Abstract
Isolates of Klebsiella pneumoniae harbouring the carbapenemase KPC may have carbapenem MICs that remain in the susceptible range, and may therefore go unrecognized. To understand the mechanisms contributing to the variability in carbapenem MICs, 20 clinical isolates, all belonging to either of two clonal groups of KPC-possessing K. pneumoniae endemic to New York City, were examined. Expression of genes encoding KPC, the porins OmpK35 and OmpK36, and the efflux pump AcrAB was examined by real-time RT-PCR. Outer-membrane profiles of selected KPC-producing isolates were examined by SDS-PAGE, and proteins were identified by matrix-assisted laser desorption/ionization mass spectrometry. The identification of SHV and TEM β-lactamases and the genomic sequences of ompK35 and ompK36 were determined by PCR and DNA sequencing, respectively. For one clonal group, carbapenem MICs increased with decreasing expression of ompK36. A second clonal group also had carbapenem MICs that correlated with ompK36 expression. However, all of the isolates in this latter group continued to produce OmpK36, suggesting that porin configuration may affect entry of carbapenems. For isolates that had the greatest expression of ompK36, carbapenem MICs tended to be lower when determined by the broth microdilution technique, and scattered colonies were seen around the Etest zones of inhibition. All of the KPC-producing isolates were highly resistant to ertapenem, regardless of ompK36 expression. In conclusion, isolates of KPC-possessing K. pneumoniae that express ompK36 tend to have lower MICs to carbapenems and therefore may be more difficult to detect by clinical laboratories. Regardless of ompK36 expression, all of the KPC producers were consistently resistant to ertapenem.
INTRODUCTION
Klebsiella pneumoniae possessing the carbapenemase KPC is now widespread in many medical centres in the north-eastern USA and is being increasingly reported worldwide (Bratu et al., 2005a; Landman et al., 2007; Queenan & Bush, 2007). The progressive spread of this β-lactamase, which is an efficient cephalosporinase and carbapenemase, can be partly explained by the difficulty clinical laboratories have in detecting isolates harbouring this enzyme. When carbapenem resistance is used as the gauge to identify KPC-possessing K. pneumoniae, a significant proportion will not be detected as they may have MICs that are below the breakpoint for resistance. In particular, automated systems may frequently misidentify these isolates as susceptible to carbapenems (Anderson et al., 2007; Bratu et al., 2005b; Tenover et al., 2006). The reason for the variability in carbapenem MICs, particularly with imipenem and meropenem, is unknown.
It appears that there may be several factors besides carbapenemases that can contribute to carbapenem resistance in isolates of K. pneumoniae. Although unusual, K. pneumoniae lacking an efficient carbapenemase can still achieve resistance to carbapenems. These isolates typically possess an underlying extended-spectrum β-lactamase (ESBL) or AmpC-type β-lactamase (Bradford et al., 1997; Cao et al., 2000; Crowley et al., 2002). In addition to a background of β-lactamase activity, alterations in porin production are an important contributor to carbapenem resistance. Many ESBL-carrying clinical isolates of K. pneumoniae lack OmpK35 (Hernández-Allés et al., 1999a), which augments resistance to ceftazidime (Doménech-Sánchez et al., 2003). The additional loss of OmpK36 leads to reduced susceptibility to cefotaxime, cephamycins and carbapenems (Ardanuy et al., 1998; Crowley et al., 2002; Doménech-Sánchez et al., 2000, 2003; Martínez-Martínez et al., 1996, 1999). Ertapenem may be particularly affected by the concomitant loss of OmpK35 and OmpK36 (Elliott et al., 2006; Jacoby et al., 2004; Mena et al., 2006; Woodford et al., 2007). Porin loss has been correlated with disruption of ompK35 and ompK36 by insertions or deletions in some isolates (Cai et al., 2008; Hernández-Allés et al., 1999b; Kaczmarek et al., 2006; Mena et al., 2006). In the small number of isolates of KPC-producing K. pneumoniae that have been studied, loss of OmpK35 (Woodford et al., 2004; Yigit et al., 2001) or OmpK36 (Cai et al., 2008) has been observed. Finally, the AcrAB efflux system has been shown to affect a wide range of antimicrobial agents, including tetracyclines, chloramphenicol, trimethoprim and fluoroquinolones (Nikaido, 1996). Whether antibiotic efflux via this system is an important contributor to carbapenem resistance is unknown.
In this report, the mechanisms contributing to carbapenem resistance in clonal groups of KPC-possessing K. pneumoniae endemic to New York City were examined.
METHODS
Twenty clinical isolates of K. pneumoniae obtained from prior surveillance studies were chosen because they belong to important blaKPC-possessing ribotypes endemic to our region (Bratu et al., 2005a, b; Landman et al., 2007). Susceptibility testing was performed using the Etest method (AB Biodisk); for isolates possessing blaKPC, susceptibility testing for the carbapenems was also performed using the broth microdilution method (CLSI, 2006). For isolates possessing blaKPC, carbapenem hydrolysis of crude cell extracts in 100 mM phosphate buffer was measured spectrophotometrically, as described previously (Woodford et al., 2004). Student's t-test was used to compare hydrolysis rates between isolates; a two-tailed P value of ≤0.05 was considered significant.
DNA amplification studies.
To identify conserved regions, genetic sequences for a ribosomal housekeeping gene and for the target genes ompK35, ompK36 and acrB were amplified and sequenced using the primers described in Table 1. DNA sequencing was performed using an automated fluorescent dye terminator sequencing system (Applied Biosystems) and analysed using blast. Plasmids were isolated by the alkaline lysis method (Sambrook & Russell, 2001). Plasmids were transformed into One Shot TOP10 Electrocomp Escherichia coli (Invitrogen) by electroporation. Plasmid DNA from the transformants was then screened for the presence of TEM, SHV and KPC β-lactamases, and identified using previously established primers and conditions (Bratu et al., 2005a; Paterson et al., 2003). Selected isolates were also examined for the presence of ACT-1 (Bradford et al., 1997).
Table 1.
Primers and probes used for PCR amplification and real-time RT-PCR studies
| Primer/probe | Sequence (5′→3′) |
|---|---|
| DNA amplification and sequencing | |
| ribofor | CAGCCACACTGGAACTGAGA |
| riboRev | TTATGAGGTCCGCTTGCTCT |
| ompK35for | AACTTATTGACGGCAGTGGC |
| ompK35rev | TTGGTAAACGATACCCACGG |
| ompK36for | GCAGTGGCATAATAAAAGGCA |
| ompK36rev | ACTGGTAAACCAGGCCCAG |
| acrBfor | TCAAACCAGGTGTGCAGGTA |
| acrBrev | TTAATACCCAGACCGGATGC |
| RT-PCR | |
| ribofor | GAAGAAGCACCGGCTAACTC |
| riborev | CACATCCGACTTGACAGACC |
| riboprobe | FAM-TGCCAGCAGCCGCGGTAATA-TAMRA |
| ompK35for | GTCTGGACCACCAATGGC |
| ompK35rev | GATCTGAGTTTCGCCTTTCA |
| ompK35probe | FAM-CCACCTATGCCCGTATCGGCC-TAMRA |
| ompK36for | GACCAGACCTACATGCGTGTA |
| ompK36rev | GTATTCCCACTGGCCGTAAC |
| ompK36probe | FAM-TGGGTTTCGCCTTTCACGCC-TAMRA |
| kpcfor | CGTGACGGAAAGCTTACAAA |
| kpcrev | AGCCAATCAACAAACTGCTG |
| kpcprobe | FAM-CTGGGCTCTGCACTGGCTGC-TAMRA |
| acrBfor | CAATACGGAAGAGTTTGGCA |
| acrBrev | CAGACGAACCTGGGAACC |
| acrBprobe | FAM-TCCTGGTTCACCTTCAGCAGGATG-TAMRA |
RT-PCR studies.
Isolates were examined by real-time RT-PCR for expression of ompK35, ompK36, blaKPC and acrB. DNase-treated RNA was obtained from late-exponential-phase cultures using an RNeasy kit (Qiagen), and 25 ng RNA was used for each sample. Primer and probe concentrations were adjusted to achieve efficiencies of 90–110 % and all experiments were performed in triplicate. The primers and probes used in these experiments are given in Table 1. Expression of each gene was normalized to that of a ribosomal housekeeping gene (rrsA). The relative expression of ompK35 and ompK36 was then calibrated against the corresponding expression by K. pneumoniae ATCC 13883 (an isolate with known expression of both porins). Expression of acrB was calibrated against a susceptible control isolate, K. pneumoniae ATCC 11296. Expression of blaKPC was initially calibrated against a previously described transformant possessing a plasmid with blaKPC-2 (Bratu et al., 2007); the clinical isolate of K. pneumoniae with the lowest expression was then considered the control. For all controls, relative expression was equal to 1.0.
Isolation and characterization of outer-membrane proteins.
Selected isolates had their outer-membrane proteins analysed by SDS-PAGE using standard techniques (Albertí et al., 1995; Hernández-Allés et al., 1999a). Briefly, isolates were grown in nutrient broth, sonicated and centrifuged. Cell membranes were obtained following centrifugation at 100 000 g, and extracted with 1 % N-lauroylsarcosine. Protein concentrations were measured using the Bradford method, and each well contained ∼4 μg total protein. Samples were boiled in Laemmli's sample buffer. SDS-PAGE was performed with a 10 % acrylamide gel, which was then stained with Coomassie blue, and selected proteins were identified using matrix-assisted laser desorption/ionization (MALDI) MS. Peptide sequences were compared with published outer-membrane proteins of K. pneumoniae.
RESULTS AND DISCUSSION
Isolates belonging to clonal group A
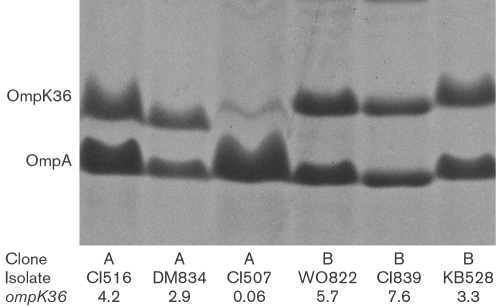
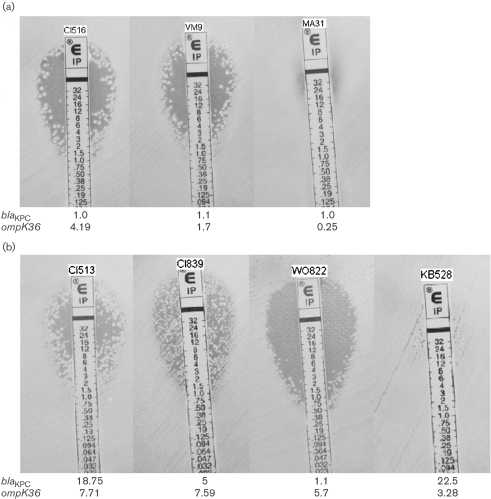
Eleven isolates belonged to clonal group A, a strain responsible for ∼85 % of blaKPC-possessing isolates in our region (Bratu et al., 2005a). Five isolates did not harbour blaKPC but did have ESBLs (either blaSHV-12-like or blaSHV-11-like) and had reduced susceptibility to ceftazidime (Table 2). The discrepancy in cephalosporin susceptibility for isolates CI504 and CI505 could not be explained by the information gathered from each isolate. All five had reduced expression of ompK35, and DNA sequencing revealed a frameshift mutation in this gene. All five expressed ompK36 and, consistent with other reports, remained susceptible to cefoxitin and carbapenems (Ardanuy et al., 1998; Crowley et al., 2002; Doménech-Sánchez et al., 2000, 2003; Martínez-Martínez et al., 1996). Six isolates in this clonal group possessed blaKPC-2 (along with other β-lactamases). Despite variable expression of ompK35, all of these isolates possessed the same frameshift mutation, and OmpK35 was not evident by SDS-PAGE (Fig. 1) and MALDI MS. DNA sequencing revealed that the ompK36 gene in this group was closely related to that found in the published genome of isolate MGH 78578. Expression of ompK36 was variable in these six isolates and correlated with findings by SDS-PAGE (Fig. 1). Isolates with evidence of OmpK36 production had lower MICs to imipenem when determined by broth microdilution (Table 2) and had scattered colonies within the Etest zone of inhibition (Fig. 2a). All six were resistant to meropenem and ertapenem (Table 2). Expression of acrB did not correlate with the MICs of the carbapenems (data not shown).
Table 2.
Presence of other β-lactamases, expression by real-time RT-PCR of blaKPC, ompK35 and ompK36 and MICs for β-lactams from clinical isolates belonging to clonal groups A and B
| Isolate | β-Lactamase(s) | Presence of blaKPC-2 | Relative expression of:* | MIC (mg l−1)† | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| blaKPC | ompK35 | ompK36 | IPM* | MEM | ERT | FOX | CAZ | FEP | |||
| Clonal group A | |||||||||||
| CI504 | SHV-11 | No | 0.54 | 4.58 | 0.19 | 0.032 | 0.064 | 0.5 | >256 | 3 | |
| CI505 | SHV-11 | No | 0.24 | 1.31 | 0.25 | 0.032 | 0.064 | 0.5 | 4 | 0.5 | |
| CI518 | SHV-12 | No | 0.13 | 1.01 | 0.5 | 0.064 | 0.047 | 0.75 | >256 | 2 | |
| VM522 | SHV-12 | No | 0.18 | 0.88 | 0.5 | 0.032 | 0.047 | 0.75 | >256 | 3 | |
| MA549 | SHV-12 | No | 0.18 | 0.87 | 0.38 | 0.047 | 0.38 | 2 | >256 | 8 | |
| CI516 | SHV-11; TEM-1 | Yes | 1 | 2.74 | 4.19 | 12 (8) | 16 (32) | 24 (32) | 48 | 128 | 16 |
| DM834 | SHV-12; TEM-1 | Yes | 1.25 | 0.54 | 2.94 | >32 (8) | >32 (>32) | >32 (>32) | 256 | >256 | 24 |
| VM9 | SHV-11; TEM-1 | Yes | 1.1 | 0.49 | 1.7 | 12 (16) | 24 (16) | 24 (>32) | 48 | >256 | 192 |
| MA31 | SHV-12 | Yes | 1 | 0.06 | 0.25 | >32 (32) | 24 (32) | >32 (>32) | 64 | >256 | 24 |
| CI507 | SHV-11; TEM-1 | Yes | 3.75 | 0.17 | 0.06 | >32 (512) | >32 (>32) | >32 (>32) | >256 | >256 | 192 |
| KC850 | SHV-12; TEM-1 | Yes | 2.5 | 0.08 | 0.004 | >32 (1024) | >32 (>32) | >32 (>32) | >256 | >256 | 128 |
| Clonal group B | |||||||||||
| CI512 | SHV-12; TEM-1 | No | 2.5 | 8.22 | 0.5 | 0.047 | 0.064 | 0.75 | >256 | 24 | |
| CI511 | SHV-12; TEM-1 | No | 1.25 | 3.11 | 0.5 | 0.125 | 0.5 | 4 | >256 | 16 | |
| KB420 | TEM-1 | No | 1.45 | 3.03 | 0.12 | 0.125 | 0.38 | 3 | 128 | 6 | |
| CI806 | SHV-12; TEM-1 | No | 1.11 | 1.81 | 0.25 | 0.125 | 0.75 | 4 | >256 | 16 | |
| CI513 | SHV-12; TEM-1 | Yes | 18.75 | 0.94 | 7.71 | 24 (8) | 6 (16) | 24 (32) | 128 | >256 | 96 |
| CI839 | SHV-12; TEM-1 | Yes | 5 | 0.37 | 7.59 | >32 (2) | >32 (>32) | >32 (>32) | 64 | >256 | 48 |
| WO822 | SHV-12; TEM-1 | Yes | 1.1 | 1.58 | 5.7 | 4 (8) | 24 (8) | >32 (>32) | 32 | >256 | 256 |
| WO555 | SHV-12; TEM-1 | Yes | 5 | 1.8 | 4.47 | >32 (32) | >32 (>32) | >32 (>32) | >256 | >256 | 96 |
| KB528 | SHV-11; TEM-1 | Yes | 22.5 | 0.16 | 3.28 | >32 (256) | >32 (>32) | >32 (>32) | >256 | >256 | 256 |
*Relative expression compared with control (=1.0).
†IPM, imipenem; MEM, meropenem; ERT, ertapenem; FOX, cefoxitin; CAZ, ceftazidime; FEP, cefepime. Carbapenem MICs in parentheses were determined by the broth microdilution method.
Fig. 1.
Outer-membrane protein analysis of isolates of K. pneumoniae by SDS-PAGE. The clonal group, isolate name and relative expression of ompK36 by real-time RT-PCR are provided for each lane. Labelled proteins were identified by MALDI MS.
Fig. 2.
Imipenem Etest images of isolates with variable expression of blaKPC and ompK36. (a) Isolates from clonal group A; (b) isolates from clonal group B. The relative expression of each gene for the isolate is given below its image.
Isolates belonging to clonal group B
Nine isolates belonged to clonal group B, a second important blaKPC-possessing strain in our region (Bratu et al., 2005a). Four ESBL-possessing isolates (lacking blaKPC) expressed ompK36 and remained susceptible to cefoxitin (and resistant to ceftazidime; Table 2). Five isolates possessed blaKPC-2. Although most had evidence of transcription of ompK35 by real-time RT-PCR, production of OmpK35 was not evident by SDS-PAGE (Fig. 1), suggesting post-transcriptional events affecting this porin. All of the isolates in this group had ample expression of ompK36, and this was also evident by SDS-PAGE (Fig. 1). Except for a 2 aa insertion (Gly-Asp) in the region of loop 3 of the porin, OmpK36 for isolates in this group was similar to that found in the susceptible control, K. pneumoniae ATCC 13883. Compared with OmpK36 found in clonal A group, the protein in this group had a 7 aa insert in the loop 6 region of the porin (in addition to several amino acid substitutions). Despite production of OmpK36 in these five isolates with blaKPC, it is noteworthy that decreasing expression of ompK36 still affected the broth microdilution MICs and Etest zones of inhibition of imipenem (Table 2, Fig. 2b). It appears that, despite OmpK36 production in this group, subtle quantitative or qualitative changes in the porin affected entry of β-lactams. It is also noteworthy that the meropenem, ertapenem and cefoxitin MICs in the group B isolates lacking blaKPC were higher than those seen in the comparable group A isolates. Therefore, it appears that the OmpK36 phenotype in the group B isolates affected entry of β-lactams. Finally, the expression of acrB did not correlate with carbapenem resistance for isolates in this group (data not shown).
There appeared to be interplay between expression of blaKPC and ompK36 that affected the MICs to imipenem. Compared with isolate WO822, isolates CI513 and CI839, which had greater expression of blaKPC and ompK36, had substantially higher imipenem MICs (Fig. 2b). Carbapenem hydrolysis studies supported the observations of the blaKPC expression studies. Compared with isolates with an expression level of 1–1.25 times the control expression of blaKPC, isolates with a ≥2.5-fold level of expression tended to have increased spectrophotometric hydrolysis rates of imipenem [0.22±0.16 vs 0.13±0.07 μg (mg protein)−1 min−1] and of meropenem [0.12±0.09 vs 0.09±0.04 μg (mg protein)−1 min−1], and significantly increased hydrolysis rates of ertapenem [0.09±0.06 vs 0.05±0.02 μg (mg protein)−1 min−1, P=0.05].
Expression of ompK35 and ompK36 was also assessed in four unrelated cephalosporin-susceptible clinical isolates. Compared with the control strain, K. pneumoniae ATCC 13883, expression of ompK35 was similar (mean 1.9 times the control, range 0.43–4.4) in these susceptible isolates. However, expression of ompK36 was increased compared with the control isolate (mean 4.4 times the control, range 2.0–5.4). Therefore, although K. pneumoniae ATCC 13883 is frequently used as a control in studies examining outer-membrane proteins, expression of the genes encoding the porins in this isolate is reduced compared with wild-type clinical isolates.
The rapid and, in some regions, unabated spread of K. pneumoniae with KPC β-lactamases has been especially troubling. Clinical laboratories have relied on carbapenem susceptibility to identify pathogens harbouring these enzymes; however, it has been well documented that many isolates with KPC β-lactamases will have MICs of carbapenems below the CLSI-defined breakpoint for susceptibility (Anderson et al., 2007; Bratu et al., 2005b; Tenover et al., 2006). In this report, involving important nosocomial strains of K. pneumoniae endemic to New York City, the MICs of carbapenems were found to be dependent on an interaction between the presence of blaKPC and expression of ompK36. It was also apparent that the OmpK36 phenotype may influence susceptibility. Isolates that abundantly expressed ompK36 tended to have lower imipenem MICs by the broth microdilution method and had scattered colonies within the Etest zone of inhibition. Consistent with other reports, the MICs of ertapenem were consistently higher than those of imipenem and meropenem for the isolates possessing blaKPC (Anderson et al., 2007; Bratu et al., 2005b). Isolates that had MICs of imipenem and meropenem in the susceptible or intermediate range continued to be frankly resistant to ertapenem. As all of our examined isolates had either diminished or defective production of OmpK35 (typical of many ESBL-possessing strains), the contribution of this porin to resistance involving ertapenem cannot be assessed in this report. However, prior studies have not found this porin to be important for the development of carbapenem resistance (Hernández-Allés et al., 1999a). Other studies have found that restoring OmpK36 and OmpK37 improved susceptibility to ertapenem in KPC-producing isolates of K. pneumoniae (Jacoby et al., 2004). However, OmpK37 is often not expressed (Doménech-Sánchez et al., 1999), and its role in carbapenem resistance in clinical isolates is unknown.
Although cephamycins retain activity against many ESBL-producing isolates of K. pneumoniae, they are often not recommended as therapeutic agents due to the development of resistance (secondary to diminished OmpK36 production). A similar caution should be exercised for KPC-producing isolates that appear susceptible to imipenem or meropenem, as downregulation of ompK36 expression will adversely affect the MICs to the carbapenems.
Acknowledgments
This study was supported by the National Institutes of Health (RO1 AI070246-01A1).
Abbreviations
ESBL, extended-spectrum β-lactamase
MALDI, matrix-assisted laser desorption/ionization
Footnotes
References
- Albertí, S., Rodríquez-Quiñones, F., Schirmer, T., Rummel, G., Tomás, J. M., Rosenbusch, J. P. & Benedí, V. J. (1995). A porin from Klebsiella pneumoniae: sequence homology, three-dimensional model, and complement binding. Infect Immun 63, 903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, K. F., Lonsway, D. R., Rasheed, J. K., Biddle, J., Jensen, B., McDougal, L. K., Carey, R. B., Thompson, A., Stocker, S. & other authors (2007). Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J Clin Microbiol 45, 2723–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardanuy, C., Liñares, J., Domínguez, M. A., Hernández-Allés, S., Benedí, V. J. & Martínez-Martínez, L. (1998). Outer membrane profiles of clonally related Klebsiella pneumoniae isolates from clinical samples and activities of cephalosporins and carbapenems. Antimicrob Agents Chemother 42, 1636–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, P. A., Urban, C., Mariano, N., Projan, S. J., Rahal, J. J. & Bush, K. (1997). Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob Agents Chemother 41, 563–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratu, S., Landman, D., Haag, R., Recco, R., Eramo, A., Alam, M. & Quale, J. (2005a). Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City. Arch Intern Med 165, 1430–1435. [DOI] [PubMed] [Google Scholar]
- Bratu, S., Mooty, M., Nichani, S., Landman, D., Gullans, C., Pettinato, B., Karumudi, U., Tolaney, P. & Quale, J. (2005b). Emergence of KPC-possessing Klebsiella pneumoniae in Brooklyn, New York: epidemiology and recommendations for detection. Antimicrob Agents Chemother 49, 3018–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratu, S., Brooks, S., Burney, S., Kochar, S., Gupta, J., Landman, D. & Quale, J. (2007). Detection and spread of Escherichia coli possessing the plasmid-borne carbapenemase KPC-2 in Brooklyn, New York. Clin Infect Dis 44, 972–975. [DOI] [PubMed] [Google Scholar]
- Cai, J. C., Zhou, H. W., Zhang, R. & Chen, G.-X. (2008). Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob Agents Chemother 52, 2014–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, V. T. B., Arlet, G., Ericsson, B.-M., Tammelin, A., Courvalin, P. & Lanbert, T. (2000). Emergence of imipenem resistance in Klebsiella pneumoniae owing to combination of plasmid-mediated CMY-4 and permeability alteration. J Antimicrob Chemother 46, 895–900. [DOI] [PubMed] [Google Scholar]
- CLSI (2006). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 7th edn. Approved Standard M7-A7. Wayne, PA: Clinical and Laboratory Standards Institute.
- Crowley, B., Benedí, V. J. & Doménech-Sánchez, A. (2002). Expression of SHV-2 β-lactamase and reduced amounts of OmpK36 porin in Klebsiella pneumoniae results in increased resistance to cephalosporins and carbapenems. Antimicrob Agents Chemother 46, 3679–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doménech-Sánchez, A., Hernández-Allés, S., Martínez-Martínez, L., Benedí, V. J. & Albertí, S. (1999). Identification and characterization of a new porin gene of Klebsiella pneumoniae: its role in β-lactam antibiotic resistance. J Bacteriol 181, 2726–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doménech-Sánchez, A., Pascual, A., Suárez, A. I., Alvarez, D., Benedí, V. J. & Martínez-Martínez, L. (2000). Activity of nine antimicrobial agents against clinical isolates of Klebsiella pneumoniae producing extended-spectrum β-lactamases and deficient or not in porins. J Antimicrob Chemother 46, 858–860. [DOI] [PubMed] [Google Scholar]
- Doménech-Sánchez, A., Martínez-Martínez, L., Hernández-Allés, S., del Carmen Conejo, M., Pascual, A., Tomás, T. M., Albertí, S. & Benedí, V. J. (2003). Role of Klebsiella pneumoniae OmpK35 porin in antimicrobial resistance. Antimicrob Agents Chemother 47, 3332–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, E., Brink, A. J., van Greune, J., Els, Z., Woodford, N., Turton, J., Warner, M. & Livermore, D. M. (2006). In vivo development of ertapenem resistance in a patient with pneumonia caused by Klebsiella pneumoniae with an extended-spectrum β-lactamase. Clin Infect Dis 42, e95–e98. [DOI] [PubMed] [Google Scholar]
- Hernández-Allés, S., Albertí, S., Alvarez, D., Doménech-Sánchez, A., Martínez-Martínez, L., Gil, J., Tomás, J. M. & Benedí, V. J. (1999a). Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology 145, 673–679. [DOI] [PubMed] [Google Scholar]
- Hernández-Allés, S., Benedí, V. J., Martínez-Martínez, L., Pascual, A., Aguilar, A., Tomás, J. M. & Albertí, S. (1999b). Development of resistance during antimicrobial therapy caused by insertion sequence interruption of porin genes. Antimicrob Agents Chemother 43, 937–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby, G. A., Mills, D. M. & Chow, N. (2004). Role of β-lactamases and porins in resistance to ertapenem and other β-lactams in Klebsiella pneumoniae. Antimicrob Agents Chemother 48, 3203–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek, F. M., Dib-Hajj, F., Shang, W. & Gootz, T. D. (2006). High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of blaACT-1 β-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin PhoE. Antimicrob Agents Chemother 50, 3396–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landman, D., Bratu, S., Kochar, S., Panwar, M., Trehan, M., Doymaz, M. & Quale, J. (2007). Evolution of antimicrobial resistance among Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae in Brooklyn, NY. J Antimicrob Chemother 60, 78–82. [DOI] [PubMed] [Google Scholar]
- Martínez-Martínez, L., Hernández-Allés, S., Albertí, S., Tomás, J. M., Venedi, V. J. & Jacoby, G. A. (1996). In vivo selection of porin-deficient mutants of Klebsiella pneumoniae with increased resistance to cefoxitin and expanded-spectrum cephalosporins. Antimicrob Agents Chemother 40, 342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Martínez, L., Pascual, A., Hernández-Allés, S., Alvarez-Díaz, D., Suárez, A. I., Tran, J., Benedí, V. J. & Jacoby, G. (1999). Roles of β-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob Agents Chemother 43, 1669–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena, A., Plasencia, V., García, L., Hidalgo, O., Ayestarán, J. I., Albertí, S., Borrell, N., Pérez, J. L. & Oliver, A. (2006). Characterization of a large outbreak by CTX-M-1-producing Klebsiella pneumoniae and mechanisms leading to in vivo carbapenem resistance development. J Clin Microbiol 44, 2831–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido, H. (1996). Multidrug efflux pumps of Gram-negative bacteria. J Bacteriol 178, 5853–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, D. L., Hujer, K. M., Hujer, A. M., Yeiser, B., Bonomo, M. D., Rice, L. B., Bonomo, R. A. & the International Klebsiella Group (2003). Extended-spectrum β-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: dominance and widespread prevalence of SHV- and CTX-M-type β-lactamases. Antimicrob Agents Chemother 47, 3554–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queenan, A. M. & Bush, K. (2007). Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev 20, 440–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. & Russell, D. W. (2001). Molecular Cloning: a Laboratory Manual, 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Tenover, F. C., Kalsi, R. K., Williams, P. P., Carey, R. B., Stocker, S., Lonsway, D., Rasheed, J. K., Biddle, D. W., McGowan, J. E., Jr & Hanna, B. (2006). Carbapenem resistance in Klebsiella pneumoniae not detected by automated susceptibility testing. Emerg Infect Dis 12, 1209–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford, N., Tierno, P. M., Jr, Young, K., Tysall, L., Palepou, M.-F. I., Ward, E., Painter, R. E., Suber, D. F., Shungu, D. & other authors (2004). Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A β-lactamase, KPC-3, in a New York Medical Center. Antimicrob Agents Chemother 48, 4793–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford, N., Dallow, J. W. T., Hill, R. L. R., Palepou, M.-F. I., Pike, R., Ward, M. E., Warner, M. & Livermore, D. M. (2007). Ertapenem resistance among Klebsiella and Enterobacter submitted in the UK to a reference laboratory. Int J Antimicrob Agents 29, 456–459. [DOI] [PubMed] [Google Scholar]
- Yigit, H., Queenan, A. M., Anderson, G. J., Doménech-Sánchez, A., Biddle, J. W., Steward, C. D., Albertí, S., Bush, K. & Tenover, F. C. (2001). Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45, 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]