Abstract
Talin is a large flexible rod-shaped protein that activates the integrin family of cell adhesion molecules and couples them to cytoskeletal actin. Its rod region consists of a series of helical bundles. Here we show that residues 1815–1973 form a 5-helix bundle, with a topology unique to talin which is optimally suited for formation of a long rod such as talin. This is much more stable than the 4-helix (1843–1973) domain described earlier and as a result its vinculin binding sequence is inaccessible to vinculin at room temperature, with implications for the overall mechanism of the talin-vinculin interaction.
Structured summary
MINT-7722300, MINT-7760951: Talin-1 (uniprotkb:P26039) and Vinculin (uniprotkb:P12003) bind (MI:0407) by molecular sieving (MI:0071)
Abbreviations: HSQC, heteronuclear single quantum coherence; SCOP, structural classification of proteins; VBS, vinculin-binding site
Keywords: Talin, Vinculin, NMR, Domain structure, Helical bundle
1. Introduction
Talin is a large cytoskeletal protein (2541 amino acid residues) that activates the integrin family of cell adhesion molecules and couples them to cytoskeletal actin [1]. It is composed of an N-terminal globular head (∼50 kDa) that interacts with the cytoplasmic tails of β-integrins [2], linked to an extended flexible rod (∼220 kDa) comprising 62 helices organized into a series of helical bundles arranged like beads on a string [3] (Fig. 1A). The rod contains a second integrin binding site [4], numerous putative binding sites for the cytoskeletal protein vinculin [3], at least two actin binding sites [5,6], and a C-terminal helix required for assembly of talin dimers [5,7]. Talin exists in both an extended and a compact auto-inhibited form, and a domain in the talin rod (residues 1655–1822) interacts with the talin head rendering it unable to bind integrin tails [8].
Fig. 1.
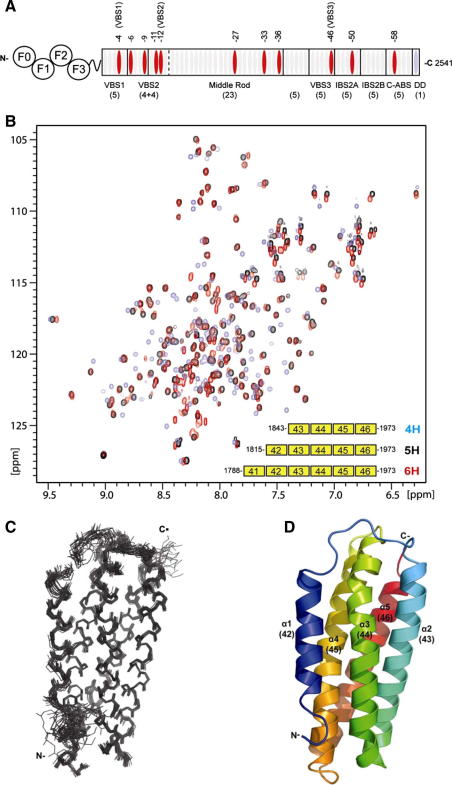
Characterisation of the talin polypeptides containing VBS3. (A) Schematic diagram of the talin molecule. The rod contains 62 predicted α-helices (ovals); the ∼11 vinculin-binding sites (VBS) are shown in red. (B) Superimposition of the 2D [1H, 15N]-heteronuclear single quantum coherence spectra of talin 1843–1973 (blue), 1815–1973 (black) and 1788–1973 (red). (Inset: Schematic of the constructs tested; the numbering corresponds to the helix number within the whole talin rod.) (C) Superimposition of the 20 lowest energy structures of talin 1815–1973 consistent with the NMR data. Only the structured region, 1820–1973, is shown, not the disordered N-terminus. (D) Ribbon drawing of a representative low-energy structure showing the overall topology of the 5-helix bundle.
While it is clear that the talin rod consists of a series of helical bundles the definition of the boundaries between these domains has not been straightforward. The first structures of the talin rod revealed a 5-helix bundle packed against a 4-helix bundle (residues 482–789) [9], and two 4-helix bundles (residues 755–889 and 1843–1973) [10,11]. Among helical bundle domains, 4-helix bundles are among the most frequently observed, with an up-down-up-down topology such as that seen in talin being particularly common (structural classification of proteins (SCOP) ID 47161: http://scop.mrc-lmb.cam.ac.uk/scop/data/scop.b.b.dg.html; [12]). However, our subsequent work showed that the domains towards the C-terminal end of the rod, spanning residues 1974–2482, are in fact 5-helix bundles [4,5,8], and this led us to re-examine the boundaries of the preceding domain. Here we show that this domain is actually a 5-helix bundle (residues 1815–1973) rather than a 4-helix bundle, and that the extra helix dramatically enhances the stability of the domain and renders the vinculin-binding site contained therein cryptic.
2. Materials and methods
2.1. Expression of recombinant talin polypeptides
The regions encoding murine talin1 residues 1788–1973 (6h), 1815–1973 (5h) and 1843–1973 (4h) were synthesized by PCR using a mouse talin1 cDNA as template, and cloned into the expression vector pet-151TOPO (Invitrogen). Talin polypeptides were expressed in Escherichia coli BL21 STAR (DE3) cultured in M9 minimal media containing 15NH4Cl and/or 13C-glucose. His-tagged talin polypeptides were purified by nickel-affinity chromatography. The His-tag was removed by cleavage with AcTEV protease (Invitrogen), and the proteins were further purified by anion-exchange.
2.2. NMR spectroscopy
NMR spectra of all the proteins were obtained at 298 K using Bruker AVANCE DRX 600 or AVANCE DRX 800 spectrometers both equipped with CryoProbes, with 1 mM protein in 20 mM sodium phosphate pH 6.5, 50 mM NaCl, 2 mM DTT, 10% (v/v) 2H2O. Spectra were processed with TopSpin (Bruker) and analysed using ANALYSIS [13]. Resonance assignment was carried out as described [14]; assignments of 1815–1973 have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu) with the accession number 15625.
2.3. Structure calculations
These were carried out as described previously [8]; structural statistics are presented in Table 1. The set of 20 lowest energy structures has been submitted to the Protein Data Bank (www.rcsb.org), accession number 2kvp. Molecular models were generated using PYMOL [15].
Table 1.
Solution structure determination of talin 1815–1973.
| Restraints | |
| Unique/ambiguous NOEs | 4490/627 |
| Intraresidue | 1557/121 |
| Sequential | 1062/113 |
| Short range (1 < [i − j] < 5) | 1051/169 |
| Long range ([i − j] > 4) | 820/224 |
| φ/ψ dihedral anglesa | 232 |
| Energies (kcal mol−1)b | |
| Total | −6784.45 ± 58.83 |
| Van Der Waals | −1454.48 ± 13.97 |
| NOE | 36.01 ± 4.09 |
| RMS deviationsb | |
| NOEs (Å) (no violations > 0.5 Å) | 0.012 ± 0.004 |
| Dihedral restraints (°) (no violations > 5°) | 0.29 ± 0.03 |
| Bonds (Å) | 0.0032 ± 0.0001 |
| Angles (°) | 0.43 ± 0.01 |
| Impropers (°) | 1.21 ± 0.05 |
| Ramachandran map analysisc | |
| Allowed regions | 96.3% |
| Additional allowed regions | 3.6% |
| Generously allowed regions | 0.0% |
| Disallowed regions | 0.2% |
| Pairwise rms difference (Å)d | |
| Residues 2301–2476 | 0.47 (0.85) |
| Secondary structure | 0.35 (0.73) |
From chemical shifts using Talos.
Calculated in ARIA 1.2 for the 20 lowest energy structures refined in water.
Obtained using PROCHECK-NMR.
For backbone atoms; value for all heavy atoms in brackets.
2.4. Circular dichroism spectroscopy
Far-UV CD spectra were recorded, using a JASCO J-715 spectropolarimeter, over the wavelength range 200–250 nm (scan rate 50 nm min−1) in a quartz cell of 0.1 cm path length with ∼25 μM protein in 20 mM sodium phosphate, pH 6.5, 50 mM NaCl.
2.5. Vinculin binding
Analytical gel filtration chromatography using Superdex-75 (10/300) GL (Amersham Biosciences) was used to measure binding of talin polypeptides to the vinculin Vd1 domain. Polypeptides were incubated at various temperatures for 30 min prior to loading onto the column, which was pre-equilibrated and eluted with 20 mM Tris pH 8.0, 150 mM NaCl, 2 mM DTT at a flow rate of 0.8 mL/min at room temperature.
3. Results and discussion
3.1. Mapping the domain boundaries of the VBS3 region
Our earlier studies of talin residues 1843–1973, which contains a vinculin-binding site referred to as VBS3, showed that its [1H, 15N]-heteronuclear single quantum coherence (HSQC) spectrum had good dispersion with peak line widths consistent with a monomeric state (Fig. 1B) and the structure (PDB ID: 2B0H) showed a well folded 4-helix bundle domain [11]. However, in the light of our subsequent demonstration that the domains on either side (residues 1655–1822, 1974–2140, 2137–2294 and 2300–2482) are all 5-helix bundles [4,5,8], we sought to establish whether residues 1843–1973 could also be part of a larger 5-helix bundle. To test this we produced a series of constructs with different boundaries containing 4-helices (1843–1973), 5-helices (1815–1973) and 6-helices (1788–1973) (Fig. 1B). Each of these polypeptides expressed well and was soluble.
Addition of a further predicted helix at the N-terminus resulted in a polypeptide (residues 1815–1973) whose spectrum showed a similar pattern of chemical shifts to that of residues 1843–1973 but with better chemical shift dispersion and similar signal intensities for all the peaks, suggesting that the 5-helix construct has a more stable fold. Addition of a sixth helix (residues 1788–1973) did not affect the signals of the 5-helix bundle but introduced a cluster of sharp signals close to the middle of the [1H, 15N]-HSQC spectrum, suggesting that the additional residues were unfolded (Fig. 1B).
3.2. Structure of talin residues 1815–1973
The solution structure of the talin domain comprising residues 1815–1973 was calculated from 5117 NOE-based distance and 232 dihedral angle restraints. The structure consists of five anti-parallel amphipathic α-helices forming a bundle with up-down-up-down-up left-handed topology (Fig. 1C and D). Helices 2–5 bear a strong resemblance to the 4-helix bundle structure previously determined (PDB ID: 2B0H [11]), with an RMSD of 1.2 Å for the region 1843–1973 and the same topology (Fig. 2A–D). The 4-helix up-down, left-handed twist bundle is a common fold (SCOP 47161), and it forms the core of this and other domains in the talin rod. However in talin, uniquely, this fold is augmented by an extra N-terminal helix (here residues 1825–1842) connected by a long (9 residue) loop that allows this helix to pack against helices 3 and 4 of the bundle. Helix-1 is relatively short compared with the other helices (15 residues compared to 27–28 residues) and sits half way down the bundle, packing against the core of the bundle by an extensive hydrophobic interface (Fig. 2C). Like the other helices, helix-1 is amphipathic and has its more hydrophobic surface buried in the bundle, packing against the hydrophobic groove between helices 3 and 4. The aromatic ring of Y1893 partially protrudes from the side of the 4-helix core of the bundle but the small side chain of A1834 in helix-1 allows close packing of the helices. This 5-helix left-handed crossover connectivity (Fig. 2A) has been observed in several other talin bundles (482–655, 1655–1822, 1974–2140 and 2137–2294; SCOP ID 109879) but, to date, has not been observed in other proteins. We show here in the case of residues 1815–1973 that the extra helix has very striking effects on the properties of the domain.
Fig. 2.
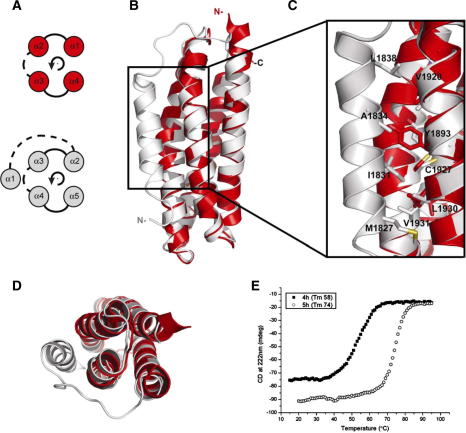
Structural comparisons between the 5-helix and the 4-helix bundles. (A) Topology diagram of the 4-helix up-and-down fold (red) and the 5-helix left-handed crossover connectivity fold (grey) common in the talin rod. Solid and dashed lines represent connecting loops on opposite ends of the helices. Helices 1–4 in the 4-helix are equivalent to helices 2–5 in the 5-helix. (B) Overlay of the structures of 1815–1973 (grey) and 1843–1973 (red) showing the similarity of the core domain and the location of the extra helix. (C) Region of the structure highlighted by the box in (A) showing the hydrophobic contacts made by helix-1 with helices 3 and 4. (D) Top down view of (B). (E) The thermal denaturation profiles for the talin rod polypeptides; profiles are shown for the 4-helix module (squares) and the 5-helix bundle (circles).
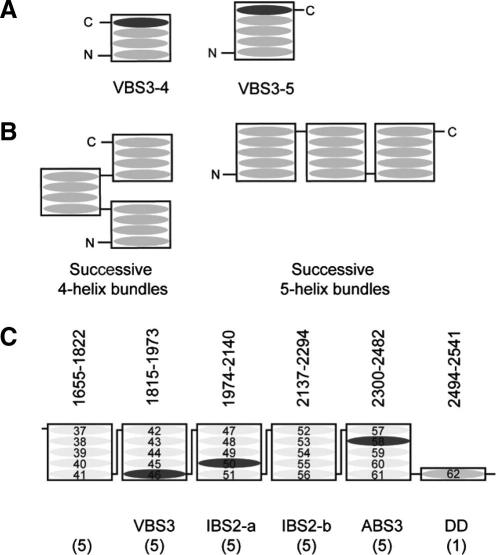
The 4-helix up-down-up-down fold results in the N- and C-termini of the domain being at the same end of the bundle (Fig. 3A), whereas with a fifth helix the N- and C-termini are at opposite ends of the bundle. This is a key difference in a long rod such as talin; it is possible to arrange 5-helix bundles in the rod with the component helices parallel to the rod axis, whereas this is not possible for 4-helix bundles (Fig. 3B and C).
Fig. 3.
(A) Schematic of the 4-helix and 5-helix bundles showing the locations of the N and C-termini. (B) Schematic of the rod structures resulting from successive 4-helix and 5-helix bundles. (C) The domain architecture of the C-terminal region of the talin rod.
This clear difference in the orientation of the helices relative to the long axis of the rod will result in significant differences in the mechanical properties of these two arrangements, which may be important for the activation of vinculin binding (see below).
3.3. Helix-1 stabilises the bundle
The 4-helix construct, residues 1843–1973, forms a stable bundle with a melting temperature of 58° (Fig. 2E). Whilst the addition of the extra helix has only minimal structural effect on the core of the bundle (Fig. 2B and D) it has a substantial effect on the stability of the bundle, the 5-helix bundle having a melting temperature of 74 °C (Fig. 2E).
3.4. The vinculin binding characteristics of the domain depend on helix-1
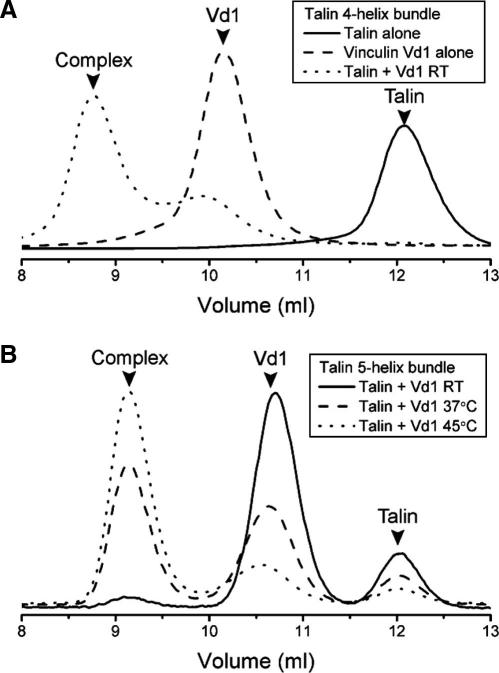
The four helix bundle containing VBS3 can bind the vinculin Vd1 domain at room temperature [11] (Fig. 4). However, since the addition of the extra helix has a large effect on the stability of the bundle we anticipated that this would affect vinculin binding, since this requires unfolding of the bundle [9–11,16,17], and indeed the 5-helix bundle requires incubation at ⩾37 °C for binding to be observed (Fig. 4). This result highlights the importance of using complete domains for vinculin binding studies, and suggests that the vinculin-binding site (VBS) in the 1815–1973 5-helix bundle is unlikely to be constitutively active in full length talin.
Fig. 4.
Vinculin Vd1 binding analysed by gel filtration. Vinculin Vd1 was incubated with (A) the talin 4-helix or (B) the 5-helix polypeptide at various temperatures, and complex formation was analysed on a gel filtration column at room temperature (RT). (A) Incubation of the 4-helix with Vd1 resulted in complex formation at room temperature (dotted line). (B) The 5-helix bundle did not bind vinculin Vd1 at room temperature (solid line), but pre-incubation of the proteins at 37 °C increased complex formation (broken line) and pre-incubation at 45 °C resulted in predominantly complexed proteins (dotted line).
The various vinculin binding sequences in talin are similar [3] and the corresponding peptides all bind tightly to vinculin in vitro. However, this and previous studies show that VBSs buried within stable 5-helix bundles are cryptic, whereas those in 4-helix bundles are constitutively active [9–11,16,18]. Experimental and theoretical studies show that vinculin binding to cryptic binding sites in talin can be activated by mechanical strain [19–22], and the present results indicate that this is likely to be essential for all the VBSs in the C-terminal part of the talin rod which are contained within 5-helix bundles (Fig. 3C). By contrast, two VBSs in the central and N-terminal part are likely to be constitutively active and may provide the initial attachment points for vinculin.
Acknowledgements
The work was supported by grants from the Wellcome Trust, the NIH Cell Migration Consortium (Grant U54 GM64346 from the National Institute of General Medical Sciences) and Cancer Research UK.
References
- 1.Critchley D.R. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Ann. Rev. Biophys. 2009;38:235–254. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- 2.Wegener K.L., Basran J., Bagshaw C.R., Campbell I.D., Roberts G.C.K., Critchley D.R., Barsukov I.L. Structural basis for the interaction between the cytoplasmic domain of the hyaluronate receptor layilin and the talin F3 subdomain. J. Mol. Biol. 2008;382:112–126. doi: 10.1016/j.jmb.2008.06.087. [DOI] [PubMed] [Google Scholar]
- 3.Gingras A.R., Ziegler W.H., Frank R., Barsukov I.L., Roberts G.C.K., Critchley D.R., Emsley J. Mapping and consensus sequence identification for multiple vinculin binding sites within the Talin rod. J. Biol. Chem. 2005;280:37217–37224. doi: 10.1074/jbc.M508060200. [DOI] [PubMed] [Google Scholar]
- 4.Gingras A.R., Ziegler W.H., Bobkov A.A., Joyce M.G., Fasci D., Himmel M., Rothemund S., Ritter A., Grossmann J.G., Patel B., Bate N., Goult B.T., Emsley J., Barsukov I.L., Roberts G.C.K., Liddington R.C., Ginsberg M.H., Critchley D.R. Structural determinants of integrin-binding to the talin rod. J. Biol. Chem. 2009;284:8866–8876. doi: 10.1074/jbc.M805937200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gingras A.R., Bate N., Goult B.T., Hazelwood L., Canestrelli I., Grossmann J.G., Liu H., Putz N.S.M., Roberts G.C.K., Volkmann N., Hanein D., Barsukov I.L., Critchley D.R. The structure of the C-terminal actin-binding domain of talin. EMBO J. 2008;27:458–469. doi: 10.1038/sj.emboj.7601965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hemmings L., Rees D.J.G., Ohanian V., Bolton S.J., Gilmore A.P., Patel B., Priddle H., Trevithick J.E., Hynes R.O., Critchley D.R. Talin contains three actin-binding sites each of which is adjacent to a vinculin-binding site. J. Cell Sci. 1996;109:2715–2726. doi: 10.1242/jcs.109.11.2715. [DOI] [PubMed] [Google Scholar]
- 7.Smith S.J., McCann R. A C-terminal dimerization motif is required for focal adhesion targeting of Talin 1 and the interaction of the Talin 1 I/LWEQ module with F-actin. Biochemistry. 2007;46:10886–10898. doi: 10.1021/bi700637a. [DOI] [PubMed] [Google Scholar]
- 8.Goult B.T., Bate N., Anthis N.J., Wegener K.L., Gingras A.R., Patel B., Barsukov I.L., Campbell I.D., Roberts G.C.K., Critchley D.R. The structure of an Interdomain complex that regulates talin activity. J. Biol. Chem. 2009;284:15097–15106. doi: 10.1074/jbc.M900078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papagrigoriou E., Gingras A.R., Barsukov I.L., Bate N., Fillingham I.J., Patel B., Frank R., Ziegler W.H., Roberts G.C.K., Critchley D.R., Emsley J. Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle. EMBO J. 2004;23:2942–2951. doi: 10.1038/sj.emboj.7600285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fillingham I., Gingras A.R., Papagrigoriou E., Patel B., Emsley J., Critchley D.R., Roberts G.C., Barsukov I.L. A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head. Structure (Camb) 2005;13:65–74. doi: 10.1016/j.str.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Gingras A.R., Vogel K.P., Steinhoff H.J., Ziegler W.H., Patel B., Emsley J., Critchley D.R., Roberts G.C.K., Barsukov I.L. Structural and dynamic characterization of a vinculin binding site in the talin rod. Biochemistry. 2006;45:1805–1817. doi: 10.1021/bi052136l. [DOI] [PubMed] [Google Scholar]
- 12.Murzin A.G., Brenner S.E., Hubbard T., Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 13.Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., Laue E.D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 14.Goult B.T., Gingras A.R., Bate N., Roberts G.C.K., Critchley D.R., Barsukov I.L. NMR assignment of the C-terminal actin-binding domain of talin. Biomol. NMR Assign. 2008;2:17–19. doi: 10.1007/s12104-007-9073-5. [DOI] [PubMed] [Google Scholar]
- 15.DeLano W.L. DeLano Scientific LLC; San Carlos, CA: 2004. The PyMOL Molecular Graphics System. [Google Scholar]
- 16.Patel B., Gingras A.R., Bobkov A.A., Fujimoto L.M., Zhang M., Liddington R.C., Mazzeo D., Emsley J., Roberts G.C.K., Barsukov I.L., Critchley D.R. The activity of the vinculin binding sites in talin is influenced by the stability of the helical bundles that make up the talin rod. J. Biol. Chem. 2006;281:7458–7467. doi: 10.1074/jbc.M508058200. [DOI] [PubMed] [Google Scholar]
- 17.Roberts G.C.K., Critchley D.R. Structural and biophysical properties of the integrin-associated cytoskeletal protein talin. Biophys. Rev. 2009;1:61–69. doi: 10.1007/s12551-009-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gingras, A.R., Patel, B., Goult, B.T., Bate, N., Kopp, P. M., Sun, N., Robson, R.M., Emsley, J., Barsukov, I.L., Roberts, G.C.K., and Critchley, D.R. (submitted for publication). The central region of talin has a unique fold and binds vinculin, actin and α-synemin. J. Biol. Chem. [DOI] [PMC free article] [PubMed]
- 19.del Rio A., Perez-Jimenez R., Liu R.C., Roca-Cusachs P., Fernandez J.M., Sheetz M.P. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hytonen V.P., Vogel V. How force might activate talin’s vinculin binding sites: SMD reveals a structural mechanism. PLOS Comput. Biol. 2008;4:e24. doi: 10.1371/journal.pcbi.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S.E., Chunsrivirot S., Kamm R.D., Mofrad M.R.K. Molecular dynamics study of talin-vinculin binding. Biophys. J. 2008;95:2027–2036. doi: 10.1529/biophysj.107.124487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S.E., Kamm R.D., Mofrad M.R.K. Force-induced activation of talin and its possible role in focal adhesion mechanotransduction. J. Biomech. 2007;40:2096–2106. doi: 10.1016/j.jbiomech.2007.04.006. [DOI] [PubMed] [Google Scholar]