long. The respective fragments generated had approximate sizes (estimated by agarose gel electrophoresis, data not shown) of ca 800 bp, in reasonable agreement with the expected sizes. The unusual structure of TaEXPB11cs1 (insert in exon 1 and a small insert in exon3) was shown to exist in genomic DNA by amplifying a DNA fragment using primers from within the respective insertions and which were predicted to generate a 1027 bp fragment. The results (Figure 4) indicated that the fragment obtained was slightly larger than 1000 bp using agarose gel electrophoresis, in agreement with the expected size. The analysis of Nulli-Tetra stocks of wheat showed that TaEXPB11cs1 was present only on chromosome 3B since the PCR product was missing when chromosome 3B was missing from the wheat line analysed (Nulli3B-Tetra3A and Nulli3B-Tetra3D lines; Figure 4).
Figure 4.
Nulli-Tetra lines for the group 3 chromosomes in Chinese Spring genetic stock lines analysed for occurrence of TaEXPB11cs1 (left panel) and TaEXPB11cs2 (right panel). A (left panel). For the chromosome assignment of TaEXPB11cs1, PCR products assaying exon 1 with one of the primers located in the insertion that characterizes TaEXPB11cs1 (see Material and Methods) were analysed on 2% agarose gels and stained with SYBR Green. The molecular markers are a 100 bp ladder (Axygen) and the product size of just over 1000 bp was as expected from the genome sequence (1027 bp). B (right panel). For the chromosome assignment of TaEXPB11cs2 (right panel), SNPs in exon 1 that differentiated the genes on chromosome 3A, 3B and 3D were assayed by direct sequencing of PCR products from primers that amplified a common section of this exon. All possible Nulli-Tetra combinations were assayed and the sequences compared to the respective sequence from the TaEXPB11wy, TaEXPB11hp and TaEXPB11cs2 genes (top of figure) in order to assign SNPs to particular chromosomes in the Nulli-Tetra combinations.
The transcribed gene, TaEXPB11cs2, was shown to be present on chromosomes 3A, 3B and 3D using the same Nulli-Tetra stocks illustrated in Figure 4, generated PCR products of the same size from the three chromosomes (data not shown). Several SNP differences between TaEXPB11cs2 on chromosomes 3A, 3B and 3D were detected by sequencing a genome PCR product directly (see for example Figure 4). Sequencing of the PCR products from the respective Nulli-Tetra lines allowed the assignment of the base pair differences to a chromosome. In Figure 4, an example is presented for exon 1 where several base positions show a mixed base-call in the lower panel (direct sequencing of genome DNA PCR product) where chromosome 3B is present (example shown is a genetic stock where only chromosome 3D is missing). In the genetic stocks where 3B is missing (Nulli 3B) the mixed base-call is resolved indicating that the base which is removed in the top panel is located on the chromosome 3B site for expansin EXBP11. Although only two examples are shown in Figure 4, all possible Nulli-Tetra combinations for 3A, 3B and 3D were analysed. Based on SNP analyses, the 3B gene was more similar to the published TaEXB11 than the genes on 3A and 3D but the relationship was not unambiguous because of the background of SNPs expected between wheat cv. Chinese Spring (source of the genome sequence) and cv. Wyuna (source of the published TaEXPB11 cDNA).
Comparative sequence analysis in the beta-expansin gene sequence from selected wheat species
The beta-expansin gene PCR products from wheat cv. Westonia, cv. Hope and cv. Wyuna (published cDNA sequence; AJ890019), and a T. spelta line were compared to TaEXPB11cs2 in Figure 5. The cv Westonia and T. spelta survey sequences were recovered from genomic DNA using primers positioned just inside the first exon and 60 bp before the end of the gene in order to amplify exon and intron sequences. The resulting sequences amplified from the DNA samples was ~1,000 bp in length and were specific as judged from direct sequencing of the PCR product. In the genomic BAC sequencing of wheat cv. Hope (R. Mago et al. 2010 in preparation), an ORF from the sequence was identified and named TaEXPB11hp as it contained the three predicted exons and was shown to have high sequence similarity to TaEXPB11wy as well as TaEXPB11cs2. Exon 3 of this TaEXPB11hp gene was truncated due to a point mutation causing a premature end to the coding sequence. Overall the nucleotide sequence similarity of TaEXPB11hp and TaEXPB11cs2 was 90%. The wheat cv. Westonia beta-expansin (TaEXPB11ws) and T. spelta (TaEXPB11sp) accessions showed 96.5% sequence similarity.
Figure 5.
Pair-wise sequence comparison of the TaEXPB11 genomic sequences of wheat cultivar Chinese Spring and an example of the 'survey' sequences (spelt-red) carried out in this study on different wheat cultivars and spelt species. The three exons of TaEXPB11cs2 are indicated below the figure (pale blue boxes, exon 1, 2 and 3 from left to right).
Transcription of the TaEXPB11cs genes
Assessing the data sets available from the NCBI Unigene EST profile (Ta.31031) indicated that two TaEXPB11 ESTs were present in libraries comprising 960,174 EST clusters. The ESTs were identified as being expressed from seed (6 transcripts per million or TPM) and flower (15 TPM). No evidence could be found for the expression of TaEXPB11cs1, using the insertion sequence in exon 1 that is unique to this gene, as a probe.
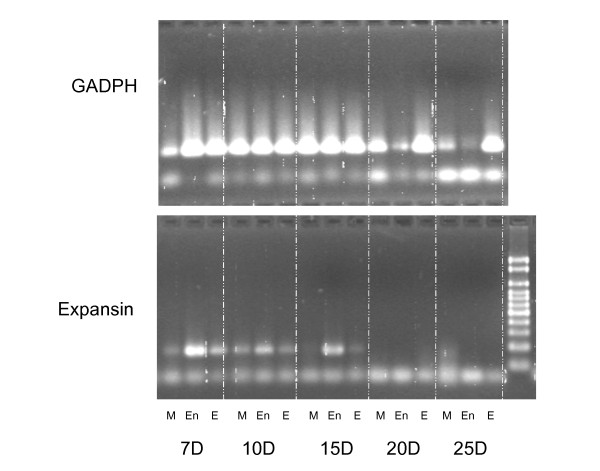
The endosperm expression of TaEXPB11cs2 was confirmed by RT-PCR (Figure 6). The tissue analysed was from developing grain with endosperm and embryo tissues separated by hand. The pericarp tissue was retained as an example of maternal tissue. In a study of the time course of expression, using tissues collected 7, 10, 15, 20 and 25 days post anthesis, strong expression was found for the three tissues at all stages of development. Quantitative analyses of the RT-PCR data indicated that the relatively lower expression for the maternal tissue suggestive in Figure 6 was not significant (data not shown). No evidence for the transcription of TaEXPB11cs1 could be found in these RT-PCR experiments consistent with the analysis of available EST databases.
Figure 6.
Gel analysis show samples from the RT-PCR (2% agarose gel) of the internal control GADPH (A, top panel) and the first exon of TaEXPB11cs2 (B, lower panel). Material was analysed 7, 10, 15, 20 and 25 days post anthesis and included maternal tissue from the developing grain (M), endosperm (En) and embryonic (Em) tissues of wheat cv Cranbrook.
Protein domain characterisation
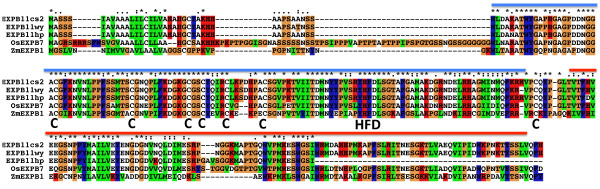
The well-characterised maize beta-expansin gene (EXPB1 in Zea mays) purified from maize pollen, and its crystal structure [30] was used to validate the two protein domains commonly identified in expansins, within the full-length TaEXPB11cs2 and TaEXPB11hp genes. Both genes contained conserved cysteine residues that create disulphide bonds between the domain folds as well as the characteristic 'HFD' motif that is the catalytic site of the distantly related family-45 endoglucanases (domain 1, GH45) [27] (Figure 7). Domain 1 is a lipoprotein A (RlpA)-like double-psi beta-barrel family domain (PF03330) commonly found at the N-terminus of pollen allergens. Domain 2 (Figure 7) is a Pollen_allerg_1 family (PF01357), grass type-2 pollen allergen domain originally characterised in timothy grass (P. pratense) [31]. The conserved residues in domain 2 outlined in [32] could also be identified although, when compared to the maize EXPB1 sequence, its amino acid sequence is shown to be more diverged (44% identity over 99 amino acids) compared to domain 1 (66% over 125 amino acids). As mentioned previously (and shown in Figure 7), domain 2 is truncated in TaEXPB11hp due to a stop codon terminating the coding sequence.
Figure 7.
Multiple sequence alignment of two full-length beta-expansin genes found in wheat genomic sequences of chromosome 3B (TaEXPB11cs1 and TaEXPB11cs2) compared to the TaEXPB11 cDNA, rice homolog OsEXPB7 and maize Zea M 1 using the ClustalX program [44]. The blue (domain 1) and red (domain 2) lines above the sequence indicate the different domains and the signature EXPB motifs (the 'HFD' and conserved cysteine residues) are indicated below the sequence.
Discussion
Expansins, a cell-wall loosening class of proteins, are a multigene family found in grass genomes that are considered to play important roles in growth and development in wheat [17]; EST mining has estimated that there exist at least 30 and 65 alpha- and beta-expansins, respectively [4] in wheat. The higher beta-expansin gene estimate compared to alpha-expansins within wheat is consistent with the estimates in other grasses such as maize [33]. Different expression characteristics suggest that the two expansin classes play different functional roles within the cell wall structure [5].
In the present study we identified and characterised three beta-expansin genes from wheat cv. Chinese Spring located in a ~357 kb region of chromosome 3BS. Only one of these genes was found to be the TaEXPB11 gene, coding for an IgE binding protein identified from the sera of patients that had undergone positive double-blind, placebo-controlled food challenge to wheat [19]. Sequence analysis of specific PCR products using TaEXPB11 primers from the wheat cultivars and a T. spelta wheat accession validated the identification of the wheat cv. Chinese Spring variant of TaEXPB11 cDNA [18], originally obtained from wheat cv. Wyuna [28]. It is proposed that sequence variants of TaEXPB11 be designated TaEXPB11wy, TaEXPB11cs, TaEXPB11ws, TaEXPB11hp. The high sequence similarity between the TaEXPB11 cDNA and TaEXPB11cs2 provided good evidence for assigning the two sequences to be alleles of the same gene. Even though a large family of expansin genes has been reported in wheat [4,17], the primers used to assay the transcripts (Figure 5) and gene locations did not provide any evidence of the presence of other copies of this particular gene elsewhere in the genome. The primers are evidently specific enough to assay only the TaEXPB11 gene category on homoeologous locations on chromosome 3A, 3B and 3D.
Utilizing SNPs identified in the present study, expansin was ruled out as a candidate gene for Sr2 resistance because re-examination of recombinant Chinese Spring-Hope lines studied by Kota et al [34] identified a recombination event between the Sr2 phenotype and TaEXPB11hp (R. Mago unpublished). We note that in the region examined, wheat cv Hope (the source of Sr2) is missing the mitochondrial DNA insert found in wheat cv Chinese Spring. This mitochondrial DNA segment encodes a gene nad7, which is a subunit of NADH dehydrogenase that is active in producing reactive oxygen species. Although this is not genetically linked to the Sr2 resistance gene, its close proximity may indicate it is part of a gene network controlling levels of active oxygen [35] (a generally accepted feature of resistance genes).
The TaEXPB11cs genes identified from the genome sequencing of the ctg11 contig on the small arm of wheat chromosome 3B could be characterized in detail. The TaEXPB11cs3 gene fragment contained only one coding exon and showed an extremely high sequence similarity to TaEXPB11cs2. This sequence similarity extended not only over its coding sequence but also the 880 bp 5' upstream from the start of the coding sequence of its only exon, as well as part of the sequence corresponding to the first intron sequence. The high sequence similarity over the coding and non-coding regions of the two sequences indicated that TaEXPB11cs3 is a very recent duplication of TaEXPB11cs2 with an estimated duplication age of 1.59 MYA. Direct genome analysis confirmed the existence of TaEXPB11cs3. The neighbouring CACTA DNA transposon 'Caspar' found within a base pair of the duplicated TaEXPB11cs3 fragment suggests the possibility that this genomic duplication was mediated by this TE or was the result of a TE-mediated recombination event [29]. While CACTA DNA transposons have not yet been shown to be involved in gene fragment duplication and the creation of chimeric genes, repetitive elements such as pack-MULEs found in multiple copies within the rice genome, have been shown to capture gene fragments and other genomic DNA to create chimeric ORFs [36].
The TaEXPB11cs1 gene appeared to be an older duplication of TaEXPB11cs2 with an estimated age of the duplication being 5.69 MYA. There was significant conserved nucleotide sequence either side of the gene coding sequences and a high level of sequence similarity between TaEXPB11cs1 and TaEXPB11cs2 intron2 sequences. The characteristic insertions within TaEXPB11cs1 (Figure 3) were validated by direct genome PCR and the pseudo-gene was shown to exist only on chromosome 3B using Nulli-Tetra mapping lines of wheat.
At the level of the SNP analysis it is clear that very similar genes exist as homoeologues on chromosomes 3A, 3B and 3D. Each of these homoeologous genes could be contributing to the mRNA assayed in Figure 6. The SNP analysis in Figure 4 is consistent with TaEXPB11cs2 representing the cDNA identified as TaEXPB11.
A well-characterised maize beta-expansin gene (EXPB1 in Zea mays) was used as a comparison with TaEXPB11cs2, to identify the particular motifs that are conserved between the protein domains. The three wheat beta-expansin genes, shown in Figure 7, contained the conserved cysteine residues that form disulphide bonds between the domain folds and the 'HFD' motif that is common in the catalytic site of the distantly related family-45 endoglucanases (GH45) [30]. They also contained many conserved residues in domain 2 outlined in [31,32], but when compared to the maize EXPB1 sequence, this allergen domain amino acids sequence is shown to be more diverged (44% identity over 99 amino acids) compared to domain 1 (66% identity over 125 amino acids), (Figure 7). Similar folding patterns were identified in TaEXPB11hp except that the amino acid sequence in the pollen allergen domain was largely missing.
Conclusions
Sequence analysis and annotation of 357 kb of chromosome 3B genomic sequence identified three beta-expansin genes, one of which was identified to be TaEXPB11, originally from a cDNA identified from wheat cv. Wyuna. Through the analysis of intron sequences of the three wheat cv Chinese Spring genes, we propose that two of these beta-expansin genes are duplications of the TaEXPB11 gene. Comparative sequence analysis with two other wheat cultivars (Westonia and Hope) and a T. spelta accession validated the identification of the wheat cv. Chinese Spring variant of TaEXPB11. EST and RT-PCR experiments confirmed the expression in maternal and grain tissues. The variation established in this gene suggests it is a clearly identifiable member of a gene family and reflects the dynamic features of the wheat genome as it adapted to a range of different environments and uses.
Methods
Wheat BAC sequencing
The sequencing of 20 T. aestivum cv. Chinese Spring BACs from ctg11 (Sr2 locus, http://urgi.versailles.inra.fr/cgi-bin/gbrowse/wheat_FPC_pub/) was carried out using a BAC-by-BAC shotgun method at 6× to 10× sequencing coverage (Wicker et al. 2010 in preparation) as well as additional 454 sequencing. The genome sequencing of wheat cv. Hope over the syntenic Sr2 region was carried out using the same BAC-by-BAC approach with sequencing carried out at 10× coverage from chromosome 3B and cultivar Hope-specific BAC library (R. Mago et al. 2010 in preparation).
E. coli-DNA-free BAC DNA were extracted with Qiagen Large-Construct Kit (QIAGEN, Cat. No. 20021) and mechanically sheared with HydroShear as recommended by ABI applied biosystems https://products.appliedbiosystems.com/ab/en/US/adirect/ab?cmd=catNavigate2&catID=604432, generating a concentrated smear ~3-5 kb in length. The sheared fragments were blunt ended with mung bean nuclease and dephosphorylated with Shrimp Alkaline Phosphatase (SAP). The short fragments were then tailed with A by PCR using standard procedures. Fragments ranging from 3-5 kb in size were isolated and ligated into a pCR4-TOPO vector and transformed into TOP10 electrocompetent cells (Invitrogen, Cat. No. K4580-01). The clones were sequenced from both directions with T3 and T7 primers using BigDye3.1 termination chemistry and run on an ABI Prism 3730 XL capillary sequencer (Applied Biosystems, Foster City, Calif., USA). Base calling, quality assessment and sequence assembly were carried out using the PHRED/PHRAP [37]. Gaps were filled by designing PCR primers located on the nearest random clone to the sequence gap. Sequencing was then performed using primer walking with additional dGTP mix and DMSO in the sequencing reaction system.
Sequence Analysis and Annotation
Repetitive DNA analysis was carried out using RepeatMasker (Smit et al. 1996-2004, http://www.repeatmasker.org) and local alignment searches using BLAST [38] against the Triticeae repetitive element (TREP) database http://wheat.pw.usda.gov/ITMI/Repeats/, gene models were identified by the use of FGENESH http://www.softberry.com/, GENSCAN [39] and GlimmerHMM [40]. Sequence homology searches were carried out using BLAST and protein domains were identified by searching the Pfam protein family database [41] as well as the conserved domains database at NCBI [42]. InterProScan [43] was run against the InterPro protein domain database [44], which also includes a signal peptide and Trans-membrane search. Sequence comparisons were carried out using DOTTER [45] and multiple sequence comparisons were carried out using the CLUSTALX [46]. Graphical display of the sequence map was produced using WICKERsoft (T Wicker, pers. comm.).
Dating of Chinese Spring gene duplications [29] were carried out by comparing two sequences using WATER (EMBOSS; http://www.ebi.ac.uk/Tools/emboss/), using a gap creation penalty of 30 and a gap extension penalty of 0.1 parameters. WICKERsoft scripts (T. Wicker, pers. comm.) were used to identify transversions (Tv) and transitions (Ti) between the two sequences, which were then used with a base-pair substitution rate of 1.3 × 10-8 [47] to identify the age of the two sequences.
Plant material and analysis
DNA from the standard Nulti-Tetra 'Chinese Spring' wheat stocks [48] (kindly provided by J Raupp, Wheat Genetics Resource Centre, Kansas State University) were used for assigning homoeologous locations (3A, 3B or 3D) for expansin genes. The DNA from these genetic stocks was amplified by primers designed from the genome sequence. Deletion lines and all other wheat varieties were kindly provided by Ms F Drake-Brockman (Department of Agriculture and Food, Western Australia). The Triticum spelta line 2255 was originally obtained from the Australian Winter Cereals Collection (Tamworth).
For RT PCR, wheat lines were grown in a glasshouse and heads tagged at anthesis. Developing grain was collected at 7, 10 and 15 days post-anthesis and three tissue types collected. After removing the embryo (one tissue), the endosperm was squeezed out (second tissue) and the remaining maternal tissue formed the third tissue that was collected. Tissue samples were frozen on dry ice and total RNA was extracted from approximately 100 mg of frozen seed tissue using the TRIzol reagent (Invitrogen). The extracted RNA was quantified using a Nanodrop ND-1000 Spectrophotometer before being used for cDNA synthesis. The cDNA was synthesised from 1 ug of total RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Changes in the expression of expansin in seed tissue were examined using real time RT-PCR. The housekeeping gene GAPDH was used as an internal standard in the RT-PCR. Each sample tested was completed in triplicate and mRNA expression levels were quantified using a Corbett RG3000 (following manufacturer instructions, using the delta-CT procedure). PCR amplification was performed in a 20 μl reaction volume containing 10 μl of Power 2 × Power SYBR Green PCR Master Mix, 1 μl of each primer (10 μM) and 1 μl of cDNA. Cycling conditions 95°C for 10 min followed by 40 cycles of 95°C for 10 sec and 57°C for 1 min. The primer sequences are listed in Table 2.
Table 2.
RT-PCR primers used for expression analysis of TaEXPB11cs2-domain 2 and a GAPDH control
| Exp Dom2 - For | GAGTCGTGGGGTTCCATCT |
| Exp Dom2 - Rev | AACTGGACGAGGGAGCTGT |
| Exp Front - For | GATCCTCTGCATCCTCGTC |
| Exp Front - Rev | GGGTGGTAAGTTGACGTTCT |
| Exp InDel - For | CACCAAAAAGCCTCCCTAC |
| Exp InDel - Rev | AGGTCATGGCAGAGAAGG |
| GAPDH - For | CGAAGCCAGCAACCTATGAT |
| GAPDH - Rev | CAAAGTGGTCGTTCAGAGCA |
Protein modelling
SignalP [49] was used to identify signal peptide sequences. Protein models of the expansin genes were created by comparing the amino acid sequence to a closely related expansin sequence using MODELLER [50] and viewing the protein databank file (PDB) output using iMol http://www.pirx.com/iMol/.
Authors' contributions
All authors read and approved the final manuscript.
JB, TW, MB, DD, RA: assembly of the genome sequence and annotation of genes and TEs
RM: identification of BACs from chromosome 3B
JJ, XK, JZ: sequencing of BAC clones
DL, WM: mapping of ISBPs to wheat genetic map and RT PCRs
FB: interpretation of data related to the different wheat lines studied
Contributor Information
James Breen, Email: jbreen@ccg.murdoch.edu.au.
Dora Li, Email: D.Li@murdoch.edu.au.
David S Dunn, Email: d.dunn@murdoch.edu.au.
Ferenc Békés, Email: Ferenc.Bekes@csiro.au.
Xiuying Kong, Email: xykong@mail.caas.net.cn.
Juncheng Zhang, Email: jczhang2008@163.com.
Jizeng Jia, Email: jzjia@mail.caas.net.cn.
Thomas Wicker, Email: wicker@botinst.unizh.ch.
Rohit Mago, Email: Rohit.Mago@csiro.au.
Wujun Ma, Email: W.Ma@murdoch.edu.au.
Matthew Bellgard, Email: mbellgard@ccg.murdoch.edu.au.
Rudi Appels, Email: rappels@ccg.murdoch.edu.au.
Acknowledgements
The authors are grateful to Catherine Feuillet, Frederic Choulet and Etienne Paux (INRA, France) for providing the BAC clones from the Minimal Tiling Path of ctg11 in the chromosome 3B physical map of hexaploid wheat. The INRA colleagues also provided Perl scripts for identifying ISBP markers. Thanks to Yasunari Ogihara (Kihara Institute for Biological Research and Graduate School of Integrated Science, Yokohama City University, Japan) for advanced access to full-length cDNAs for wheat and Jun-Hong Ma (SABC, Murdoch University, Australia) for laboratory work. Thanks also to Wolfgang Spielmeyer (CSIRO, Canberra, Australia) and Hana Šimková (Institute of Experimental Botany, Czech Republic) for the chromosome 3B Hope BAC library. Finally, thanks to Paula Moolhuijzen and all technical staff at the CCG for Bioinformatics support. Molecular Plant Breeding Co-operative Research Centre (MPBCRC) and Murdoch University generously funded this work. The collaborations that formed the basis of the research were made possible by the International Wheat Genome Sequencing Consortium (IWGSC).
References
- Gupta PK, Mir RR, Mohan A, Kumar J. Wheat genomics: present status and future prospects. International journal of plant genomics. 2008. p. 896451. [DOI] [PMC free article] [PubMed]
- Stein N. Triticeae genomics: advances in sequence analysis of large genome cereal crops. Chromosome Res. 2007;15:21–31. doi: 10.1007/s10577-006-1107-9. [DOI] [PubMed] [Google Scholar]
- Paux E, Sourdille P, Salse J, Saintenac C, Choulet F, Leroy P, Korol A, Michalak M, Kianian S, Spielmeyer W. A physical map of the 1-gigabase bread wheat chromosome 3B. Science. 2008;322:101–104. doi: 10.1126/science.1161847. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu D, Zhang H, Gao H, Guo X, Wang D, Zhang X, Zhang A. The alpha- and beta-expansin and xyloglucan endotransglucosylase/hydrolase gene families of wheat: molecular cloning, gene expression, and EST data mining. Genomics. 2007;90:516–529. doi: 10.1016/j.ygeno.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Enzymes and other agents that enhance cell wall extensibility. Annual review of plant biology. 1999;50:391–417. doi: 10.1146/annurev.arplant.50.1.391. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- Huang J, Takano T, Akita S. Expression of alpha-expansin genes in young seedlings of rice (Oryza sativa L.) Planta. 2000;211:467–473. doi: 10.1007/s004250000311. [DOI] [PubMed] [Google Scholar]
- Darley CP, Forrester AM, McQueen-Mason SJ. The molecular basis of plant cell wall extension. Plant Mol Biol. 2001;47:179–195. doi: 10.1023/A:1010687600670. [DOI] [PubMed] [Google Scholar]
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S. Activation of the Indole-3-Acetic Acid-Amido Synthetase GH3-8 Suppresses Expansin Expression and Promotes Salicylate- and Jasmonate-Independent Basal Immunity in Rice. Plant Cell. 2008;20(1):228–40. doi: 10.1105/tpc.107.055657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ. Two endogenous proteins that induce cell wall extension in plants. Plant Cell. 1992;4:1425–1433. doi: 10.1105/tpc.4.11.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpat AB, Waugh M, Sullivan JP, Gonzales M, Frisch D, Main D, Wood T, Leslie A, Wing RA, Wilkins TA. Functional genomics of cell elongation in developing cotton fibers. Plant Mol Biol. 2004;54:911–929. doi: 10.1007/s11103-004-0392-y. [DOI] [PubMed] [Google Scholar]
- An C, Saha S, Jenkins JN, Scheffler BE, Wilkins TA, Stelly DM. Transcriptome profiling, sequence characterization, and SNP-based chromosomal assignment of the EXPANSIN genes in cotton. Mol Genet Genomics. 2007;278:539–553. doi: 10.1007/s00438-007-0270-9. [DOI] [PubMed] [Google Scholar]
- Vogler H, Caderas D, Mandel T, Kuhlemeier C. Domains of expansin gene expression define growth regions in the shoot apex of tomato. Plant Mol Biol. 2003;53:267–272. doi: 10.1023/B:PLAN.0000006999.48516.be. [DOI] [PubMed] [Google Scholar]
- Shcherban TY, Shi J, Durachko DM, Guiltinan MJ, McQueen-Mason SJ, Shieh M, Cosgrove DJ. Molecular cloning and sequence analysis of expansins--a highly conserved, multigene family of proteins that mediate cell wall extension in plants. Proc Natl Acad Sci USA. 1995;92:9245–9249. doi: 10.1073/pnas.92.20.9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Choi D, Kende H. Expansins: ever-expanding numbers and functions. Curr Opin Plant Biol. 2001;4:527–532. doi: 10.1016/S1369-5266(00)00211-9. [DOI] [PubMed] [Google Scholar]
- Talbott LD, Ray PM. Molecular Size and Separability Features of Pea Cell Wall Polysaccharides: Implications for Models of Primary Wall Structure. Plant Physiol. 1992;98:357–368. doi: 10.1104/pp.98.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Ni Z, Zhang Y, Yao Y, Wu H, Sun Q. Isolation and characterization of 18 genes encoding alpha- and beta-expansins in wheat (Triticum aestivum L.) Mol Genet Genomics. 2005;274:548–556. doi: 10.1007/s00438-005-0029-0. [DOI] [PubMed] [Google Scholar]
- Jin Y, Tashpulatov AS, Katholnigg H, Heberle-Bors E, Touraev A. Isolation and characterisation of two wheat beta-expansin genes expressed during male gametophyte development. Protoplasma. 2006;228:13–19. doi: 10.1007/s00709-006-0176-0. [DOI] [PubMed] [Google Scholar]
- Weichel M, Vergoossen NJ, Bonomi S, Scibilia J, Ortolani C, Ballmer-Weber BK, Pastorello EA, Crameri R. Screening the allergenic repertoires of wheat and maize with sera from double-blind, placebo-controlled food challenge positive patients. Allergy. 2006;61:128–135. doi: 10.1111/j.1398-9995.2006.00999.x. [DOI] [PubMed] [Google Scholar]
- Kwasniewski M, Szarejko I. Molecular cloning and characterization of beta-expansin gene related to root hair formation in barley. Plant Physiol. 2006;141:1149–1158. doi: 10.1104/pp.106.078626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zheng B, Mao C, Qi X, Liu F, Wu P. Analysis of transcripts that are differentially expressed in three sectors of the rice root system under water deficit. Mol Genet Genomics. 2004;272:433–442. doi: 10.1007/s00438-004-1066-9. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Bedinger P, Durachko DM. Group I allergens of grass pollen as cell wall-loosening agents. Proc Natl Acad Sci USA. 1997;94:6559–6564. doi: 10.1073/pnas.94.12.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox B, Suphioglu C. Environmental and molecular biology of pollen allergens. Trends Plant Sci. 1996;1:156–164. doi: 10.1016/S1360-1385(96)80051-3. [DOI] [Google Scholar]
- Freidhoff LR, Ehrlich-Kautzky E, Grant JH. A study of the human immune response to Lolium perenne (rye) pollen and its components, Lol p I and Lol p II (rye I and rye II). I. Prevalence of reactivity to the allergens and correlations among skin test, IgE antibody, and IgG antibody data. J Allergy Clin Immunol. 1986;78:1190–1201. doi: 10.1016/0091-6749(86)90271-X. [DOI] [PubMed] [Google Scholar]
- De Marino S, Morelli MA, Fraternali F, Tamborini E, Musco G, Vrtala S, Dolecek C, Arosio P, Valenta R, Pastore A. An immunoglobulin-like fold in a major plant allergen: the solution structure of Phl p 2 from timothy grass pollen. Structure. 1999;7:943–952. doi: 10.1016/S0969-2126(99)80121-X. [DOI] [PubMed] [Google Scholar]
- Paux E, Legeai F, Guilhot N, Adam-Blondon AF, Alaux M, Salse J, Sourdille P, Leroy P, Feuillet C. Physical mapping in large genomes: accelerating anchoring of BAC contigs to genetic maps through in silico analysis. Funct Integr Genomics. 2007. [DOI] [PubMed]
- Cosgrove DJ, Li LC, Cho HT, Hoffmann-Benning S, Moore RC, Blecker D. The growing world of expansins. Plant Cell Physiol. 2002;43:1436–1444. doi: 10.1093/pcp/pcf180. [DOI] [PubMed] [Google Scholar]
- Clarke BC, Hobbs M, Skylas D, Appels R. Genes active in developing wheat endosperm. Funct Integr Genomics. 2000;1:44–55. doi: 10.1007/s101420000008. [DOI] [PubMed] [Google Scholar]
- Wicker T, Keller B. Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res. 2007;17:1072–1081. doi: 10.1101/gr.6214107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yennawar NH, Li LC, Dudzinski DM, Tabuchi A, Cosgrove DJ. Crystal structure and activities of EXPB1 (Zea m 1), a beta-expansin and group-1 pollen allergen from maize. Proc Natl Acad Sci USA. 2006;103:14664–14671. doi: 10.1073/pnas.0605979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffer S, Valenta R, Vrtala S, Susani M, van Ree R, Kraft D, Scheiner O, Duchêne M. Complementary DNA cloning of the major allergen Phl p I from timothy grass (Phleum pratense); recombinant Phl p I inhibits IgE binding to group I allergens from eight different grass species. J Allergy Clin Immunol. 1994;94:689–698. doi: 10.1016/0091-6749(94)90176-7. [DOI] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ. The expansin superfamily. Genome Biol. 2005;6:242. doi: 10.1186/gb-2005-6-12-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Meeley RB, Cosgrove DJ. Analysis and expression of the alpha-expansin and beta-expansin gene families in maize. Plant Physiol. 2001;126:222–232. doi: 10.1104/pp.126.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota R, Spielmeyer W, McIntosh RA, Lagudah ES. Fine genetic mapping fails to dissociate durable stem rust resistance gene Sr2 from pseudo-black chaff in common wheat (Triticum aestivum L.) Theor Appl Genet. 2006;112:492–499. doi: 10.1007/s00122-005-0151-8. [DOI] [PubMed] [Google Scholar]
- Dutilleul C, Garmier M, Noctor G, Mathieu C, Chétrit P, Foyer CH, de Paepe R. Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell. 2003;15(5):1212–26. doi: 10.1105/tpc.009464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167:GC1–10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Current protocols in bioinformatics/editoral board, Andreas D Baxevanis [et al] 2002;Chapter 2(Unit 2.3) doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA. 2004;101:12404–12410. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourdille P, Singh S, Cadalen T, Brown-Guedira GL, Gay G, Qi L, Gill BS, Dufour P, Murigneux A, Bernard M. Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.) Funct Integr Genomics. 2004;4:12–25. doi: 10.1007/s10142-004-0106-1. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]