Abstract
A mesophilic, aerobic, facultatively chemolithoautotrophic bacterium, designated strain EPR70T, was isolated from hydrothermal fluids from diffuse-flow vents on the East Pacific Rise at ° 50′ N 10 ° 17′ W. Cells were Gram-negative rods, approximately 0.8–1.0 μm long and 0.3–0.5 μm wide. Strain EPR70T grew at 20–40 °C (optimum 30–35 °C), 1–25 % NaCl (optimum 2.5 %) and pH 5.0–7.5 (optimum pH 5.5). The shortest generation time observed for strain EPR70T was 42 min. Growth occurred under aerobic chemolithoautotrophic conditions in the presence of thiosulfate and CO2. Strain EPR70T grew heterotrophically with acetate or n-alkanes as sole carbon and energy sources, and in complex artificial seawater medium. Nitrate was not used as an electron acceptor. The G+C content of the genomic DNA was 64 mol%. Phylogenetic analysis of the 16S rRNA gene indicated that this organism is a member of the class Gammaproteobacteria, with Salinisphaera shabanensis E1L3AT as its closest relative (94 % sequence similarity). On the basis of phylogenetic analyses based on 16S rRNA, rbcL and alkB genes and physiological analysis, it is proposed that the organism represents a novel species within the genus Salinisphaera, for which the name Salinisphaera hydrothermalis sp. nov. is proposed. The type strain is EPR70T (=DSM 21483T =JCM 15514T).
Deep-sea hydrothermal vents are located in tectonically active areas of the sea floor and release either high-temperature fluids (focused-flow vents) or moderate-temperature fluids (diffuse-flow vents). At deep-sea vents, seawater interacts with volcanic rocks at high temperature and becomes enriched in a variety of reduced chemical species, which represent a constant flux of electron donors for microbial oxidations. In the absence of light, chemosynthetic micro-organisms oxidize these reduced chemical species (predominantly sulfur compounds) and mediate the primary production of organic carbon (Jannasch, 1995; McCollom & Shock, 1997).
Typically, representatives of the genera Thiomicrospira and Thiobacillus have been isolated when diffuse-flow hydrothermal fluids were inoculated in culture media for the enrichment of chemosynthetic, aerobic, sulfide-, sulfur- and thiosulfate-oxidizing bacteria (Ruby et al., 1981; Ruby & Jannasch, 1982; Jannasch et al., 1985; Durand et al., 1993; Teske et al., 2000; Takai et al., 2004). Heterotrophic bacteria of the genera Marinobacter, Vibrio, Pseudoalteromonas and Halomonas, among others, have also been isolated routinely from hydrothermal vent samples (Raguénès et al., 1997; Teske et al., 2000; Kaye et al., 2004; Vetriani et al., 2005; Simon-Colin et al., 2008) and their abundance in vent fluids collected from the Pacific Ocean was estimated to be up to 28 % of the total micro-organisms (Kaye & Baross, 2000).
Here, we report the isolation of three strains of mesophilic, halotolerant, facultatively chemolithoautotrophic, thiosulfate-oxidizing gammaproteobacteria from deep-sea hydrothermal vents located on the East Pacific Rise (EPR) at ° N and the characterization of one of these strains.
Hydrothermal fluid samples from diffuse-flow vents were collected from the EPR ( ° 50′ N 10 ° 17′ W) at a depth of approximately 2500 m during an oceanographic expedition aboard the R/V Atlantis in April 2004. The fluids were collected using titanium samplers operated by the manipulator of the DSV Alvin, immediately above the venting source and 1 m above the source. On the surface, samples were transferred promptly to the ship's laboratory and subsamples were stored at 4 °C. Primary enrichment cultures were initiated immediately after sample collection in growth medium designed to enrich for chemolithoautotrophic, thiosulfate-oxidizing bacteria. The medium used was a modification of medium 142 (http://www.dsmz.de), referred to as 142−A, which was composed of (l−1): NaCl, 25.0 g; (NH4)2SO4, 1.0 g; MgSO4 . 7H2O, 1.5 g; CaCl2 . 2H2O, 0.42 g; KCl, 0.64 g; NaHCO3, 0.046 g; K2HPO4, 0.05 g. Two millilitres of a 0.5 % phenol red solution was added to 1 l medium as a pH indicator. Following sterilization, the medium was supplemented with 20 mM Na2S2O3, 1 μM vitamin B12, 1 ml mixed vitamin solution 141 (http://www.dsmz.de) and 1 ml trace-element solution SL10 (http://www.dsmz.de). For isolation of single colonies, liquid cultures were inoculated on Petri dishes containing medium 142−A solidified with 15 g Noble agar l−1 (Sigma). Stocks for long-term storage were prepared by adding 150 μl sterile glycerol (Fisher Scientific) to 850 μl culture grown overnight, and were stored at –80 °C.
Heterotrophic growth was determined by transferring 100 μl of an overnight culture from medium 142−A to medium 142+A, which was depleted of NaHCO3 and supplemented with 10 mM sodium acetate (Sigma). Growth was also tested in artificial seawater (ASW) medium (l−1: NaCl, 24 g; KCl, 0.7 g; MgCl2, 7.0 g; yeast extract, 3.0 g; peptone, 2.5 g), in low-strength ASW (LS ASW) medium (modified ASW containing 0.1 g yeast extract and 0.5 g peptone l−1) and in ASW minimal medium (ASW MM) (l−1: NaCl, 23.6 g; KCl, 0.64 g; MgCl2 . 6H2O, 4.53 g; MgSO4 . 7H2O, 5.94 g; CaCl2 . 2H2O, 1.3 g) Na2HPO4 . 7H2O, 43.0 mg; NaNO3, 0.22 g; NH4Cl, 0.65 g) supplemented with dodecane (C12H26) in the vapour phase as the only carbon and energy source. After autoclaving, ASW MM was supplemented with 1 μM vitamin B12, 1 ml trace-element solution SL-10 and 1 ml mixed vitamin solution 141.
Growth rates (μ; h−1) were estimated as described previously (Vetriani et al., 2004). Unless specified otherwise, growth ranges and optimal growth conditions were determined in LS ASW medium. To determine the optimal growth temperature of the new isolate, cultures were incubated between 10 and 45 °C (at 5 °C intervals). All other experiments were carried out at 35 °C, the optimal growth temperature. To determine optimal salt requirements, the concentration of NaCl was varied between 1.0 and 25 % (w/v). The influence of pH on growth was determined between pH 4.5 and 8.0 by using the following buffers at a concentration of 10 mM: acetate at pH 4.5 and 5.0; MES at pH 5.5 and 6.0; PIPES at pH 6.5, 7.0 and 7.5; and Tris at pH 8.0. Anaerobic growth with nitrate as an electron acceptor was tested in ASW medium supplemented with 7.3 mM KNO3 under a N2 atmosphere.
Catalase activity was determined as described previously (Vetriani et al., 2004) and the presence of cytochrome c, a component of the cytochrome oxidase system, was determined according to the protocol described by Kovacs (1956). Escherichia coli K-12 and Pseudomonas aeruginosa were used as negative and positive controls for the cytochrome oxidase test, respectively.
A Biolog GN2 MicroPlate test panel was used to compare the carbon utilization/oxidation profiles of the new isolate with that of the reference strain, Salinisphaera shabanensis DSM 14853T. Confluent growth of both strains was obtained on solid ASW medium overnight. Cells were collected using a sterile cotton swab and resuspended in 15 ml salt solution (l−1: NaCl, 23.5 g; MgCl2 . 6H2O, 10.6 g). The cell suspension was adjusted to an OD600 of 0.3±0.05, supplemented with 5 mM sodium thioglycolate and dispensed (in 150 μl aliquots) to each well of two Biolog GN2 MicroPlates, which were incubated at 35 °C. A change in colour, indicative of the oxidation of the substrate, was monitored for 48 h.
For direct counts, cells were stained routinely with 0.1 % acridine orange and visualized with an Olympus BX 60 microscope with an oil-immersion objective lens (UplanF1 ×100/1.3). Cells for ultrathin sections and for platinum shadowing were prepared as described previously (Vetriani et al., 2004). Motility was determined by phase-contrast microscopy. Gram staining was performed as described elsewhere (Holt et al., 1994).
Genomic DNA was extracted from cells collected by centrifugation using the UltraClean Microbial DNA isolation kit, according to the manufacturer's instructions (MoBio Laboratories). The full-length sequence of the 16S rRNA gene was selectively amplified from the genomic DNA by PCR, cloned, sequenced and subjected to phylogenetic analysis as described previously (Vetriani et al., 2004; Voordeckers et al., 2005).
The genes encoding the enzymes ribulose-1,5-bisphosphate carboxylase/oxygenase form I and II (RuBisCO) and alkane hydroxylase (AlkB, involved in the oxidation of hydrocarbons) were amplified from the genomic DNA of the novel strains and S. shabanensis DSM 14853T. A 500 bp fragment of the gene encoding form I RuBisCO (rbcL/cbbL) from S. shabanensis DSM 14853T was amplified as described previously (Nanba et al., 2004), and its sequence was determined. Internal primers ss rbcLF (5′-GGTCTATGAAAGCGCTCAAGG-3′) and ss rbcLR (5′-ATCCATTTCGAGATCACGCGG-3′) were designed based on the rbcL sequence from S. shabanensis DSM 14853T (using the IDT OligoAnalyser 3.1 program; http://www.idtdna.com), and were used to amplify a 400 bp fragment of the rbcL gene from the three novel strains. The PCR protocol used to amplify the rbcL genes was 5 min at 94 °C followed by 30 cycles of 45 s at 94 °C, 1 min at 50 °C and 45 s at 72 °C, ending with a final extension of 20 min at 72 °C. The PCR for the amplification of a 1040 bp fragment of the gene encoding the form II RuBisCO (cbbM) was carried out as described previously (Elsaied et al., 2007). A 550 bp fragment of the gene encoding alkane hydroxylase (alkB) was amplified selectively from the genomic DNA of S. shabanensis DSM 14853T and of two of the novel strains by PCR, as described previously (Smits et al., 1999). The sequences for rbcL and alkB were determined for both strands and were translated into the respective amino acid sequences using EMBOSS Transeq (http://www.ebi.ac.uk/emboss/transeq). The amino acid sequences were aligned with clustal_x v. 1.8 (Thompson et al., 1997) and adjusted manually using SeaView (Galtier et al., 1996). Phylogenetic distances were calculated using the Observed Divergence matrix and the neighbour-joining method was used to evaluate tree topologies. phylo_win was used to plot tree topologies (Galtier et al., 1996) and their robustness was tested by bootstrap analysis with 1000 resamplings. The determination of total DNA base composition (mol% G+C) was carried out by HPLC (Mesbah et al., 1989).
Enrichment cultures for mesophilic, chemolithoautotrophic, thiosulfate-oxidizing bacteria were obtained by inoculating 10 ml medium 142−A with 1 ml fluid from three different samples collected on the EPR. The temperature of the fluids at the time of collection was 2.5, 6 and 13 °C, respectively. Cultures were incubated at 30 °C. A change in colour of the pH indicator present in the medium, suggesting growth, was observed after 1 or 2 days from the beginning of the incubation and then confirmed by direct cell counting. Three independent cultures showed consistent growth after repeated transfers and were purified by successive isolations of single colonies on solidified medium. The resulting pure cultures were designated strains EPR70T, EPR71 and EPR72. Preliminary phylogenetic analysis of the 16S rRNA gene sequence indicated that the three strains were closely related (sequence identity 99 %). Strain EPR70T was selected for further characterization.
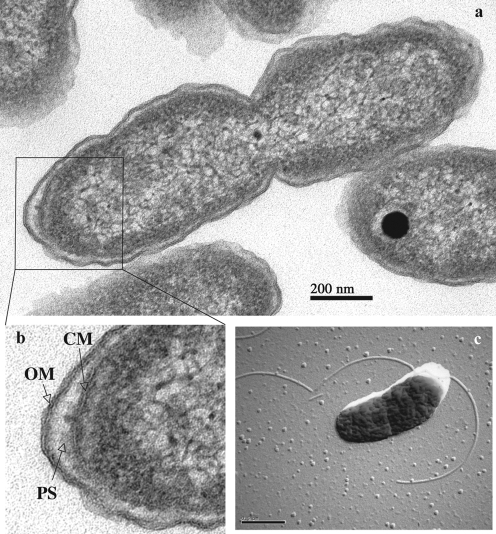
Cells of EPR70T were short rods, 0.8–1.0 μm long and 0.3–0.5 μm wide, and divided by constriction (Fig. 1a). Cells stained Gram-negative. The cell envelope of EPR70T included a cytoplasmic membrane surrounded by a periplasmic space and an outer membrane (Fig. 1b). The organism was motile and possessed one or more flagella, which were observed in electron micrographs of platinum-shadowed cells (Fig. 1c). The presence of endospores was not observed.
Fig. 1.
(a) Electron micrograph of a thin section of cells of strain EPR70T. (b) Details of the cell envelope of strain EPR70T. OM, Outer membrane; PS, periplasmic space; CM, cytoplasmic membrane. (c) Electron micrograph of a platinum-shadowed cell of strain EPR70T showing the presence of flagella. Bars, 200 nm (a) and 0.5 μm (c).
Under strictly autotrophic conditions, EPR70T, EPR71 and EPR72 oxidized Na2S2O3 and acidified the culture medium. Heterotrophic growth occurred when medium 142 was depleted of Na2S2O3 and NaHCO3 and supplemented with 10 mM acetate; under these conditions, strain EPR70T alkalified the culture medium. EPR70T, EPR71 and EPR72 also grew well heterotrophically in ASW, LS ASW and ASW MM supplemented with dodecane as the sole carbon and energy source. Strain EPR70T grew at temperatures between 20 and 40 °C, with optimal growth at 35 °C. No growth was detected at 10 or 45 °C. EPR70T grew at NaCl concentrations between 1.0 and 25.0 % (w/v) with an optimum at 2.5 % (w/v). Growth occurred between pH 5.0 and 7.5 with an optimum at pH 5.5. No growth was observed at pH 4.5 or 8.0. The shortest generation time of strain EPR70T in LS ASW was 42 min (Supplementary Fig. S1, available in IJSEM Online). For comparison, the generation time of strain EPR70T grown under optimal conditions of temperature, salinity and pH and with CO2 as the sole carbon source (in medium 142) was 231 min. Strain EPR70T was a fully aerobic organism, and growth did not occur in medium 142 supplemented with 5 % oxygen (v/v) or in ASW medium with 7.3 mM nitrate.
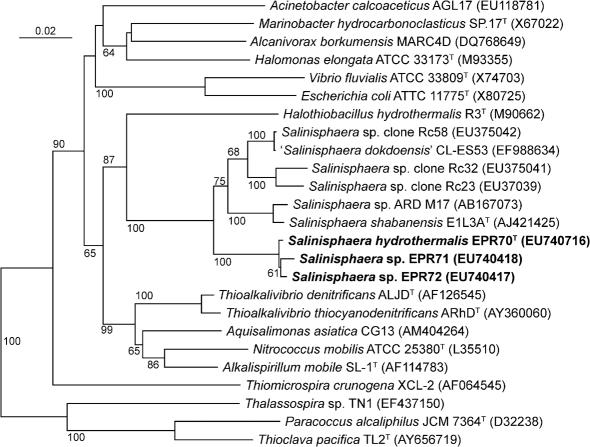
Phylogenetic analysis of the 16S rRNA gene sequence, carried out using the neighbour-joining method, placed EPR70T, EPR71 and EPR72 (98–99 % sequence identity between each sequence) in a unique cluster within the class Gammaproteobacteria (Fig. 2). The closest relatives of these strains were S. shabanensis E1L3AT, a moderate halophile which was isolated from the brine–seawater interface in the Shaban Deep in the Red Sea (94 % sequence identity to the 16S rRNA gene of strain EPR70T) and which is the only formally described member of this genus (Antunes et al., 2003), Salinisphaera sp. strain ARD M17, isolated from deep-sea water from the Knipovich Ridge, in the Arctic Ocean (94 % sequence identity to strain EPR70T) (T. Okamoto and T. Naganuma, unpublished; GenBank accession no. AB167073), and ‘Salinisphaera dokdoensis’ CL-ES53, isolated from the East Sea of Korea (95 % sequence identity to strain EPR70T) (B. C. Cho and others, unpublished; EF988634). While the type species of the genus Salinisphaera, S. shabanensis, was originally described as a heterotroph (Antunes et al., 2003), our study demonstrated that this organism also grew chemolithoautotrophically by thiosulfate oxidation and with n-alkanes as the sole carbon and energy source (Table 1). Interestingly, 16S rRNA gene sequences related to Salinisphaera species were retrieved from the microbial community attached to hydrocarbon-contaminated rocks along the Spanish shoreline, represented in Fig. 2 by clones Rc23, Rc32 and Rc58 (J. Alonso-Gutierrez and others, unpublished).
Fig. 2.
Phylogenetic position of strains EPR70T, EPR71 and EPR72, according to their 16S rRNA gene sequences. The neighbour-joining tree was constructed using phylo_win. Bootstrap values higher than 50 % are indicated. Bar, 2 % estimated base substitutions.
Table 1.
Differentiating features of strain EPR70T and S. shabanensis E1L3AT
Both strains are catalase-positive. Data for S. shabanensis E1L3AT were taken from Antunes et al. (2003) unless indicated.
| Feature | Strain EPR70T | S. shabanensis E1L3AT |
|---|---|---|
| Morphology | Small rods | Cocci |
| Cell size (μm) | 0.8–1.0×0.3–0.5 | 0.7–1.2 (diameter) |
| Oxidase | Negative | Positive |
| Temperature for growth (°C) | ||
| Range | 20–40 | 5–42 |
| Optimum | 30–35 | 30–37 |
| Salinity for growth (% NaCl) | ||
| Range | 1–25 | 1–28 |
| Optimum | 2.5 | 10 |
| pH for growth | ||
| Range | 5.0–7.5 | 4.0–8.0 |
| Optimum | 5.5 | 6.5–7.5 |
| Chemolithoautotrophic growth | Yes | Yes* |
| Growth on dodecane | Yes | Yes* |
| Anaerobic growth in ASW plus nitrate | No | Yes |
| DNA G+C content (mol%) | 64.0 | 61.8 |
*Data for S. shabanensis DSM 14853T obtained in this study.
High bootstrap values supported the branching topology of the EPR strains relative to the other strains (Fig. 2). The G+C content of the genomic DNA of strain EPR70T, determined by HPLC analysis of deoxyribonucleosides, was 64.0 mol%, while that of S. shabanensis E1L3AT was 61.8 mol% (Antunes et al., 2003).
Comparative analyses of strain EPR70T and S. shabanensis DSM 14853T revealed morphological (rods and cocci, respectively) and physiological differences (Table 1). In particular, EPR70T had lower salinity and pH optima than S. shabanensis DSM 14853T (2.5 vs 10 % NaCl and pH 5.5 vs ∼pH 7.0, respectively), suggesting specific adaptations to the slightly acidic vent fluids, and strain EPR70T could not grow anaerobically in the presence of nitrate as a terminal electron acceptor (Table 1). The metabolic fingerprints of strain EPR70T and S. shabanensis DSM 14853T, determined using a Biolog assay on a GN2 MicroPlate, showed that both bacteria were able to oxidize a wide range of carbon sources, but some differences were evident (Table 2). For instance, EPR70T oxidized sugars and sugar alcohols preferentially, while S. shabanensis preferred amino acid derivatives.
Table 2.
Comparative metabolic profiles of strain EPR70T and S. shabanensis DSM 14853T
Both strains oxidized Tweens 20 and 80, d-mannitol, d-mannose, xylitol, pyruvic acid methyl ester, succinic acid monomethyl ester, γ-hydroxybutyric acid, dl-lactic acid, succinic acid and l-alanine. Data were obtained in this study using the Biolog GN2 MicroPlate.
| Carbon source | Strain EPR70T | S. shabanensis DSM 14853T |
|---|---|---|
| Glycogen | − | + |
| Adonitol | + | − |
| d-Arabitol | + | − |
| i-Erythritol | + | − |
| d-Fructose | + | − |
| d-Galactose | + | − |
| α-d-Glucose | + | − |
| l-Rhamnose | + | − |
| d-Sorbitol | + | − |
| cis-Aconitic acid | − | + |
| Formic acid | − | + |
| α-Ketobutyric acid | + | − |
| α-Ketoglutaric acid | − | + |
| Bromosuccinic acid | − | + |
| Succinamic acid | + | − |
| l-Alanyl glycine | − | + |
| l-Asparagine | − | + |
| l-Aspartic acid | − | + |
| l-Glutamic acid | − | + |
| Glycyl l-aspartic acid | − | + |
| Glycyl l-glutamic acid | − | + |
| l-Proline | + | − |
| γ-Aminobutyric acid | + | − |
| Glycerol | + | − |
In order to investigate the carbon-fixation pathway in the EPR strains and in S. shabanensis DSM 14853T, we carried out PCR amplification of the genes encoding both RuBisCO forms I (rbcL/cbbL) and II (cbbM) for all strains, and a product was obtained only for the rbcL/cbbL gene (encoding RuBisCO form I). The amino acid identity among the RuBisCO sequences of the three EPR strains was 98–99 %, while the sequence identity between these enzymes and the RuBisCO from S. shabanensis DSM 14853T ranged from 94 to 95 %. Phylogenetic analysis of the amino acid sequence of the form I RuBisCO of strains EPR70T, EPR71 and EPR72 and S. shabanenesis DSM 14853T, carried out using the neighbour-joining method, showed that these sequences formed a unique cluster related to other form I, type C enzymes (Supplementary Fig. S2; Xu & Tabita, 1996). The closest relatives of the RuBisCO from the EPR strains and S. shabanenesis DSM 14853T were the enzymes of the methylotrophic bacterium Methylibium petroleiphilum PM1T and of the ammonia-oxidizing bacteria Nitrosospira multiformis ATCC 25196T and Nitrosococcus oceani ATCC 19707T (Supplementary Fig. S2). It is worth noting that the RuBisCO enzymes from the EPR strains were also related to a group of sequences retrieved from natural microbial communities associated with the plumes of black smokers located in the Western Pacific arc hydrothermal vent system, represented in Supplementary Fig. S2 by clones ICS1 and ICP1 (Elsaied et al., 2007).
Since both the EPR strains and S. shabanensis DSM 14853T can use n-alkanes as their sole carbon and energy source, we investigated the presence in these organisms of the alkB gene, which encodes alkane hydroxylase (AlkB), an enzyme that catalyses the first step in the oxidation of hydrocarbons. Phylogenetic analysis of the amino acid sequence of the AlkB enzyme from strains EPR70T, EPR71 and S. shabanensis DSM 14853T placed these enzymes in a discrete cluster related to the alkane hydroxylase of Nocardia farcinia IFM 10152 (Supplementary Fig. S3). The identity between the AlkB amino acid sequences of EPR70T and EPR71 was 90 %, while the sequence identities of the enzymes from the two EPR strains and the alkane hydroxylase from S. shabanensis DSM 14853T were 62 and 63 %, respectively.
Physiological and phylogenetic analyses indicated that strain EPR70T and S. shabanensis DSM 14853T are not related at the species level, and therefore EPR70T represents a new species within the genus Salinisphaera, for which we propose the name Salinisphaera hydrothermalis sp. nov.
At deep-sea hydrothermal vents, micro-organisms must adapt to highly dynamic environmental conditions, where there are fluctuations in temperature, salinity and nutrient availability (Karl, 1995). The presence of the rbcL/cbbL gene, which encodes RuBisCO form I, in all the Salinisphaera strains suggests strongly that these bacteria use the Calvin–Benson–Bassham cycle to fix CO2. The metabolic versatility of S. hydrothermalis, which can fix CO2 and use a wide range of organic carbon sources, may be an advantage for its growth and survival in these environments. In particular, the ability of S. hydrothermalis to grow autotrophically, to oxidize n-alkanes (which are enriched in hydrothermal fluids; Brault et al., 1988) and to grow optimally at a slightly acidic pH suggests that this bacterium is particularly well suited to thrive in the moderate-temperature fluids that are typically emitted by diffuse-flow deep-sea hydrothermal vents.
Emended description of the genus Salinisphaera Antunes et al. 2003
Cells are Gram-negative cocci or short rods, possessing one or more flagella. Mesophilic, halotolerant and catalase-positive. Aerobic or facultatively anaerobic. Growth occurs chemolithoautotrophically with thiosulfate as an electron donor and oxygen as an electron acceptor, heterotrophically on n-alkanes (dodecane) as the sole carbon and energy source or on complex media. Known strains have been found in marine environments. The type species is Salinisphaera shabanensis Antunes et al. 2003.
Description of Salinisphaera hydrothermalis sp. nov.
Salinisphaera hydrothermalis (hy.dro.ther.ma′lis. N.L. fem. adj. hydrothermalis hydrothermal, pertaining to a hydrothermal vent).
Cells are small rods (0.8–1.0 μm long, 0.3–0.5 μm wide), which are motile by means of one or more flagella. Obligate aerobe. Oxidase-negative. Growth occurs at 20–40 °C (optimum 30–35 °C), 1–25 % NaCl (optimum 2.5 %) and pH 5.0–7.5 (optimum pH 5.5). The shortest generation time observed is 42 min. Growth occurs under aerobic, chemolithoautotrophic conditions in the presence of thiosulfate and CO2. Heterotrophic growth occurs with acetate or n-alkanes as sole carbon and energy sources and in complex ASW medium. Nitrate is not used as an electron acceptor. The DNA G+C content of the type strain is 64.0 mol% as determined by the HPLC method.
The type strain is EPR70T (=DSM 21483T =JCM 15514T), which was isolated from diffuse-flow hydrothermal vent fluids collected from the East Pacific Rise at ° 50′ N.
Supplementary Material
Acknowledgments
We wish to thank Amber G. Jensen for excellent technical assistance. We thank the crew of the R/V Atlantis and the crew and pilots of the DSV Alvin for their skilled operation at sea. This research was supported by NSF grant OCE 03-27353 to C. V. and R. A. L., NSF grant MCB 04-56676 to C. V., NSF grant EAR 04-33793 to T. B., and by NIH R25GM-58389 and an NSF Graduate Research Fellowship to M. C.-M.
Abbreviations
EPR, East Pacific Rise
RuBisCO, ribulose-1,5-bisphosphate carboxylase/oxygenase
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequences of strains EPR70T, EPR72 and EPR71 are EU740416–EU740418. The accession numbers for the partial rbcL gene sequences from strains EPR70T, EPR71 and EPR72 and S. shabanensis DSM 14853T are EU740422–EU740425 and those for the partial alkB gene sequences of strain EPR70T, S. shabanensis DSM 14853T and strain EPR71 are EU740419–EU740421.
Graphs showing the effects of temperature, pH and NaCl concentration on growth of strain EPR70T and neighbour-joining trees based on amino acid sequences of RuBisCO form I and AlkB are available as supplementary material with the online version of this paper.
References
- Antunes, A., Eder, W., Fareleira, P., Santos, H. & Huber, R. (2003). Salinisphaera shabanensis gen. nov., sp. nov., a novel, moderately halophilic bacterium from the brine-seawater interface of the Shaban Deep, Red Sea. Extremophiles 7, 29–34. [DOI] [PubMed] [Google Scholar]
- Brault, M., Simoneit, B. R. T., Marty, J. C. & Saliot, A. (1988). Hydrocarbons in waters and particulate material from hydrothermal environments at the East Pacific Rise, 1 ° N. Org Geochem 12, 209–219. [Google Scholar]
- Durand, P., Reysenbach, A. L., Prieur, D. & Pace, N. (1993). Isolation and characterization of Thiobacillus hydrothermalis sp. nov., a mesophilic obligately chemolithotrophic bacterium isolated from a deep-sea vent in Fiji Basin. Arch Microbiol 159, 39–44. [Google Scholar]
- Elsaied, H. E., Kimura, H. & Naganuma, T. (2007). Composition of archaeal, bacterial, and eukaryal RuBisCO genotypes in three Western Pacific arc hydrothermal vent systems. Extremophiles 11, 191–202. [DOI] [PubMed] [Google Scholar]
- Galtier, N., Gouy, M. & Gautier, C. (1996). SeaView and phylo_win, two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci 12, 543–548. [DOI] [PubMed] [Google Scholar]
- Holt, J. G., Krieg, N. R., Sneath, P. H. A., Staley, J. T. & Williams, S. T. (1994). Bergey's Manual of Determinative Bacteriology, 9th edn. Baltimore: Williams & Wilkins.
- Jannasch, H. W. (1995). Microbial interactions with hydrothermal fluids. In Seafloor Hydrothermal Systems: Physical, Chemical, Biological and Geological Interactions, pp. 273–296. Edited by S. E. Humphris, R. A. Zierenberg, L. S. Mullineaux & R. E. Thomson. Washington, DC: American Geophysical Union.
- Jannasch, H. W., Wirsen, C. O., Nelson, D. C. & Robertson, L. A. (1985). Thiomicrospira crunogena sp. nov., a colorless sulfur-oxidizing bacterium from a deep-sea hydrothermal vent. Int J Syst Bacteriol 35, 422–424. [Google Scholar]
- Karl, D. M. (1995). Ecology of free-living, hydrothermal vent microbial communities. In The Microbiology of Deep-Sea Hydrothermal Vents, pp. 35–124. Edited by D. M. Karl. Boca Raton, FL: CRC Press.
- Kaye, J. Z. & Baross, J. A. (2000). High incidence of halotolerant bacteria in Pacific hydrothermal-vent and pelagic environments. FEMS Microbiol Ecol 32, 249–260. [DOI] [PubMed] [Google Scholar]
- Kaye, J. Z., Marquez, M. C., Ventosa, A. & Baross, J. A. (2004). Halomonas neptunia sp. nov., Halomonas sulfidaeris sp. nov., Halomonas axialensis sp. nov. and Halomonas hydrothermalis sp. nov.: halophilic bacteria isolated from deep-sea hydrothermal-vent environments. Int J Syst Evol Microbiol 54, 499–511. [DOI] [PubMed] [Google Scholar]
- Kovacs, N. (1956). Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature 178, 703. [DOI] [PubMed] [Google Scholar]
- McCollom, T. M. & Shock, E. L. (1997). Geochemical constraints on chemolithoautotrophic metabolisms by microorganisms in seafloor hydrothermal systems. Geochim Cosmochim Acta 61, 4375–4391. [DOI] [PubMed] [Google Scholar]
- Mesbah, M., Premachandran, U. & Whitman, W. B. (1989). Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39, 159–167. [Google Scholar]
- Nanba, K., King, G. M. & Dunfield, K. (2004). Analysis of facultative lithotroph distribution and diversity on volcanic deposits by use of the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase. Appl Environ Microbiol 70, 2245–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raguénès, G., Christen, R., Guezennec, J., Pignet, P. & Barbier, G. (1997). Vibrio diabolicus sp. nov., a new polysaccharide-secreting organism isolated from a deep-sea hydrothermal vent polychaete annelid, Alvinella pompejana. Int J Syst Bacteriol 47, 989–995. [DOI] [PubMed] [Google Scholar]
- Ruby, E. G. & Jannasch, H. W. (1982). Physiological characteristics of Thiomicrospira sp. strain L-12 isolated from deep-sea hydrothermal vents. J Bacteriol 149, 161–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby, E. G., Wirsen, C. O. & Jannasch, H. W. (1981). Chemolithotrophic sulfur-oxidizing bacteria from the Galapagos rift hydrothermal vents. Appl Environ Microbiol 42, 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Colin, C., Raguénès, G., Cozien, J. & Guezennec, J. G. (2008). Halomonas profundus sp. nov., a new PHA-producing bacterium isolated from a deep-sea hydrothermal vent shrimp. J Appl Microbiol 104, 1425–1432. [DOI] [PubMed] [Google Scholar]
- Smits, T. H. M., Röthlisberger, M., Witholt, B. & van Beilen, J. B. (1999). Molecular screening for alkane hydroxylase genes in Gram-negative and Gram positive strains. Environ Microbiol 1, 307–317. [DOI] [PubMed] [Google Scholar]
- Takai, K., Hirayama, H., Nakagawa, T., Suzuki, Y., Nealson, K. H. & Horikoshi, K. (2004). Thiomicrospira thermophila sp. nov., a novel microaerobic, thermotolerant, sulfur-oxidizing chemolithomixotroph isolated from a deep-sea hydrothermal fumarole in the TOTO caldera, Mariana Arc, Western Pacific. Int J Syst Evol Microbiol 54, 2325–2333. [DOI] [PubMed] [Google Scholar]
- Teske, A., Brinkhoff, T., Muyzer, G., Moser, D. P., Rethmeier, J. & Jannasch, H. W. (2000). Diversity of thiosulfate-oxidizing bacteria from marine sediments and hydrothermal vents. Appl Environ Microbiol 66, 3125–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997). The clustal_x windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetriani, C., Speck, M. D., Ellor, S. V., Lutz, R. A. & Starovoytov, V. (2004). Thermovibrio ammonificans sp. nov., a thermophilic, chemolithotrophic, nitrate-ammonifying bacterium from deep-sea hydrothermal vents. Int J Syst Evol Microbiol 54, 175–181. [DOI] [PubMed] [Google Scholar]
- Vetriani, C., Chew, Y. S., Miller, S. M., Yagi, J., Coombs, J., Lutz, R. A. & Barkay, T. (2005). Mercury adaptation among bacteria from a deep-sea hydrothermal vent. Appl Environ Microbiol 71, 220–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordeckers, J. W., Starovoytov, V. & Vetriani, C. (2005). Caminibacter mediatlanticus sp. nov., a thermophilic, chemolithoautotrophic, nitrate-ammonifying bacterium isolated from a deep-sea hydrothermal vent on the Mid-Atlantic Ridge. Int J Syst Evol Microbiol 55, 773–779. [DOI] [PubMed] [Google Scholar]
- Xu, H. H. & Tabita, F. R. (1996). Ribulose-1,5-bisphosphate carboxylase/oxygenase gene expression and diversity of Lake Erie planktonic microorganisms. Appl Environ Microbiol 62, 1913–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.