Abstract
Continuous cell renewal in mouse epidermis is at the expense of a pool of pluripotent cells that lie in a well defined niche in the hair follicle known as the bulge. To identify mechanisms controlling hair follicle stem cell homeostasis, we developed a strategy to isolate adult bulge stem cells in mice and to define their transcriptional profile. We observed that a large number of transcripts are underexpressed in hair follicle stem cells when compared to non-stem cells. Importantly, the majority of these downregulated genes are involved in cell cycle. Using bioinformatics tools, we identified the E2F transcription factor family as a potential element involved in the regulation of these transcripts. To determine their functional role, we used engineered mice lacking Rb gene in epidermis, which showed increased expression of most E2F family members and increased E2F transcriptional activity. Experiments designed to analyze epidermal stem cell functionality (i.e.: hair regrowth and wound healing) imply a role of the Rb-E2F axis in the control of stem cell quiescence in epidermis.
Electronic supplementary material
The online version of this article (doi:10.1007/s12015-010-9139-0) contains supplementary material, which is available to authorized users.
Keywords: Epidermis, Hair follicle, Stem cells, CD34, E2F, Microarrays, Rb, Genomic profile
Introduction
Continually renewing tissues maintain their homeostasis at the expense of stem cells (SC) with the properties to self-renew and differentiate into the cell lineages that constitute their tissue of origin. Such characteristics are sustained by the niches where the SC reside, which provide specialized environments. SC functionality is critical for regenerating tissues damaged during injury [1]. In addition, the relevance of SC in pathology is highlighted by multiple observations pointing to them as a possible origin of aggressive metastatic tumors [2]. These particular properties, as well as their potential use in regenerative medicine, have led many groups to analyze and characterize different adult SC populations. In consequence, multiple approaches, including functional genomics, have been carried out to define specific biochemical and genetic determinants that define the actual SC attributes.
The control of stem cell properties in multiple tissues is linked to cell cycle, according to the cell quiescence demonstrated by most adult stem cells. The Rb family members (Rb1, Rbl1 and Rbl2) are negative regulators of cell cycle progression and have also been involved in the regulation of stem cell biology, including lineage specification of several types of stem cells [3]. The Rb family functions in cell cycle control are mostly mediated by their ability to interact with E2F transcription factors, while their modulation of stem cell lineage specification may involve the regulation of tissue-specific transcription factors (reviewed in (3)). The E2F family of transcription factors comprises eight members (E2F1-8) and two related proteins (DP1 and DP2), essential for E2Fs binding to DNA (reviewed in (4)). Different functions have been assigned to E2Fs, E2F1 to −3 are considered activators and E2F4 and −5 repressors of target genes, in an Rb family (or pocket protein family)-dependent manner. E2F6 to −8 lack the Rb family binding domain, being considered pocket protein-independent repressors [5]. Importantly, increased E2F activity due to inactivation of the pRb pathway has been found in most tumors and is associated to the increased proliferation in cancer cells [6].
The epidermis is the primary barrier that protects our bodies from the environment thus facing permanent wearing. To maintain its integrity this epithelium undergoes continuous cell renewal throughout the life of the individual at the expense of a small population of adult SC. In mouse epidermis, SC reside in a well established niche termed the bulge, a specialized portion of the hair follicle [7]. Hair follicles periodically undergo cycles of growth (anagen), regression (catagen) and rest (telogen) [8]. During the resting phase of the hair cycle (telogen), hair follicle SC (HFSC) are located at the bottom of the follicle, close to a specialized group of mesenchymal cells termed the dermal papilla (DP) and, at the anagen onset, DP cells activate HFSC forming a highly proliferative hair germ (HG). As the germ grows a proliferative compartment of transient amplifying cells (matrix cells) surround the DP at the base. The matrix cells terminally differentiate to produce the inner/outer root sheath and the hair shaft. When anagen ends, matrix cells apoptose and as a consequence the lower follicle degenerates and regresses (catagen), by this moment the DP is again located near the bulge and the sebaceous gland, resting until a new cycle starts. The source of cells that feeds these cycles comes from the bulge [9, 10]; however, bulge SC can also contribute to regenerate interfollicular epidermis after injury [9, 11] and sebaceous gland [12]. Therefore, HFSC are multipotent progenitors capable of differentiating in all the cell lineages of the epidermis.
To determine which genes and signaling pathways operate within the HFSC, several groups have performed transcriptional profiling on isolated bulge cells [13–16]. Isolation procedures are mostly based on the specific expression of SC markers and/or the slow-cycling properties of these cells. Initial microarray experiments were designed to characterize cells that divide infrequently in the adult epidermis of transgenic mice expressing a repressible GFP marked histone [13]; however, hair follicle stem cells may not be as quiescent as initially thought [17, 18]. Other approaches took advantage of the restricted expression of cytokeratin 15 (K15) to mouse and human HFSC [19, 20]. Again, due to the intracellular nature of K15, it was necessary to develop transgenic mice that express GFP under the control of the K15 promoter to isolate viable cell populations with promoter activity [14]. Direct isolation of HFSC circumventing the use of transgenic mice was possible using fluorescence-activated cells sorting (FACS) of CD34 positive cells. CD34 is a cell surface glycoprotein that has been shown to be distinctively expressed in the hair follicle bulge in murine epidermis [21, 22] where it may serve as an adhesion molecule, but its precise function is yet unknown. In mouse skin, CD34 expression is needed for tumor development [23], and CD34 positive populations isolated from epidermal tumors contained cells with tumor-initiating properties [24], supporting the notion that the HFSC population could represent an important target in the process of tumor development.
Gene expression studies in HFSC have led to the identification of genes and molecular pathways involved in SC maintenance, which in some cases have been validated through genetic approaches in transgenic mice [18, 25–27]. Similarly, the factors regulating HFSC gene expression are beginning to be elucidated and some transcription factors, identified in transcriptional profiling studies, have shown to be involved in the function and maintenance of this population, as it is the case of NFATc1 [25], Tcf3 [27, 28], Lhx2 [26] and Sox9 [29].
We have used magnetic-activated cell sorting (MACS), as a novel way to isolate HFSC based on their CD34 cell surface expression [30], to develop a CD34+ HFSC signature that is robust and contains more genes than in previously reported datasets. This has allowed us to undertake the study of the regulation of gene expression in HFSC, in addition to the classical gene expression analyses. Our data suggest that the Rb/E2F families could be important regulators of HFSC gene expression. In agreement, functional data using mice deficient for Rb1 gene in epidermis, suggest that pRb ablation, which led to increased E2F activity, results in the dysregulation of stem cell quiescence.
Materials and Methods
Mice and Histological Procedures
Mice expressing GFP from the cytokeratin 15 (Krt1-15) gene promoter (Krt1-15-EGFP) were obtained from Jackson Laboratory (Jax Mice, Bar Harbour, MN). Mice lacking Rb1 gene in epidermis, through tissue specific ablation using K14Cre, have been previously described [31]. All the animal experiments were approved by the Animal Ethical Committee (CEEA) and conducted in compliance with Centro de Investigaciones Energéticas, Medioambietales y Tecnológicas (CIEMAT) guidelines. For histological analysis, dorsal skin samples from wild type B6D2F1 mice were fixed in 70% ethanol and embedded in paraffin prior to sectioning. Sections were stained and processed as described [32]. Antibodies used were anti-K15 (Neomarkers, Lab Vision, Fremont, CA), anti-CD34 (clone RAM 34, eBioscience, San Diego, CA), anti cytokeratin 5 (Covance, Emeryville, CA) and anti-BrdU (Roche, Indianapolis, IN). FITC, TexasRed or peroxidase conjugated secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA). Control slides were obtained by replacing primary antibodies with PBS or preimmune sera (data not shown).
Isolation of Hair Follicle Stem Cells Using Magnetic Cell Sorting (MACS)
Keratinocyte isolation from mice back skin, immunofluorescent and magnetic labeling followed by MACS isolation of the stem population have been previously described [30]. Briefly, adult mice in telogen (resting) phase of the hair cycle were used. Dorsal hair was removed and skin was placed on trypsin overnight at 4°C. Keratinocytes were collected scraping the epidermis with a scalpel and cell viability was determined by trypan blue exclusion. We confirmed that in the isolation procedure contamination with dermal cells is negligible [30]. Cells were labeled (typically 1 μg antibody per 106 cells) with anti-mouse CD34 (eBioscience), anti-rat FITC (Jackson ImmunoResearch) and anti-FITC MicroBeads (Miltenyi Biotech, Auburn, CA). CD34-magnetic labeled (CD34+) and CD34 unlabelled fractions (CD34−) were separated using LS columns and LS column adapters (Miltenyi Biotec) following the manufacturer’s guidelines. Purity of the isolated populations was assessed by FACS and by direct visualization under the microscope (FITC fluorescence) and typically exceeded 90%.
Flow Cytometry
For analysis of CD34 and cytokeratin 15 expression we used transgenic mice that express EGFP under the control of the Krt1-15 promoter. Keratinocyte populations isolated from Krt1-15-EGFP mice were labeled with anti-CD34 (eBioscience), washed in FACS Buffer (PBS containing 5% fetal bovine serum), stained with anti-rat PE (Jackson ImmunoResearch) and assessed for GFP and PE fluorescence. Cell preparations from wild type, Rb F19/F19 and Rb F19/F19;K14Cre mice were used to analyze CD34 and Itgα6 surface expression. CD34 was stained as described and anti-Itga6 antibody directly conjugated to FITC (BD Pharmingen, Franklin Lakes, NJ). Cells were analyzed in an EPICS XL flow cytometer (Coulter Electronics, Hialeah, FL). Dead cells and debris were excluded from the analysis.
RNA Isolation From CD34+ Epidermal Cells and Microarray Analyses
Total RNA was prepared from CD34+ and CD34—keratinocytes obtained from pools of at least six mice per MACS isolation procedure [30]. A total of three isolation procedures were performed. RNA extracted (RNAEasy, Qiagen) from each fraction (CD34+ / CD34−) was assessed for its quality using Agilent 2100 Bioanalyzer (Agilent Technologies) and hybridized to two Affimetrix Mouse 430 2.0 arrays, to ensure reproducibility of the process. This resulted in a total of 12 chips (6 CD34+ / 6 CD34−). Microarray data are accessible at NCBI Gene Expression Omnibus, http:www.ncbi.nlm.nih.gov/geo/, accession number GSE19448. Microarray normalization was done using RMA (http://rmaexpress.bmbolstad.com) [33]. Differential expression analysis was done using Ttest (p-val < 0.01, fold change>2).
Transcription factor consensus binding motifs search in genes specifically deregulated in CD34+ versus CD34−cells was done using Gene Set Enrichment Analysis software (GSEA, http://www.broadinstitute.org/gsea/) [34], using a collection of 615 motif gene sets, including i) gene sets that contain genes that share a cis-regulatory motif that is conserved across the human, mouse, rat, and dog genomes, and ii) gene sets that contain genes that share a transcription factor binding site defined in the TRANSFAC (version 7.4, http://www.gene-regulation.com/) database.
MatInspector (http://www.genomatix.de/products/MatInspector/) [35] was used to identify putative transcription factor binding sites in the promoter region of specific HFSC genes.
Comparative Microarray Dataset Studies
In order to find similar expression patterns between our MACS-purified CD34+ HFSC cells and published studies, we performed Gene Set Enrichment Analysis using our HFSC signature as the gene set, and the full microarray datasets of the other analyses. Log2 expression raw data of microarray analyses of skin stem cells of either mouse or human species was obtained online (see Table 1).
Table 1.
Currently published datasets for the molecular profile of the hair follicle stem cells. aDataset accession number to Gene Expression Omnibus, http:www.ncbi.nlm.nih.gov/geo; N-LCM: navigated laser capture microdissection; LRCs: label-retaining cells
| Authors, year Accession | Platform Chip | Probe sets | Species | Selection marker / method | GSEA using MACS CD34+ HFSC signature | FDR q-value |
|---|---|---|---|---|---|---|
| C. Lorz et al. | Affymetrix | ∼ 45,000 | Mouse | CD34 / MACS | – | – |
| GSE19448a | Mouse430_2 | |||||
| V. Greco et al. 2009 | Affymetrix | ∼ 22,000 | Mouse | K14GFP-CD34 / | 452 | 0.0002 |
| GSE15185a | MOE430A_2 | FACS | ||||
| C.S. Trempus et al., 2007 | Agilent | ∼ 22,000 | Mouse | CD34 / FACS | 783 | <0.0001 |
| GSE7690a | ||||||
| M. Ohyama et al., 2006 | Affymetrix | ∼ 22,000 | Human | LRCs / N-LCM | 334 | <0.0001 |
| GSE3419a | HG_U133A | |||||
| R.J. Morris et al., 2004 | Affymetrix | ∼ 12,000 | Mouse | K15GFP / FACS | 495 | <0.0001 |
| GSE1096a | MG_U74Av | |||||
| T. Tumbar et al., 2004 | Affymetrix | ∼ 12,000 | Mouse | LRCs / FACS | 331 | <0.0001 |
| http://www.rockefeller.edu/labheads/fuchs/intro.php | MG_U74Av2 |
Label-Retaining Cell (LRC) Analysis
Ten-day old Rb F19/F19 and Rb F19/F19;K14Cre pups were injected with bromodeoxyuridine (BrdU) (20 μl of a 12,5 mg/ml dilution in NaCl 0,9%) every 12 h for a total of 4 injections. Skin sections were collected 75 days after the last injection and BrdU incorporation was measured as the percentage of hair follicles containing positive cells. Four different animals of Rb F19/F19 and Rb F19/F19;K14Cre were used to count at least 100 follicles.
Primary Keratinocytes Studies
Primary keratinocytes were obtained from newborn mice skin and cultured as previously described [36]. Cells pooled from 5 to 8 different mice of each genotype were used for luciferase experiments or real time RT-PCR. E2F activity was measured in primary mouse keratinocytes transfected using Superfect (Qiagen) with pSV-Renilla (0,1 μg Promega) and pE2F-Luc (2.4 μg, a generous gift of Dr. X. Lu) as previously described [31]. Luciferase activity, measured with the Dual Luciferase Reporter Assay system (Promega) was determined as the ratio of firefly to Renilla luciferase activity. Transfections were performed in triplicate, and the mean and standard error were calculated for each condition. Two independent transfection experiments were performed. Gene expression levels for each E2F family member was determined upon reverse transcription of 1 μg of total RNA using Transcriptor first strand cDNA synthesis kit (Roche, Basel, Switzerland). cDNA PCR amplification and analysis was done using Platinum® SYBR® Green qPCR SuperMix-UDG (Invitrogen, Carlsbad, CA) and Rotor Gene 3000 systems (Corbett Research, Cambridge UK). PCR primer sequences for E2F genes were previously reported [37]. The expression values were first normalized using the 18 S ribosomal RNA (Universal 18 S rRNA kit, Ambion) and subsequently normalized to Rb F19/F19 primary keratinocytes.
Wound Healing and Hair Regrowth
Two-month old Rb F19/F19 and Rb F19/F19;K14Cre mice (n = 4 each) were anesthetized, shaven and 4 mm skin punch (4 per mouse) were made on the back. Wound healing was monitored by external caliper measurements every day. Data (mean ± SE) are shown as the percentage of initial maximal wound diameter as a function of time after wounding. To monitor the histological process of wound healing, another cohort of Rb F19/F19 and Rb F19/F19;K14Cre mice (n = 8 each) was used. In this case animals were sacrificed at different time points after wounding and the wound excised and fixed for analysis of the histology, BrdU incorporation, keratin 5 expression and collagen remodeling using sirius red staining. Mice were injected i.p. with 100 μg/g of body weight BrdU (Sigma Aldrich, St. Louis, MO) 1 h before they were sacrificed to analyze BrdU incorporation on the healing fronts.
To induce hair regrowth, two-month old Rb F19/F19 and Rb F19/F19;K14Cre mice (n = 10 each) were shaven and depilatory cream was applied to the back skin. Animals were sacrificed at different time points after depilation and monitored for hair growth. Depilated skin was processed for histology and at least 5 sections (2 cm length) of each mouse were analyzed for the presence of anagen hairs.
Results
Magnetic-Activated Cell Sorting of CD34 Positive Hair Follicle Stem Cells
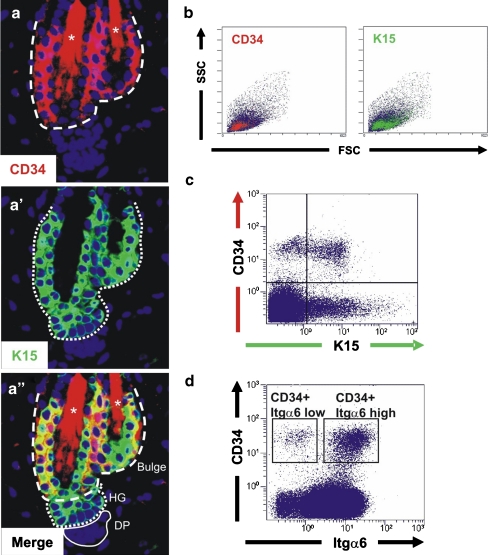
To date, a variety of molecular markers and strategies have been employed to isolate hair follicle stem cells; particularly, the finding that the cell surface marker CD34 is enriched in hair follicle bulge stem cells [21] has eased direct studies on live cells avoiding the use of transgenic mice. Immunofluorescence and flow cytometry analysis showed partial overlap between CD34 and K15 expression in resting hair follicles (Fig. 1a–c), where K15 is present not only in bulge SC but also in cells of the hair germ, a small bulge-derived cell cluster that contributes to the formation of the new hair but shows less proliferative potential in vitro [16, 38]. Detailed FACS analysis of this CD34 population revealed the existence of two subsets of CD34+ keratinocytes with differential expression of the cell surface integrin alpha 6 (Itgα6) (Fig. 1d). Itgα6 mediates attachment to the basal lamina, thus these two CD34+ subpopulations have been dubbed basal (Itgα6 high) and suprabasal bulge cells (Itgα6 low) [22]. Despite differences in Itgα6 surface expression both populations behave like true follicle stem cells: they display similar self-renewal capacity, proliferation potential and ability to terminally differentiate in vitro; and are fairly quiescent in vivo [22]. At present, CD34 is the best characterized HFSC marker.
Fig. 1.
CD34 is a marker of follicular stem cells. Immunofluorescent labeling for CD34 a and K15 (a´) HFSC markers in a telogen hair follicle. In the merged image (a´´), the region double labeled with CD34 and K15 (dashed line) corresponds to the bulge, where true hair follicle stem cells reside. The region positive only for K15 (dotted line) corresponds to the hair germ (HG). Dermal papilla (DP) thin continuous line; * autofluorescence from hair shaft. b & c) Flow cytometry dot plots of backskin keratinocytes preparations from telogen transgenic mice that express GFP under the control of the K15 promoter. b Forward and side scatter analysis (FSC and SSC) reveal that CD34 and K15 positive cell populations are partially overlapping. c Analysis of CD34 surface expression and K15-GFP epifluorescence reveals keratinocyte populations positive for both markers (upper right quadrant), only positive for K15-GFP (lower right quadrant), and a CD34+ population that shows no detectable activation of the K15 promoter (upper left quadrant). d CD34+ keratinocytes show different level of Itgα6 expression
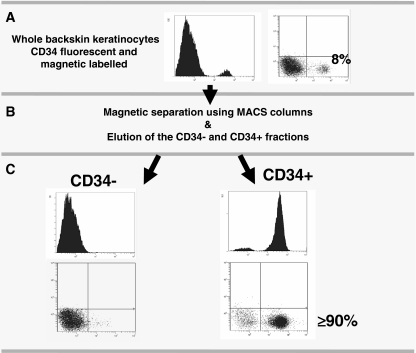
Based on the expression levels of the aforementioned markers, other groups have isolated follicle stem cells for transcriptional profiling using fluorescence-activated cell sorting (FACS) (Table 1). We decided to isolate CD34+ HFSC from non-genetically manipulated mice in the resting phase of the hair cycle using a magnetic-activated cell sorting (MACS) based method [30] to perform the gene expression analysis. This isolation procedure is efficient, fast and cell friendly, facilitating the recovery of high quality RNA. Purity of MACS isolated cells can be assessed by FACS analysis and is fairly similar to that obtained using FACS-sorting (Fig. 2). To our knowledge, this is the first time that MACS has been used to isolate CD34+ stem cells from the epidermis to perform genomic analyses.
Fig. 2.
Magnetic cell sorting (MACS) as a method for HFSC isolation. a Whole backskin keratinocytes preparations are fluorescent and magnetic labelled for the bulge surface marker CD34. In telogen mice the CD34+ population represents about 8% of the total. b Keratinocytes are passed through a column in the presence of a magnetic field and the CD34 negative fraction is collected; the column is then removed from the magnetic field and the CD34 positive fraction is eluted. Due to fluorescent label pre-and post-sort fractions can be assessed for CD34 enrichment by FACS and immunofluorescence. CD34+ population is represented by the peak at the right (not detectable in the CD34−fraction) in the FACS histogram plots and by the events that fall in the lower left quadrant of the FACS dot plots. Adapted from C. Lorz et al., Epidermal Cells: Methods and Protocols [30]
Analysis of the Hair Follicle Stem Cell signature
We performed comparative gene expression analyses on CD34+ HFSC versus CD34−cells and identified 2,297 differentially expressed probes with statistical significance p < 0.01 and a expression change (up or down) above 2-fold (Supplementary Table S1). Among them, there are many genes, such as Tcf3, Tcf4, Nfatc1, Lgr5 and Lhx2, that are well characterized as HFSC markers and are involved in the functionality of these cells (Fig. 3a). Interestingly, our HFSC signature contains more underexpressed (1,634 probe sets out of 2,297) than overexpressed genes, this feature is in agreement with previous studies [15, 39], and may indicate the status of a transcriptionally quiescent cell. Also, this HFSC signature contains more differentially expressed genes than previous datasets [13–16, 39], this is likely due to the combination of i) MACS cell isolation procedure that yields good-quality RNA, and ii) the use of the most comprehensive expression array available for our analysis.
Fig. 3.
Gene expression analysis of HFSC. a Heat map showing differential regulation of known hair follicle stem cell markers in CD34+ HFSC isolated using MACS. b Gene Ontology analysis [68, 69] reveals overrepresented Biological Processes within the differentially regulated genes in HFSC (overexpressed genes in red, uderexpressed genes in green). Note that cell cycle related genes are among the most frequently downregulated genes in HFSC, while the expression of cell communication/adhesion genes is enriched in HFSC
Currently existing HFSC gene signatures have been developed using different species (human, mouse), selection markers, platforms, expression arrays and statistical software. To compare our HFSC signature with others we applied Gene Set Enrichment Analysis (GSEA). This analysis searches whether genes present in our 2,297-probe HFSC signature are also specifically regulated in bulge stem cells from previously reported datasets, including human (Table 1). The results from the GSEA showed that genes in our HFSC signature are significantly present in stem cells of all datasets (FDR q-val <0.0002), thus validating our experimental approach to isolate bulge stem cells based on the expression of the CD34 marker. Selecting for those genes enriched in HFSC from all datasets, we obtained a group of 156 genes (69 up / 87 down) characteristic of murine stem cells, termed moHFSC signature, and a subgroup of 59 genes (25 up / 34 down) that are also common to human hair follicle stem cells (Supplementary Table S2), termed hu-moHFSC gene signature. This gene list could represent bona fide molecular markers of skin stem cells in mice and humans. Interestingly, many of the genes common to all datasets are involved in cell cycle regulation.
We performed functional analyses of the HFSC-signature genes, and we searched for significant enrichment in Gene Ontology Biological Processes (GOBP) terms and KEGG pathways which are relevant for HFSC identity (Fig. 3b, Tables 2 and S3). We found that the most significantly enriched GOBP terms and pathways contain genes involved in cell cycle (Fig. S1). In particular, there is an extensive underexpression of genes implicated in cell cycle progression in the HFSC population, consistent with the essentially non-proliferative status of these cells. The identification of p53 and apoptosis as important pathways in HFSC is also a novel finding. Other pathways identified in our analysis, such as Wnt and Hedgehog/Bmp, have been previously reported to be regulated in HFSC [40–42], and our data add new genes regulated within these pathways which functionality may be also relevant.
Table 2.
The KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis tool in DAVID [58, 59] was used to search for the most relevant pathways present in the 2,297-probe HFSC signature. The table shows genes over-or underexpressed within each pathway in this cell population. NR: non-relevant. Default cutoff for functional annotation charts is 1.0E-1 (0.1)
| Pathway | Genes (UP/DOWN) | p-value | Gene symbol | p-value UP/DOWN | |
|---|---|---|---|---|---|
| Cell Cycle | 37 (1 / 36) | 1.4E-7 | UP | Crebbp | NR |
| DOWN | Bub1, Bub1b, Cdc2a, Cdc6, Cdc7, Cdc20, Chek1, Ccna2, Ccnb1, Ccnb2, Ccnd1, Ccnd2, Ccne2, Cdk2, Cdk4, Cdk6, Cdkn1a, Dbf4, Fzr1, Hdac2, Mcm2, Mcm3, Mcm4, Mcm5, Mcm6, Mcm7, Pcna, Rbl1, Rbl2 , Trp53, Sfn, Ywhab, Ywhah, Ywhaq, Ywhaz, Wee1 | 8.3E-10 | |||
| Wnt signaling | 35 (13 / 22) | 9.3E-4 | UP | Crebbp, Ctbp2, Daam1, Fzd1, Fzd2, Fzd7, Nfatc5, Nfatc1, Nkd1, Sfrp1, Tcf3, Tcf4, Wif1 | 1.0E-3 |
| DOWN | Axin2, Chp, Csnk2a1, Ccnd1, Ccnd2, Map3k7, Nfatc3, Prickle1, Prkcb1, Prkacb, Rock1, Siah1a, Siah1b, Senp2, Tcf7, Tbl1x, Trp53, Wnt10a, Wnt3, Wnt3a, Wnt5a, Wnt7b | 9.9E-2 | |||
| p53 signaling | 19 (0 / 19) | 1.8E-3 | UP | – | – |
| DOWN | Casp8, Cdc2a, Chek1, Ccnb1, Ccnb2, Ccnd1, Ccnd2, Ccne2, Cdk2, Cdk4, Cdk6,Cdkn1a, Igfbp3, Pmaip1, Rrm2, Siah1a, Siah1b, Sfn, Trp53 | 9.2E-5 | |||
| Hedgehog signaling | 14 (6 / 8) | 2.2E-2 | UP | Bmp6, Gli1, Gli2, Gli3, Gas1, Hhip | 1.8E-2 |
| DOWN | Bmp4, Csnk1d, Prkacb, Wnt10a, Wnt3, Wnt3a, Wnt5a, Wnt7b | NR | |||
| Adherens junctions | 17 (5 / 12) | 4.3E-2 | UP | Actn1, Crebbp, Fgfr1, Tcf3, Tcf4 | NR |
| DOWN | Acp1, Actb, Cdh1, Csnk2a1, Mapk1, Map3k7, Pvrl2, Pvrl3, Ptpn6, Tcf7, Tgfbr2, Wasl | NR | |||
| Cell communication | 24 (13/11) | 6.2E-2 | UP | Fn1, Krt6a, Krt75, Col1a1, Col1a2, Col3a1, Col4a4, Col5a1, Col5a2, Col6a1, Col6a2, Krt24, Tnc | 1.9E-4 |
| DOWN | Krt79, Actb, Dsg2, Gjb4, Itga6, Itgb4, Krt14, Lmna, Col4a6, Krt77, Thbs2 | NR | |||
| Apoptosis | 18 (3/15) | 6.8E-2 | UP | Bcl2, Capn2, Akt3 | NR |
| DOWN | Capn1, Casp8, Chp, Dffa, Il1rap, Il1r1, Irak3, Nfkbia, Nfkb2, Pik3cb, Prkar1a, Prkacb, Ripk1, Tradd, Trp53 | 7.1E-2 |
Analysis of Gene Expression Regulation in Hair Follicle Stem Cells
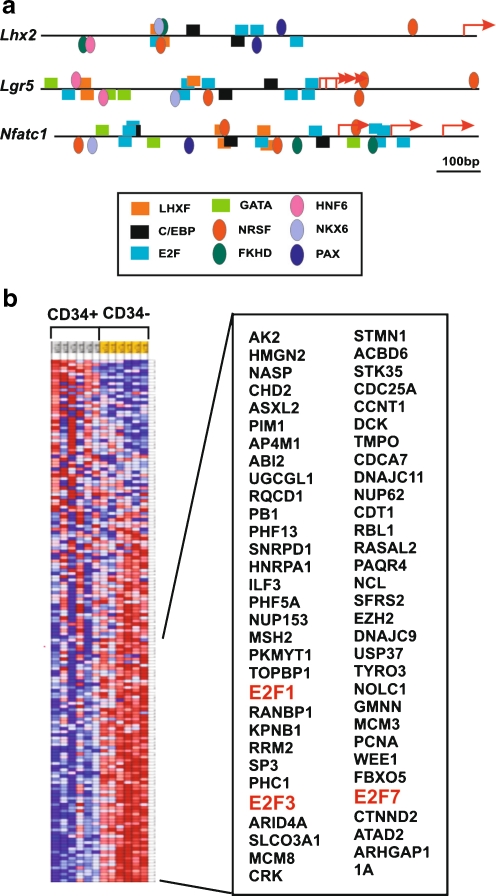
Differential expression analyses have demonstrated a strong deregulation between CD34+ HFSC and CD34−mouse skin cells. We wanted to analyze possible transcription factors responsible for the expression differences between these two cell populations. For this purpose we searched for statistically significant enrichment in specific transcription factor sequence motifs within the promoters of genes overexpressed and underexpressed within the HFSC compartment using GSEA. This kind of analyses requires a critical number of genes to work with, and most of the previously reported datasets [13, 14, 39] were not large enough to undertake this study. Tables 3 and 4 show the analyses results, with the most significantly enriched transcription factor gene set motifs. Candidate regulators of the genes overexpressed in HFSC are homeobox, paired box and forkhead transcription factors (HFSC ON, Table 3); in fact it has been shown for some members of these families their role as regulators of stem cell homeostasis [26, 43, 44]. Using MatInspector [35], we studied the presence of binding sites in genes that are known HFSC markers and also regulate the biological properties of these cells, and found that the 5′ ends proximal to the transcriptional initiation sites of Lhx2, Lgr5 and Nfatc1 are loaded with potential binding sites for the above mentioned transcription factors (Fig. 4a). Similar observations were obtained using CD34, Sox9, Tcf3 and Tcf4 genes (not shown). We also found potential involvement of C/EBPs in regulating HFSC gene expression. We have recently reported that transcription factors of this family modulate epidermal terminal differentiation and loss of C/EBPα/β induces stem cell gene expression signatures in the epidermis, thus suggesting that they may act to restrict the epidermal stem cell compartment [45].
Table 3.
Transcription factor motif gene sets enriched in the promoter region of the genes upregulated in the 2,297-probe HFSC signature as described in MSigDB (GSEA molecular signatures database). aHFSC ON: genes overexpressed in HFSC that contain within their promoter region a binding motif for that transcription factor; Total, refers to the total number of targets in the expression array. NES: Normalized enrichment score; FDR: False discovery rate
| Gene or family | Motif gene sets | aHFSC ON/ Total | NES | FDR q-val |
|---|---|---|---|---|
| Homeobox | YTAATTAA_LHX3_01 | 40 / 138 | 2.06 | 0.010 |
| S8_01 | 53 / 179 | 1.96 | 0.016 | |
| POU6F1_01 | 41 / 178 | 1.86 | 0.019 | |
| POU3F2_02 | 46 / 187 | 1.87 | 0.020 | |
| HNF6_Q6 | 49 / 182 | 1.91 | 0.021 | |
| LHX3_01 | 51 / 171 | 1.84 | 0.021 | |
| CDX2_Q5 | 42 / 184 | 1.87 | 0.022 | |
| NKX62_Q2 | 52 / 171 | 1.79 | 0.027 | |
| CHX10_01 | 38 / 173 | 1.67 | 0.058 | |
| OCT1_02 | 42 /157 | 1.68 | 0.061 | |
| Paired box | PAX4_04 | 30 / 157 | 2.01 | 0.013 |
| PAX8_B | 10 / 73 | 1.66 | 0.057 | |
| Forkhead box | HFH4_01 | 38 / 152 | 2.11 | 0.016 |
| FOXJ2_01 | 25 / 138 | 1.66 | 0.056 | |
| FOXD3_01 | 40 / 159 | 1.84 | 0.021 | |
| RE1-silencing transcription factor | NRSF_01 | 45 / 71 | 1.90 | 0.019 |
| GATA binding | GATA1_04 | 57 / 177 | 1.82 | 0.022 |
| GATA_Q6 | 40 / 142 | 1.64 | 0.060 | |
| CCAAT/enhancer binding protein gamma | CEBPGAMMA_Q6 | 21 / 184 | 1.75 | 0.034 |
| T-cell acute lymphocytic leukemia 1 Transcription factor 3 | TAL1BETAE47_01 | 44 / 177 | 1.67 | 0.056 |
Table 4.
Transcription factor motif gene sets enriched in the promoter region of the genes downregulated in the 2,297-probe HFSC signature as described in MSigDB (GSEA molecular signatures database). aHFSC OFF: genes underexpressed in HFSC that contain within their promoter region a binding motif for that transcription factor; Total, refers to the total number of targets in the expression array. NES: Normalized enrichment score, for repressed genes in a given dataset (HFSC) NES values are negative; FDR: False discovery rate
| Gene or family | Motif gene sets | aHFSC OFF / Total | NES | FDR q-val |
|---|---|---|---|---|
| E2F transcription factor | E2F_Q4 | 75 / 165 | −2.35 | <0.0001 |
| E2F_Q3_01 | 80 / 166 | −2.17 | <0.0001 | |
| E2F1_Q6 | 68 / 163 | −2.14 | <0.0001 | |
| E2F1DP1RB_01 | 68 / 160 | −2.23 | <0.0001 | |
| RB, Retinoblastoma | E2F4DP2_01 | 67 / 162 | −1.87 | 0.003 |
| DP1, E2F dimerization partner 1 | E2F1DP2_01 | 67 / 162 | −1.86 | 0.003 |
| DP2, E2F dimerization partner 2 | E2F4DP1_01 | 66 / 167 | −1.82 | 0.004 |
| E2F1DP1_01 | 67 / 162 | −1.85 | 0.003 | |
| SGCGSSAAA_E2F1DP2_01 | 52 / 117 | −2.00 | 0.001 | |
| GTGACGY_E4F1_Q6 | 193 / 447 | −2.00 | 0.001 | |
| Nuclear respiratotry factor | NRF2_01 | 93 / 165 | −2.26 | <0.0001 |
| GABP_B | 98 / 159 | −2.17 | 0.001 | |
| NRF1_Q6 | 76 / 159 | −1.98 | 0.001 | |
| ETS transcription factor | TEL2_Q6 | 74 / 156 | −2.11 | <0.0001 |
| ETS_Q4 | 83 / 178 | −2.07 | <0.0001 | |
| ELK1_01 | 87 / 176 | −1.97 | 0.001 | |
| ELK1_02 | 93 / 170 | −1.78 | 0.006 | |
| CETS1P54_01 | 72 / 176 | −1.76 | 0.007 | |
| Yin yang transcription factor 1 | GCCATNTTG_YY1_Q6 | 148 / 296 | −1.87 | 0.003 |
Fig. 4.
Analysis of gene expression regulation in HFSC. a Schematic representation of the 5’ ends proximal to the transcriptional initiation sites of Lhx2, Lgr5 and Nfatc1 showing the potential binding sites for the transcription factors mentioned, as obtained using MatInspector software [35]. b Heat map of the expression pattern of genes with E2F-binding sites within their promoters (overexpressed genes in red, underexpressed genes in blue). Note that the majority of the genes are underexpressed in CD34+ HFSC. Highlighted in red are E2F transcription factors
On the other hand, our results demonstrate a strong underexpression of E2F-dependent genes within the HFSC compartment, confirmed by the presence of various E2F-related gene set motifs (HFSC OFF, Table 4). More specifically, from 165 genes containing E2F binding sites within the expression array, 75 of them are specifically underexpressed in the CD34+ HFSC compartment (Fig. 4b), while the expression of most of the rest do not significantly distinguish between CD34+ and CD34−cells. We observed that, among the potentially E2F-regulated genes, E2F1, E2F3, and E2F7 are underexpressed in CD34+ HFSC. This suggests that the low expression of these E2F-dependent genes is not due to an active E2F-mediated repression, but rather to the relatively low expression level of these transcription factors.
Rb-Dependent Pathway Regulates Stem Cell Quiescence in Epidermis
The above reported findings led us to investigate the possible relevance of E2Fs in HFSC maintenance. Different E2Fs are present in epidermis where they exert specific roles in morphogenesis, differentiation and pathogenesis [46, 47]; however, the inherent complexity of this system, composed by at least ten different members with activator and repressor activities (see Introduction), makes it difficult to analyze their specific functions due to potential redundancy and functional compensation. In this regard, we have observed no evident phenotype in the skin of mice lacking E2F1 or E2F4 (CC, MS and JMP, unpublished data) in the absence of external stimuli, whereas overexpression of these proteins in epidermis leads to hyperplasic phenotype and different susceptibility to skin carcinogenesis protocols [48, 49].
Therefore, to analyze the relevance of E2Fs in HFSC homeostasis, we turned to a different approach using mice bearing epidermal specific ablation of the Rb gene using Cre-loxP technology. These mice display hyperplasia and hyperkeratosis in the epidermis with increased proliferation and aberrant expression of differentiation markers [31]. We found that in Rb-deficient keratinocytes there is a significant overexpression of all E2F family members compared to wild-type cells, with the exception of E2F4, which appeared repressed (Fig. 5a). Such altered expression is accompanied by increased overall E2F activity as measured by luciferase reporter studies (Fig. 5b). Next, we studied how Rb deficiency and subsequent E2F activation affected HFSC homeostasis. The overall size of the HFSC population, measured as the percentage of cells showing CD34/Itgα6 positive phenotype in FACS analyses, is similar in Rb-deficient and control mice (Fig. 5c). In LRC experiments, Rb-deficient mice show a significant reduction in the number of hair follicles labeled 75 days after BrdU injection (Fig. 5d). BrdU-positive bulge cells appear faintly labeled in Rb-deficient mice (Fig. 5e´) as compared to the strong label displayed in control mice (Fig. 5e); also, we observed that, in Rb-deficient hair follicles, BrdU-labeled cells appeared usually in clusters rather than isolated cells (Fig. 5e and e´). Altogether, our data suggest an increased proliferation/division of the HFSC population in Rb-deficient mice leading to a dilution of the BrdU label. However, since Rb deficiency also induces disengagement between proliferation and differentiation in epidermal keratinocytes [31], we cannot rule out the possibility that a similar imbalance may occur during the differentiation of Rb-deficient HFSC, thus perturbing the functionality of these cells.
Fig. 5.
pRb regulates stem cell quiescence. a qRT-PCR analysis of the expression of the different E2F members in Rb-deficient keratinocytes. Values were normalized with respect to each E2F member expression in control keratinocytes. b Luciferase assay showing increased E2F activity in Rb-deficient keratinocytes. The data were normalized to Renilla activity and correspond to three independent experiments. They are shown as mean fold induction ± standard error. c Summary of four independent FACS experiments showing the percentage of CD34+/Itgα6+ cells from control and Rb-deficient mice. d Quantification of BrdU-positive label retaining cells (LRC) after a 75-day chase in control and Rb-deficient mice (Rbf/f; K14Cre). Data come from the study of five different mice, 2–4 independent sections from each animal were analyzed counting at least 100 follicles per section (mean ± standard error). (e and e´) Representative skin sections showing BrdU-positive cells in follicles from LRC experiments. Insets show higher magnification of BrdU-positive labeled cells. Bars = 150 μm; Rbf/f, control mice; Rbf/f; K14Cre, Rb-deficient mice
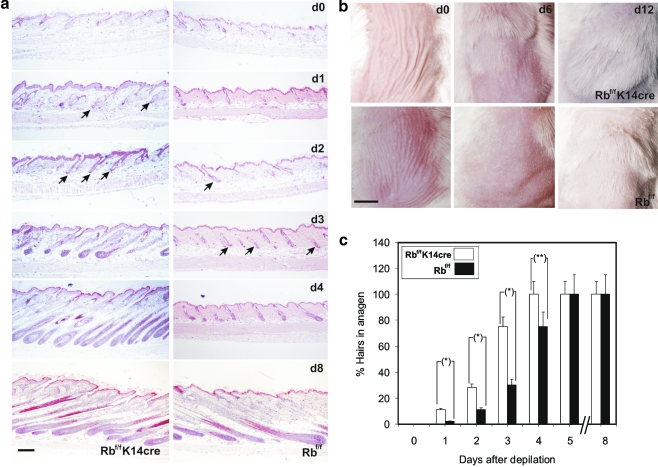
To explore the consequences of pRb ablation in HFSC behavior, we conducted functional studies. External stimuli, such as wounding or hair depilation, activate bulge stem cells to migrate upwards to contribute to epidermal regeneration, or downwards to regrow new hair. Time-course experiments analyzing hair regrowth showed accelerated anagen induction in Rb-deficient mice when compared to control littermates (Fig. 6a and c), and only by day 8 after depilation a similar appearance was observed between Rb-deficient and control adult mice. This quickening in anagen induction resulted in evident faster hair regrowth in mutant mice even up to 12 days after depilation (Fig. 6b).
Fig. 6.
pRb ablation in epidermis accelerates hair regrowth. a Representative skin sections from control and Rb-deficient mice at different time points after induction of hair regrowth by depilation. Note premature anagen induction in Rb-deficent mice. Incipient anagen hairs are denoted by arrows. Bar = 250 μm. b Representative macroscopic appearance of dorsal skin sections from control and Rb-deficient mice at different time points after depilation. Note the presence of massive hair regrowth in mutant mice 6 days after depilation. Bar = 1 cm. c Histogram summarizing anagen induction data obtained from histological analyses as those shown in (a). Data in c come from the quantification of 5 sections (2 cm length) of 10 mice of each genotype and are shown as mean ± SD. Statistical significances analyzed by two tailed tTest were (*) p ≤ 0.001, (**) p ≤ 0.005. Rbf/f, control mice; Rbf/f; K14Cre, Rb-deficient mice
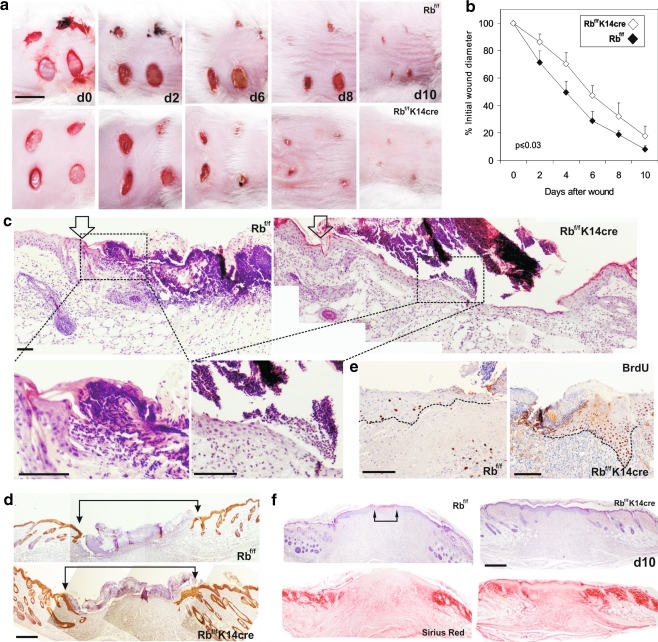
When the process of wound healing was analyzed, we observed a faster wound closure in Rb-deficient as compared to control mice (Fig. 7a and b). Histological analyses demonstrate that this is accompanied by a faster re-epithelialization in mutant mice (Fig. 7c and d). We also observed an increased cell proliferation at wound margins of mutant mice as compared to control (Fig. 7e). Finally, in the remodeling phase of wound healing, sirius red staining revealed a mild increase in the maturation of collagen fibers in Rb-deficient mice compared to control (Fig. 7f). These data are in agreement with the reported delayed in wound healing in E2F1 null mice, associated with substantially reduced rates of re-epithelialization without alterations in epidermal stratification and remodeling [50].
Fig. 7.
pRb ablation in epidermis accelerates skin wound healing. a Representative skin sections from control and Rb-deficient mice at different time points after wounding. Note the accelerated wound closure in mutant mice. Bar = 1 cm. b Plot summarizing the quantitative analysis of wound closure obtained from 4 different wounds induced in 4 different mice of each genotype. Data are shown as mean ± SE. c Haematoxylin-eosin stained sections of control and Rb-deficient mice showing the wound margins (denoted by arrows). Higher magnifications of the squared areas show the re-epithelialization front. d Keratin 5 immunostaining also demonstrates advanced re-epithelialization in pRb-deficient mice. Arrows denote initial wound margins. e BrdU-stained sections showing increased cell proliferation at the wound margins in Rb-deficient mice. Dashed lines denote the dermal-epidermal borders. F) Hematoxylin-eosin and sirius red staining of representative sections 10 days after wounding in control and Rb-deficient mice. The maturation of collagen fibers, denoted by intense red staining is slightly accelerated in Rb-deficient mice. Wound margins are still detected in control mice (arrows). Bars in c, d, e, f = 150 µm; Rbf/f, control mice; Rbf/f; K14Cre, Rb-deficient mice
Overall, these data suggest that Rb loss results in increased stem cell proliferation, without an apparent loss of the HFSC pool or overt failure of stem cell function. Increased expression and transcriptional activity of E2Fs due to pRb depletion might indicate a functional role of this family of transcription factors in the maintenance of epidermal stem cell functionality; however, we cannot rule out the possibility that some of the observed effects can be driven by the interaction of pRb with other proteins [3]. Ongoing studies, in which the epidermal specific ablation of pRb is combined with defined E2F deficiencies, will help to discern such possibilities.
Discussion
The essential role of SC in tissue homeostasis and their possible involvement in multiple pathologies, have posed numerous efforts aimed to understand the specific and differential characteristics of this cell population. We have combined magnetic-activated cell sorting, as an isolation procedure, with functional genomic approaches to study the regulation of adult HFSC gene expression. Our findings point to a possible role of E2F transcription factors in the regulation of epidermal stem cell exit from quiescence.
In epidermis, several groups have reported gene expression profiling studies using FACS and/or sophisticated transgenic models [13–16, 39]. We have used MACS as an approachable and cell-friendly system to isolate CD34+ hair follicle bulge stem cells. In the resulting molecular signature we find upregulation of multiple genes that have been previously characterized as HFSC markers, such as Tcf3, Tcf4, Nfatc1, Lgr5 and Lhx2. We have validated this HFSC-signature through comparative analysis with existing ones. Using GSEA, we have found that a relevant number of genes identified by others are significantly present in our signature. An overall comparison of the gene expression profile revealed that ∼70% of the differentially expressed genes are downregulated in the stem cell population, indicating that HFSC are less transcriptionally active than other epidermal cell populations; this feature is shared with other HFSC genomic analyses [15, 39]. Comparative analyses have allowed us also to identify genes with common expression patterns in all the published genomic datasets of HFSC, including mouse and human species. The mo-huHFSC signature represents a statistically robust list of genes over- or under-expressed in HFSC that could be useful for future analysis of HFSC markers, and highlights the importance of cell cycle regulation in this cell type.
Analysis of the molecular pathways in HFSC revealed that cell cycle control is the most relevant category enriched in this cell population, and may partly reflect the relative quiescence of these cells. We observed that genes implicated in cell cycle progression (cyclins and cyclin-dependent kinases) or mechanistically involved in DNA replication (Mcm genes, PCNA), as well as potential inhibitors of cell cycle progression, such as Cdkn1a, Rbl1 and Rbl2, are downregulated in HFSC. We have previously reported that, in the absence of p107 and p130 (Rbl1 and Rbl2 gene products, respectively), epidermal differentiation, including hair formation, is severely compromised [51] and Wnt signaling is altered [52]. Furthermore, the epidermal ablation of Rbl1 dramatically aggravates the phenotype observed in Rb-deficient mice [31] and confers susceptibility to spontaneous [53] and chemically induced tumorigenesis [54]. Altogether, these data suggest that the members of the Rb family could be playing a central role in epidermal stem cell activation.
Also, we have found downregulation of Trp53 and p53-dependent genes, including the aforementioned Cdkn1a. This is an apparent controversy with the widely accepted view that HFSC must possess robust mechanisms to detect possible genetic abnormalities and preclude appearance of pathologies such as tumors. However, since the CD34+ HFSC population is a fairly quiescent cell population, it cannot be excluded that transient activation of the p53-dependent pathway may occur during the process of HFSC activation to prevent expansion of genomic alterations, or during the process of hair regression as previously reported [55]. In this regard, we have observed that mice lacking p53 in epidermis develop spontaneous tumors arising from hair follicles [56] associated with altered centrosome number and localization [57]. Importantly, upon a chemical carcinogenesis challenge, the absence of Rb results in decreased tumor formation due to p53 activation [58] and the simultaneous absence of Rb and Trp53 results in accelerated tumor formation compared with mouse bearing only the epidermal ablation of Trp53 [56]. Also, array data suggest that HFSC are relatively protected from apoptosis, as they show upregulation of anti-apoptotic or survival genes such as Bcl2 and Akt3, and downregulation of pro-apoptotic genes such as Trp53, Casp8, Dffa, Tradd or Ripk1. These observations may indicate that both Rb-and p53-dependent pathways could be involved in HFSC homeostasis during physiological or induced activation processes and merit future investigation.
The use of MACS and a large expression array has led to the development of a HFSC signature larger than previously reported ones [13, 14, 39]. This signature has allowed us perform a comprehensive bioinformatics analysis to characterize potential transcription factors regulating gene expression in HFSC. This kind of studies is significantly relevant only when the number of genes within a geneset or molecular signature is large enough. We observed that genes upregulated in HFSC contain potential binding sites for homeobox, paired-box and forkhead transcription factors. Some of them have been previously involved in the homeostasis of embryonic and adult stem cell populations, including follicle stem cells [26, 43, 44, 59–61], suggesting that a molecular circuitry common to multiple stem cells may exist. We also found potential involvement of C/EBPs in regulating HFSC gene expression. In this regard, we have recently reported that transcription factors of this family modulate epidermal terminal differentiation and they may act to restrict the epidermal stem cell compartment, since loss of C/EBPα/β induces stem cell gene expression signatures in the epidermis [45].
Our studies show the presence of E2F sites among a considerable number of genes underexpressed in HFSC. In this regard, it is important to note that the actual number of E2F-regulated genes is likely to be even higher than revealed in our study, taking in account the fact that E2F, in addition to its consensus binding sites, is capable of binding many gene targets that do not have typical E2F-consensus binding sites [62]. This led us to investigate the possible involvement of E2Fs in the regulation of epidermal stem cell homeostasis; however, in vivo analysis is not straightforward due to the complexity of this transcription factor family, which comprises ten identified members with different activities (activation or repression) on gene transcription [4–6]. Therefore, we took advantage on the fact that the Rb ablation leads to increased expression of most E2F genes with a subsequent increase in E2F activity. We observed that the absence of pRb did not seem to alter the distribution of CD34+/Itgα6+ population in the skin. On the other hand, there was a significant reduction of the LRC population in the hair follicles of Rb-deficient mice, which frequently displayed cell clusters with decreased BrdU immunoreactivity. This label dilution points to an increased rate of HFSC proliferation and/or division in Rb-deficient mice. Another non-excluding explanation is that Rb gene ablation may affect the equilibrium between proliferation and differentiation in this cell population. In this regard, we have previously shown that epidermal keratinocytes lacking pRb display defective differentiation, and once the cells are committed into the differentiation program they are still able to proliferate [31, 37]. Future studies using CD34 purified cells from these mutant mice, or in vitro proliferation experiments, will help to clarify such possibilities.
In addition, our functional analysis did not reveal defective stem cell functionality or activation, but rather a more efficient wound healing and hair regrowth. Both are highly dynamic processes that require activation of epidermal stem cells and their interaction with dermal cells. Our data may suggest that Rb deficiency reduces the level of stem cell quiescence while maintaining the normal homeostatic functions and stemness of the epidermal SC population. This could be explained because these processes require that, upon HFSC activation, cell division proceeds; a mechanism that is facilitated by the absence of pRb and the subsequent increase in E2F activity [63]. However, we cannot exclude the possibility that E2F-independent mechanisms triggered by Rb deficiency are partially responsible for these effects, in this sense it has been shown that pRb binds and regulates transcription factors, inhibitors of differentiation and chromatin modifiers to promote differentiation [64–67]. Nonetheless, the report on impaired wound healing in E2F1-null mice, due to defective cell proliferation and thus re-epithelialization [50], would reinforce the relevance of the Rb-E2F axis in maintaining appropriate stem cell proliferation in epidermis. In addition, our findings showing that Rb-deficient mice display increased E2F expression and activity and our expression data showing that many HFSC genes are E2F-targets, suggest that the consequences of pRb loss on stem cell quiescence are likely to be E2F-mediated.
In summary, we have applied a magnetic-based method to isolate CD34+ hair follicle stem cells to perform gene expression studies. We obtained a robust molecular signature that reveals the importance of the regulation of cell cycle related genes in this cell population. Analysis of the regulation of gene expression suggests that homebox, paired-box, forkhead and E2F transcription factors are important regulators of HFSC gene expression, and in vivo experiments using Rb-deficient mice point to a possible role of Rb/E2F in HFSC activation. The analysis of the different available E2F deficient mice, or the different combinations among them and with Rb family-deficient mice, will undoubtedly help to discern their possible specific functions in HFSC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(XLS 1108 kb)
(DOC 34 kb)
(XLS 37 kb)
(GIF 4.15 mb)
Acknowledgements
Grant support: Ministerio de Ciencia e Innovación (MICINN) grants SAF2008-0121, SAF2007-66227 and PS-010000-2008-7, Comunidad Autónoma de Madrid Oncocycle Program grant S2006/BIO-0232, and Ministerio de Sanidad y Consumo grant ISCIII-RETIC RD06/0020. JMA is a MICINN fellow, FPI-CIEMAT programme.
Conflict of interest
The authors declare no potential conflicts of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- SC
Stem cells
- HFSC
Hair follicle stem cells
- DP
Dermal papilla
- HG
Hair germ
- K15
cytokeratin 15
- Itgα6
Integrin alpha 6
- MACS
magnetic-activated cell sorting
- FACS
fluorescence-activated cell sorting
- LRC
label-retaining cells
- BrdU
bromodeoxyuridine
Footnotes
Ramón García-Escudero and Carmen Segrelles contributed equally to this work.
References
- 1.Fuchs E, Horsley V. More than one way to skin. Genes and Development. 2008;22(8):976–985. doi: 10.1101/gad.1645908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nature Reviews Cancer. 2009;9(4):265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 3.Galderisi U, Cipollaro M, Giordano A. The retinoblastoma gene is involved in multiple aspects of stem cell biology. Oncogene. 2006;25(38):5250–5256. doi: 10.1038/sj.onc.1209736. [DOI] [PubMed] [Google Scholar]
- 4.van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nature Reviews Molecular Cell Biology. 2008;9(9):713–724. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- 5.Lammens T, Li J, Leone G, De Veylder L. Atypical E2Fs: new players in the E2F transcription factor family. Trends in Cell Biology. 2009;19(3):111–118. doi: 10.1016/j.tcb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nature Reviews Cancer. 2009;9(11):785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotsarelis G. Epithelial stem cells: a folliculocentric view. Journal of Investigative Dermatology. 2006;126(7):1459–1468. doi: 10.1038/sj.jid.5700376. [DOI] [PubMed] [Google Scholar]
- 8.Alonso L, Fuchs E. The hair cycle. Journal of Cell Science. 2006;119(Pt 3):391–393. doi: 10.1242/jcs02793. [DOI] [PubMed] [Google Scholar]
- 9.Taylor G, Lehrer MS, Jensen PJ, Sun TT, Lavker RM. Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell. 2000;102(4):451–461. doi: 10.1016/S0092-8674(00)00050-7. [DOI] [PubMed] [Google Scholar]
- 10.Oshima H, Rochat A, Kedzia C, Kobayashi K, Barrandon Y. Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell. 2001;104(2):233–245. doi: 10.1016/S0092-8674(01)00208-2. [DOI] [PubMed] [Google Scholar]
- 11.Ito M, Liu Y, Yang Z, et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nature Medicine. 2005;11(12):1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- 12.Horsley V, O’Carroll D, Tooze R, et al. Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell. 2006;126(3):597–609. doi: 10.1016/j.cell.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tumbar T, Guasch G, Greco V, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303(5656):359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris RJ, Liu Y, Marles L, et al. Capturing and profiling adult hair follicle stem cells. Nature Biotechnology. 2004;22(4):411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 15.Trempus CS, Dang H, Humble MM, et al. Comprehensive microarray transcriptome profiling of CD34-enriched mouse keratinocyte stem cells. Journal of Investigative Dermatology. 2007;127(12):2904–2907. doi: 10.1038/sj.jid.5700917. [DOI] [PubMed] [Google Scholar]
- 16.Greco V, Chen T, Rendl M, et al. A two-step mechanism for stem cell activation during hair regeneration. Cell stem cell. 2009;4(2):155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waghmare SK, Bansal R, Lee J, Zhang YV, McDermitt DJ, Tumbar T. Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells. Embo Journal. 2008;27(9):1309–1320. doi: 10.1038/emboj.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaks V, Barker N, Kasper M, et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nature Genetics. 2008;40(11):1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 19.Lyle S, Christofidou-Solomidou M, Liu Y, Elder DE, Albelda S, Cotsarelis G. The C8/144B monoclonal antibody recognizes cytokeratin 15 and defines the location of human hair follicle stem cells. Journal of Cell Science. 1998;111(Pt 21):3179–3188. doi: 10.1242/jcs.111.21.3179. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Lyle S, Yang Z, Cotsarelis G. Keratin 15 promoter targets putative epithelial stem cells in the hair follicle bulge. Journal of Investigative Dermatology. 2003;121(5):963–968. doi: 10.1046/j.1523-1747.2003.12600.x. [DOI] [PubMed] [Google Scholar]
- 21.Trempus CS, Morris RJ, Bortner CD, et al. Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. Journal of Investigative Dermatology. 2003;120(4):501–511. doi: 10.1046/j.1523-1747.2003.12088.x. [DOI] [PubMed] [Google Scholar]
- 22.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118(5):635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 23.Trempus CS, Morris RJ, Ehinger M, et al. CD34 expression by hair follicle stem cells is required for skin tumor development in mice. Cancer Research. 2007;67(9):4173–4181. doi: 10.1158/0008-5472.CAN-06-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malanchi I, Peinado H, Kassen D, et al. Cutaneous cancer stem cell maintenance is dependent on beta-catenin signalling. Nature. 2008;452(7187):650–653. doi: 10.1038/nature06835. [DOI] [PubMed] [Google Scholar]
- 25.Horsley V, Aliprantis AO, Polak L, Glimcher LH, Fuchs E. NFATc1 Balances Quiescence and Proliferation of Skin Stem Cells. Cell. 2008;132(2):299–310. doi: 10.1016/j.cell.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhee H, Polak L, Fuchs E. Lhx2 maintains stem cell character in hair follicles. Science. 2006;312(5782):1946–1949. doi: 10.1126/science.1128004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen H, Merrill BJ, Polak L, et al. Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nature Genetics. 2009;41(10):1068–1075. doi: 10.1038/ng.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen H, Rendl M, Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127(1):171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 29.Nowak JA, Polak L, Pasolli HA, Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell stem cell. 2008;3(1):33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorz C, Segrelles C, Garin M, Paramio JM. Isolation of adult mouse stem keratinocytes using magnetic cell sorting (MACS) Methods in molecular biology. (Clifton, NJ) 2010;585:1–11. doi: 10.1007/978-1-60761-380-0_1. [DOI] [PubMed] [Google Scholar]
- 31.Ruiz S, Santos M, Segrelles C, et al. Unique and overlapping functions of pRb and p107 in the control of proliferation and differentiation in epidermis. Development. 2004;131(11):2737–2748. doi: 10.1242/dev.01148. [DOI] [PubMed] [Google Scholar]
- 32.Segrelles C, Moral M, Lorz C, et al. Constitutively Active Akt Induces Ectodermal Defects and Impaired Bone Morphogenetic Protein Signaling. Molecular Biology of the Cell. 2008;19(1):137–149. doi: 10.1091/mbc.E07-08-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics (Oxford, England) 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cartharius K, Frech K, Grote K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics (Oxford, England) 2005;21(13):2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 36.Paramio JM, Segrelles C, Ruiz S, et al. The ink4a/arf tumor suppressors cooperate with p21cip1/waf in the processes of mouse epidermal differentiation, senescence, and carcinogenesis. Journal of biological chemistry. 2001;276(47):44203–44211. doi: 10.1074/jbc.M105650200. [DOI] [PubMed] [Google Scholar]
- 37.Lara MF, Garcia-Escudero R, Ruiz S, et al. Gene profiling approaches help to define the specific functions of retinoblastoma family in epidermis. Molecular Carcinogenesis. 2008;47(3):209–221. doi: 10.1002/mc.20376. [DOI] [PubMed] [Google Scholar]
- 38.Ito M, Kizawa K, Hamada K, Cotsarelis G. Hair follicle stem cells in the lower bulge form the secondary germ, a biochemically distinct but functionally equivalent progenitor cell population, at the termination of catagen. Differentiation. 2004;72(9–10):548–557. doi: 10.1111/j.1432-0436.2004.07209008.x. [DOI] [PubMed] [Google Scholar]
- 39.Ohyama M, Terunuma A, Tock CL, et al. Characterization and isolation of stem cell-enriched human hair follicle bulge cells. Journal of Clinical Investigation. 2006;116(1):249–260. doi: 10.1172/JCI26043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobielak K, Stokes N, de la Cruz J, Polak L, Fuchs E. Loss of a quiescent niche but not follicle stem cells in the absence of bone morphogenetic protein signaling. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(24):10063–10068. doi: 10.1073/pnas.0703004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, He XC, Tong WG, et al. Bone morphogenetic protein signaling inhibits hair follicle anagen induction by restricting epithelial stem/progenitor cell activation and expansion. Stem Cells. 2006;24(12):2826–2839. doi: 10.1634/stemcells.2005-0544. [DOI] [PubMed] [Google Scholar]
- 42.Adolphe C, Narang M, Ellis T, Wicking C, Kaur P, Wainwright B. An in vivo comparative study of sonic, desert and Indian hedgehog reveals that hedgehog pathway activity regulates epidermal stem cell homeostasis. Development. 2004;131(20):5009–5019. doi: 10.1242/dev.01367. [DOI] [PubMed] [Google Scholar]
- 43.Guo Y, Costa R, Ramsey H, et al. The embryonic stem cell transcription factors Oct-4 and FoxD3 interact to regulate endodermal-specific promoter expression. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(6):3663–3667. doi: 10.1073/pnas.062041099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tothova Z, Gilliland DG. FoxO transcription factors and stem cell homeostasis: insights from the hematopoietic system. Cell stem cell. 2007;1(2):140–152. doi: 10.1016/j.stem.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 45.Lopez RG, Garcia-Silva S, Moore SJ, et al. C/EBPalpha and beta couple interfollicular keratinocyte proliferation arrest to commitment and terminal differentiation. Nature Cell Biology. 2009;11(10):1181–1190. doi: 10.1038/ncb1960. [DOI] [PubMed] [Google Scholar]
- 46.Ivanova IA, D’Souza SJ, Dagnino L. Signalling in the epidermis: the E2F cell cycle regulatory pathway in epidermal morphogenesis, regeneration and transformation. International journal of biological sciences. 2005;1(2):87–95. doi: 10.7150/ijbs.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paramio JM, Segrelles C, Casanova ML, Jorcano JL. Opposite functions for E2F1 and E2F4 in human epidermal keratinocyte differentiation. Journal of biological chemistry. 2000;275(52):41219–41226. doi: 10.1074/jbc.M004973200. [DOI] [PubMed] [Google Scholar]
- 48.Pierce AM, Schneider-Broussard R, Gimenez-Conti IB, Russell JL, Conti CJ, Johnson DG. E2F1 has both oncogenic and tumor-suppressive properties in a transgenic model. Molecular and Cellular Biology. 1999;19(9):6408–6414. doi: 10.1128/mcb.19.9.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang D, Russell JL, Johnson DG. E2F4 and E2F1 have similar proliferative properties but different apoptotic and oncogenic properties in vivo. Molecular and Cellular Biology. 2000;20(10):3417–3424. doi: 10.1128/MCB.20.10.3417-3424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D'Souza SJ, Vespa A, Murkherjee S, Maher A, Pajak A, Dagnino L. E2F-1 is essential for normal epidermal wound repair. Journal of biological chemistry. 2002;277(12):10626–10632. doi: 10.1074/jbc.M111956200. [DOI] [PubMed] [Google Scholar]
- 51.Ruiz S, Segrelles C, Bravo A, et al. Abnormal epidermal differentiation and impaired epithelial-mesenchymal tissue interactions in mice lacking the retinoblastoma relatives p107 and p130. Development. 2003;130(11):2341–2353. doi: 10.1242/dev.00453. [DOI] [PubMed] [Google Scholar]
- 52.Ruiz S, Segrelles C, Santos M, Lara MF, Paramio JM. Functional link between retinoblastoma family of proteins and the Wnt signaling pathway in mouse epidermis. Developmental Dynamics. 2004;230(3):410–418. doi: 10.1002/dvdy.20065. [DOI] [PubMed] [Google Scholar]
- 53.Lara MF, Santos M, Ruiz S, et al. p107 acts as a tumor suppressor in pRb-deficient epidermis. Molecular Carcinogenesis. 2008;47(2):105–113. doi: 10.1002/mc.20367. [DOI] [PubMed] [Google Scholar]
- 54.Santos M, Ruiz S, Lara MF, et al. Susceptibility of pRb-deficient epidermis to chemical skin carcinogenesis is dependent on the p107 allele dosage. Molecular Carcinogenesis. 2008;47(11):815–821. doi: 10.1002/mc.20426. [DOI] [PubMed] [Google Scholar]
- 55.Botchkarev VA, Komarova EA, Siebenhaar F, et al. p53 Involvement in the control of murine hair follicle regression. American Journal of Pathology. 2001;158(6):1913–1919. doi: 10.1016/S0002-9440(10)64659-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martinez-Cruz AB, Santos M, Lara MF, et al. Spontaneous squamous cell carcinoma induced by the somatic inactivation of retinoblastoma and Trp53 tumor suppressors. Cancer Research. 2008;68(3):683–692. doi: 10.1158/0008-5472.CAN-07-3049. [DOI] [PubMed] [Google Scholar]
- 57.Martinez-Cruz AB, Santos M, Garcia-Escudero R, et al. Spontaneous tumor formation in Trp53-deficient epidermis mediated by chromosomal instability and inflammation. Anticancer Research. 2009;29(8):3035–3042. [PubMed] [Google Scholar]
- 58.Ruiz S, Santos M, Lara MF, Segrelles C, Ballestin C, Paramio JM. Unexpected roles for pRb in mouse skin carcinogenesis. Cancer Research. 2005;65(21):9678–9686. doi: 10.1158/0008-5472.CAN-05-1853. [DOI] [PubMed] [Google Scholar]
- 59.Re RN, Cook JL. The physiological basis of intracrine stem cell regulation. American Journal of Physiology. 2008;295(2):H447–H453. doi: 10.1152/ajpheart.00461.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rinn JL, Wang JK, Liu H, Montgomery K, van de Rijn M, Chang HY. A systems biology approach to anatomic diversity of skin. Journal of Investigative Dermatology. 2008;128(4):776–782. doi: 10.1038/sj.jid.5700986. [DOI] [PubMed] [Google Scholar]
- 61.Lei H, Juan AH, Kim MS, Ruddle FH. Identification of a Hoxc8-regulated transcriptional network in mouse embryo fibroblast cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(27):10305–10309. doi: 10.1073/pnas.0603552103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rabinovich A, Jin VX, Rabinovich R, Xu X, Farnham PJ. E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Research. 2008;18(11):1763–1777. doi: 10.1101/gr.080622.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chong JL, Wenzel PL, Saenz-Robles MT, et al. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature. 2009;462(7275):930–934. doi: 10.1038/nature08677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skapek SX, Pan YR, Lee EY. Regulation of cell lineage specification by the retinoblastoma tumor suppressor. Oncogene. 2006;25(38):5268–5276. doi: 10.1038/sj.onc.1209710. [DOI] [PubMed] [Google Scholar]
- 65.Lasorella A, Noseda M, Beyna M, Yokota Y, Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407(6804):592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- 66.Miyake S, Sellers WR, Safran M, et al. Cells degrade a novel inhibitor of differentiation with E1A-like properties upon exiting the cell cycle. Molecular and Cellular Biology. 2000;20(23):8889–8902. doi: 10.1128/MCB.20.23.8889-8902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG., Jr Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Molecular cell. 2005;18(6):623–635. doi: 10.1016/j.molcel.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 68.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 69.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biology. 2003;4(5):P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS 1108 kb)
(DOC 34 kb)
(XLS 37 kb)
(GIF 4.15 mb)