Abstract
Interleukin 2 (IL-2)- and IL-10-knockout mice develop spontaneous colitis under conventional but not germ-free conditions, suggesting that commensal bacteria play an important role in the pathogenesis of colitis. However, interactions between commensal bacteria and colonic epithelial cells have not been fully investigated. We therefore assessed the ability of various commensal bacteria and probiotics to adhere to and invade colonic epithelial cells. Effects of the bacteria on production of proinflammatory cytokines were also measured. Commensal bacteria, including mucosal organisms isolated from ulcerative colitis (UC) patients, such as Fusobacterium varium, reported as a possible pathogen in UC, Bacteroides vulgatus, Escherichia coli and Clostridium clostridioforme, as well as their type strains and probiotics, were assessed for their ability to adhere to and invade colonic epithelial cells using two cell lines, SW-480 and HT-29. Our experiments employed co-incubation, a combination of scanning and transmission electron microscopy and recovery of bacteria from infected-cell lysates. F. varium and several other commensal bacteria, but not probiotics, adhered to colonic epithelial cells and invaded their cytoplasm. ELISA and real-time PCR revealed that the host cells, particularly those invaded by F. varium, showed significant increases in IL-8 and TNF-α concentrations in supernatants, with elevation of IL-8, TNF-α, MCP-1 and IL-6 mRNAs. Furthermore, IL-8 and TNF-α expression and nuclear phosphorylated NF-κB p65 expression could be immunohistochemically confirmed in inflamed epithelium with cryptitis or crypt abscess in UC patients. Certain commensal bacteria can invade colonic epithelial cells, activating early intracellular signalling systems to trigger host inflammatory reactions.
INTRODUCTION
The aetiology of ulcerative colitis (UC) is still not understood in detail, although several possible mechanisms have been proposed. Since IL-2- and IL-10-knockout mice develop spontaneous colitis under routine but not germ-free conditions, commensal bacteria may play an important role in the pathogenesis of colitis (Sadlack et al., 1993; Sellon et al., 1998). There is growing evidence that numbers of mucosa-associated bacteria, especially anaerobic Bacteroides species and aerobic members of the Enterobacteriaceae (Escherichia coli), are dramatically increased in inflammatory bowel diseases, while numbers of lactic acid bacilli are decreased (Swidsinski et al., 2002; Guarner et al., 2002; Mylonaki et al., 2005). Consistent with these observations, we have found significantly increased detection rates of Fusobacterium varium in actively inflamed colonic mucosa of patients with UC and F. varium-induced specific antibodies in serum (Ohkusa et al., 2002). Furthermore, an enema with butyric acid produced by F. varium can induce apoptosis in murine colonic epithelium, resulting in crypt abscess formation and UC-like lesions, as well as induce apoptosis in an in vitro cell culture (Ohkusa et al., 2003; Yoshida et al., 2006).
Recent studies have revealed that intestinal epithelial cells, the first host cells to come in contact with enteric pathogens, secrete a chemotactic mediator in response to pathogenic bacterial attachment or entry. It is well known that enteropathogenic bacteria invade these epithelial cells and stimulate IL-8 production (Eckmann et al., 1993a; Aihara et al., 1997; Vitiello et al., 2004; Singer & Sansonetti, 2004). Evidence has been presented that intestinal epithelial cells have the capacity to express and secrete several cytokines constitutively (Eckmann et al., 1993b; Jung et al., 1995; Weinstein et al., 1997; Kuhara et al., 2000; Sartor, 2004) when stimulated by pathogenic bacteria. Studies on critical constituents of the innate immune response, such as Toll-like receptors and nucleotide binding oligomerization domain isoforms, also suggest that bacterial infections may contribute to intestinal inflammation (Hoshino et al., 1999; Ohkusa et al., 2004). However, it is currently unclear whether epithelial cells secrete cytokines in response to nonpathogenic commensal bacteria or probiotics. In the present study, we found that the production of cytokines by colonic epithelial cells appears to be an important primary event in host defence, by analysing an in vitro model system in which commensal bacteria attach to or penetrate epithelial cells. Furthermore, we defined secretion of inflammatory cytokines and mRNA expression employing co-culture experiments. Moreover, we tested immunohistochemical expression of inflammatory cytokines and activated NF-κB in the inflamed epithelium in patients with UC.
METHODS
Bacterial strains and attachment and invasion assays.
As listed in Table 1, we used four commensal bacterial strains, F. varium, E. coli, Bacteroides vulgatus and Clostridium clostridioforme, isolated from patients with UC (Ohkusa et al., 2002); four type strains, E. coli JCM 1649T, B. vulgatus JCM 5826T, C. clostridioforme JCM 1291T and F. varium ATCC 8501T (Burke & Axon, 1988; Matsuda et al., 2000); and two probiotics, Lactobacillus johnsonii (La1 strain; Nestec collection, Lausanne, Switzerland) and Lactobacillus delbrueckii subsp. bulgaricus (LB-021001; Meiji Dairies). Bacteria were harvested for attachment and invasion assays after incubation on GAM agar plates (Nissui Chemicals) under aerobic conditions with 5 % CO2 or under anaerobic conditions (anaerobic chamber) with 10 % CO2, 5 % H2 and 85 % N2 for 2–4 days at 37 °C. They were then suspended at 1×108 cells ml−1 in a 1 : 1 mixture of Dulbecco's modified Eagle's medium (DMEM; Cellgro Mediatech) and Ham's F-12 medium (Cellgro Mediatech).
Table 1.
Bacterial attachment to and invasion of SW-480 cells
The assays were carried out as described in the text. Shown here are the mean results of assays with standard deviations. Each experiment was carried out in duplicate.
| Strain | Attachment (%)* | SEM | Invasion (%)* | TEM |
|---|---|---|---|---|
| F. varium 113 | 0.034±0.008† | 2+ | 0.014±0.004† | 2+ |
| F. varium ATCC 8501T | 0.029±0.019† | 2+ | 0.008±0.003† | 2+ |
| E. coli R-1 | 0.070±0.032† | + | 0.0003±0.00001 | − |
| E. coli JCM 1649T | 0.030±0.014† | + | 0.0002±0.0001 | + |
| B. vulgatus 90 | 0.018±0.004† | + | 0.004±0.001 | + |
| B. vulgatus JCM 5826T | 0.013±0.004† | + | 0.002±0.001 | + |
| C. clostridioforme 94 | 0.003±0.002 | + | 0.003±0.001 | + |
| C. clostridioforme JCM 1291T | 0.002±0.001 | + | 0 | − |
| L. johnsonii La1 | 0.023±0.023 | + | 0.0004±0.0001 | − |
| L. delbrueckii subsp. bulgaricus LB-021001 | 0.008±0.004 | − | 0.00006±0.00002 | − |
*The attachment and invasion levels were expressed as the percentage of bacteria retrieved following cell lysis relative to the total number of bacteria initially added.
†P <0.05 by ANOVA when compared with L. delbrueckii subsp. bulgaricus LB-021001.
Bacterial attachment and invasion assays were conducted according to the methods of Han & Miller (1997) with modifications for intestinal bacteria. We chose experiments on SW-480 and HT-29 cells (SW-480, ATCC CCL228; HT-29, ATCC HTB-38), commonly studied human intestinal epithelial cell lines, because they exhibit a highly LPS-responsive phenotype and surface expression of the TLR4 protein (Suzuki et al., 2003). Human colonic epithelial cells were grown to near confluence (3.3–4.0×105 cells per well) in 6-well culture plates before infection with the bacteria in a 1 : 1 mixture of DMEM and Ham's F-12 medium containing 10 % fetal bovine serum. Bacteria were added at a m.o.i. of 1000 (c.f.u.) and incubated under 5 % CO2 at 37 °C. For the attachment assay, supernatants were collected and stored at −80 °C for ELISAs after co-incubation with bacteria for 1 h. Then, after washing the SW-480 monolayers four times, total cell-associated bacteria were counted on GAM agar plates after cell lysis with water for the attachment assay. For the invasion assay, the monolayers were washed twice with sterile PBS after co-incubation with bacteria for 3 h and incubated in fresh medium containing imipenem (50 μg ml−1) for an additional 1 h to kill extracellular bacteria. After washing twice, the monolayers were further incubated for 1 h in fresh medium. The supernatant was then collected and stored at −80 °C for ELISA. The monolayers were rinsed again with PBS and lysed with sterile water. Internalized bacteria were counted on agar plates (GAM agar plates for B. vulgatus, F. varium, Lactobacillus johnsonii and L. delbrueckii subsp. bulgaricus, and trypticase soy agar plates for E. coli). Preliminary experiments confirmed that bacterial viability was unaffected by the water treatment during cell lysis. The levels of attachment and invasion were expressed as the percentage of bacteria retrieved following cell lysis relative to the total number of bacteria initially added.
Scanning and transmission electron microscopy (SEM and TEM) analysis.
For attachment, the SW-480 monolayers were replaced with tert-butyl alcohol, freeze-dried, coated with an osmium plasma coater, and analysed by SEM. For invasion, monolayers were fixed with 2 % glutaraldehyde in 0.1 M Na-cacodylate buffer and osmium tetroxide, followed by dehydration and epon embedding. Ultrathin sections were stained with uranyl acetate and lead citrate and analysed by TEM. In experiments in which adherence and invasion levels were determined, 30 observation fields obtained at a magnification of ×5000 were randomly chosen and the number of attached or invading bacteria was semiquantified as: +, bacteria found in 1–5/30 fields; and 2+, bacteria found in >6/30 fields.
Detection of secreted cytokines in supernatants.
To determine the amounts of IL-8, IL-6, MCP-1, TNF-α and IL-18 secreted by SW-480 or HT-29 cells co-incubated with bacteria for 1 h or 1–3 h, ELISAs were conducted using commercial assay kits (IL-8 EASIA, BioSource Europe; QuantiGlo Human IL-6 Immunoassay, R&D Systems; Quantikine Human MCP-1 Immunoassay, R&D Systems; QuantiGlo Human TNF-α Immunoassay, R&D Systems; Human IL-18 ELISA kit, Medical & Biological Lab.), according to the manufacturers' protocols. All experiments were performed in triplicate and repeated at least three times. Results are shown as mean±standard deviation (sd).
Semiquantitative real-time PCR.
Real-time (TaqMan) RT-PCR for mRNA was conducted as described by Mitchell et al. (2003). Briefly, RNA was prepared, as described above, from monolayered SW-480 or HT-29 cells infected with the test bacteria during a 1 h or 1–3 h incubation. Total RNA was isolated using an RNeasy Mini kit (Qiagen), and DNase treatment was performed using an RNase-Free DNase Set (Qiagen). Reverse transcription with up to 4 μg total RNA was carried out in a total volume of 20 μl containing 250 pmol random primer, 100 U SuperScript II RNase H-Reverse Transcriptase (Invitrogen) in 50 mM Tris/HCl (pH 8.3), 40 mM KCl, 6 mM MgCl2, 1 mM DTT and 10 mM dNTPs, following the manufacturer's instructions. Initially, a total RNA solution mixed with random primer was heated at 70 °C for 10 min and immediately chilled on ice, and the other reagents were then added. First-strand cDNAs were obtained after 50 min at 42 °C and 5 min at 98 °C.
Primers and TaqMan probes for target genes (IL-8, IL-6, MCP-1, TNF-α and IL-18) were obtained from TaqMan Gene Expression Assays and an internal reference gene (β-actin) from Pre-Developed TaqMan Assay Reagents (Applied Biosystems). Each of the TaqMan probes was labelled with a reporter dye (FAM or VIC) situated at the 5′-end of the oligonucleotide and a quencher dye (MGB) located at the 3′-end. The TaqMan Gene Expression Assay numbers are Hs00174131_m1 for IL-6, Hs00174103_m1 for IL-8, Hs00155517_m1 for IL-18, Hs00234140_m1 for MCP-1, Hs00174128_m1 for TNF-α and 4326315E for β-actin. Quantification of target genes (IL-8, IL-6, MCP-1, TNF-α and IL-18) and the reference gene (β-actin) was performed in 96-well plates with ABI PRISM 7700 Sequence Detection System (ABI) data collection, and analyses were performed using the machine software. The PCR was carried out in a final volume of 25 μl containing cDNA equivalent to 10–100 ng total RNA, 12.5 μl 2× TaqMan Universal PCR Master Mix and 1.25 μl 20× TaqMan Expression Assay reagent. Thermal cycler conditions were 50 °C for 2 min and 95 °C for 10 min, followed by 50 cycles at 95 °C for 15 s and 60 °C for 1 min. The comparative CT method of data analysis was used to analyse the data. CT is the PCR cycle at which an increase in reporter fluorescence above the baseline level is first detected. The target gene CT and internal reference gene CT were calculated for each sample. ΔCT is the difference in CT between the target and reference genes; ΔΔCT is the difference in ΔCT for the sample and the calibrator sample. The amount of target gene expression, normalized to an internal reference and relative to the calibrator sample, is given by 2−ΔΔCT. Each sample was analysed in triplicate and each assay was repeated three times. Results are shown as mean±standard error (se).
Immunohistochemistry for clinical samples.
For immunohistochemistry of cytokines and NF-κB in rectal mucosae of patients with UC, surgically removed rectums were obtained from patients with colorectal cancers (10 cases), inactive UC (12 cases) and active UC (7 cases). The active and inactive phases of UC were defined using Seo's complex integrated disease activity indices for clinical UC activity (Seo et al., 1992). Immunohistochemical staining of new sets of serial sections (4 μm in thickness) from formalin-fixed, paraffin-embedded blocks was achieved using an EnVision+ kit (Dako), as in our previous study (Mikami et al., 2003; Mitsuhashi et al., 2005). The primary antibodies and their working dilutions were as follows: mouse monoclonal anti-TNF-α (clone 28401, ×50; R&D Systems), rabbit anti-human IL-8 (×100; MONOSAN), mouse anti-NF-κB subunit p65 (clone 20, ×500; BD Transduction Laboratory) and rabbit anti-phosphor-NF-κB p65 (Ser276) antibodies (×100; Cell Signaling Technology). Briefly, after a 15 min microwave pretreatment in citrate buffer (pH 6.0, 0.01 M) for retrieval of the four antigens, sections were incubated with the primary antibodies for 1 h at room temperature. Nuclei were counterstained with methyl-green solution to facilitate histopathological assessment. Crypts bearing at least two positive epithelia were considered to be positive. Two full-length crypts adjacent to the muscularis mucosa were randomly selected and observed in normal mucosa more than 2 cm from the tumour in colorectal cancer cases and mucosa in inactive UC, with neither cryptitis nor crypt abscess. Crypts with cryptitis or crypt abscess were randomly selected from seven active UC cases and observed.
Ethics.
This work using pathological samples was approved by our Kitasato University Medical School and University Hospital Ethics Committee.
Statistical analysis.
Data are presented as means±se or sd and range. Statistical analysis was performed using repeated measures ANOVA and least significant difference post hoc tests for multiple comparisons. The χ2 test was applied to compare the frequency of immunopositive crypts. Differences were considered statistically significant at P <0.05.
RESULTS AND DISCUSSION
Bacterial attachment and invasion
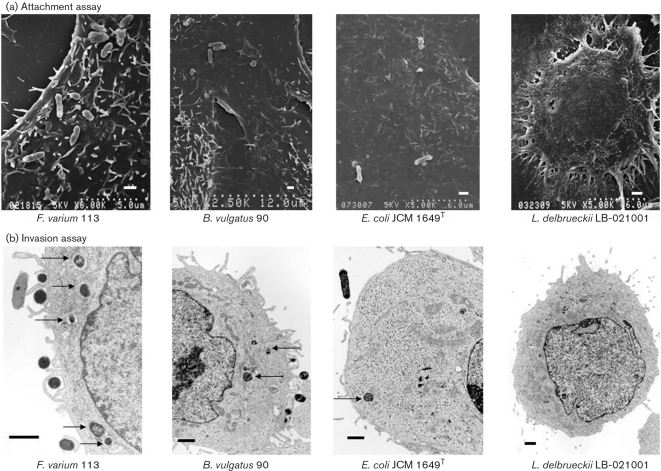
To measure bacterial attachment to and invasion of SW-480 cells, SEM and TEM as well as counting on agar plates (recovery of bacteria) after cell lysis were employed (Fig. 1). Counting on agar plates after cell lysis showed that the attachment levels of F. varium 113 and ATCC 8501T, E. coli R-1 and JCM 1649T, B. vulgatus 90 and JCM 5826T, C. clostridioforme 94 and JCM 1291T, and L. johnsonii La were approximately 2–10-fold higher than that of L. delbrueckii subsp. bulgaricus LB-021001 by the co-incubation method. SEM revealed F. varium 113 and ATCC 8501T to be more adhesive than other bacterial strains to these cells (Table 1; Fig. 1a).
Fig. 1.
Adherence and invasion of commensal bacteria and probiotics in SW-480 cells (SEM and TEM). (a) The scanning electron micrographs show attachment of bacteria, with the exception of L. delbrueckii subsp. bulgaricus LB-021001, to SW-480 cells. F. varium 113 was more adhesive than the other bacterial strains tested. Bar, 1 μm. (b) Commensal bacteria, F. varium 113 and B. vulgatus 90, isolated from patients with UC, and E. coli JCM 1649T adhere to SW-480 cells and invade their cytoplasm (arrows), while the probiotic L. delbrueckii subsp. bulgaricus does not enter the cells (TEM photograph; bar, 1 μm).
In the invasion assay, TEM revealed that E. coli R-1, C. clostridioforme JCM 1291T and the two probiotics did not invade SW-480 cells. E. coli JCM 1649T, B. vulgatus 90 and JCM 5826T, and C. clostridioforme 94 showed weak invasion. F. varium 113 and ATCC 8501T strains showed much greater invasion of SW-480 cells, both by counts on agar plates and TEM (Fig. 1b). The present study revealed using the co-incubation method that F. varium and several other commensal bacteria, but not probiotics, adhered to colonic epithelial cells and invaded their cytoplasm.
Cytokine production by bacterial attachment and invasion
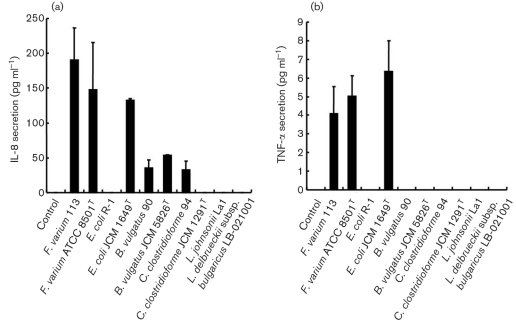
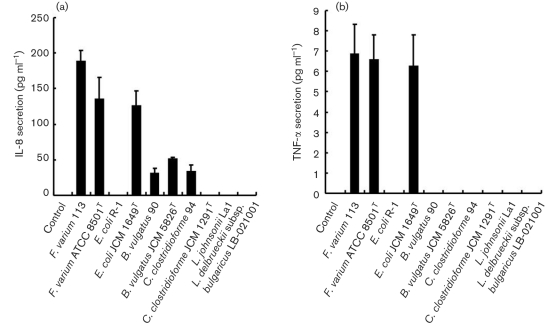
Following the above described results, to determine the production of cytokines upon infection with commensal bacteria and probiotics, the levels of IL-8, IL-6, MCP-1, TNF-α and IL-18 released by SW-480 and HT-29 in the bacterial attachment assay were measured in culture supernatants. However, concentrations of the cytokines IL-8 (<16 pg ml−1), IL-6 (<1.5 pg ml−1), MCP-1 (<31.2 pg ml−1), TNF-α (<0.6 pg ml−1) and IL-18 (<15.6 pg ml−1) were all below their detection limits. For the invasion assay, significantly increased IL-8 production by SW-480 (Fig. 2a) and HT-29 (Fig. 3a) was induced by F. varium 113 (mean±sd; 95 % confidence interval, not shown) (SW-480, 190.5±18.8 pg ml−1; HT-29, 188.8±15.0 pg ml−1, respectively), F. varium ATCC 8501T (135.3±30.5; 147.6±68.3), E. coli JCM 1649T (132.5±6.1; 125.3±21.7), B. vulgatus JCM 5826T (53.2±1.37; 50.7±3.24), B. vulgatus 90 (35.1±11.7; 30.3±7.8) and C. clostridioforme 94 (32.8±12.7; 33.3±9.5). Noninvasive bacteria, i.e. E. coli R-1, C. clostridioforme JCM 1291T, L. johnsonii La1 and L. delbrueckii subsp. bulgaricus LB-021001, failed to stimulate IL-8 secretion.
Fig. 2.
Production of cytokines upon infection with commensal bacteria and probiotics in SW-480 cells. (a) IL-8. In the invasion assay, F. varium 113 stimulated the greatest IL-8 production, followed by F. varium ATCC 8501T, E. coli JCM 1649T, B. vulgatus JCM 5826T, B. vulgatus 90 and C. clostridioforme 94. Since the IL-8 levels at the 95 % confidence interval for these bacteria were above those of the controls (<16 pg ml−1), these bacteria significantly stimulated IL-8 production by SW-480. The IL-8 levels with F. varium 113, ATCC 8501T and E. coli JCM 1649T were significantly higher than those with B. vulgatus 90 and C. clostridioforme 94 (P <0.01). (b) TNF-α. In the invasion assay, commensal bacterial strains, i.e. E. coli JCM 1649T and F. varium 113 and ATCC 8501T, induced TNF-α production by SW-480 cells. E. coli JCM 1649T stimulated the greatest TNF-α production, followed by F. varium ATCC 8501T and F. varium 113. Since the TNF-α levels at the 95 % confidence interval for the bacteria were above those of the controls, these bacteria significantly stimulated TNF-α production by SW-480 cells.
Fig. 3.
Production of cytokines upon infection with commensal bacteria and probiotics in HT-29 cells. (a) IL-8. In the invasion assay, F. varium 113 stimulated the greatest IL-8 production, followed by F. varium ATCC 8501T, E. coli JCM 1649T, B. vulgatus JCM 5826T, C. clostridioforme 94 and B. vulgatus 90. Since the IL-8 levels at the 95 % confidence interval for these bacteria were above those of the controls (<16 pg ml−1), these bacteria significantly stimulated IL-8 production by HT-29 cells. The IL-8 levels with F. varium 113 and ATCC 8501T and E. coli JCM 1649T were significantly higher than those with B. vulgatus 90 and C. clostridioforme 94 (P <0.0001). (b) TNF-α. In the invasion assay, commensal bacterial strains, i.e. F. varium 113, ATCC 8501T and E. coli JCM 1649T, induced TNF-α production by HT-29 cells. F. varium 113 stimulated the greatest TNF-α production, followed by F. varium ATCC 8501T and E. coli JCM 1649T. Since the TNF-α levels at the 95 % confidence interval for the bacteria were above those of the controls, these bacteria significantly stimulated TNF-α production by HT-29 cells.
In the invasion assay, TNF-α production by SW-480 (Fig. 2b) and HT-29 cells (Fig. 3b) was clearly induced by E. coli JCM 1649T (mean±sd; 95 % confidence interval, not shown) (SW-480, 5.1±0.6 pg ml−1; HT-29, 6.4±1.6 pg ml−1), F. varium ATCC 8501T (5.1±0.7; 6.6±1.3) and F. varium 113 (4.2±0.7; 6.9±1.1). However, IL-6, MCP-1 and IL-18 were below their detection limits for all of the bacteria tested.
mRNA expression of cytokines by bacterial attachment and invasion
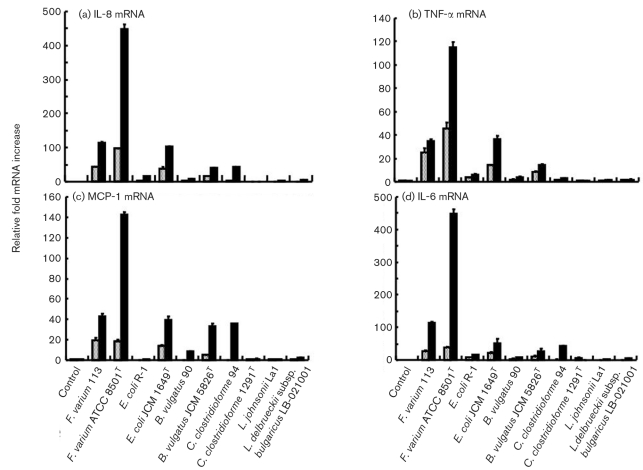
Following production of cytokines, real-time RT-PCR was conducted to measure the induction of cytokine mRNAs upon infection with commensal bacteria and probiotics. The bacteria tested, with the exception of C. clostridioforme JCM 1291T and L. johnsonii La1 strains in the attachment assay, and commensal bacteria, with the exception of C. clostridioforme JCM 1291T and probiotics in the invasion assay with SW-480 (Fig. 4a), and with the exception of L. johnsonii La1 strains in the attachment assay with HT-29 cells (Fig. 5a), markedly stimulated IL-8 mRNA expression. In the invasion assays, IL-8 mRNA expression was stimulated by F. varium ATCC 8501T (relative fold increase, mean±se; 95 % confidence intervals not shown) (SW-480, 447.9±13.4; HT-29, 130.3±7.2, respectively), F. varium 113 (115.1±2.8; 152.7±8.3) and E. coli JCM 1649T (102.9±0.5; 119.4±4.2). Since the 95 % confidence intervals for IL-8 mRNA levels in cells infected with all bacterial strains were above those of the controls, these bacteria were confirmed to stimulate IL-8 mRNA expression in SW-480 and HT-29 cells. However, the stimulated IL-8 mRNA expression levels varied with the bacterial strains, and noninvasive bacteria, i.e. E. coli R-1, C. clostridioforme JCM 1291T, L. johnsonii La1 and L. delbrueckii subsp. bulgaricus LB-021001, slightly stimulated IL-8 mRNA expression.
Fig. 4.
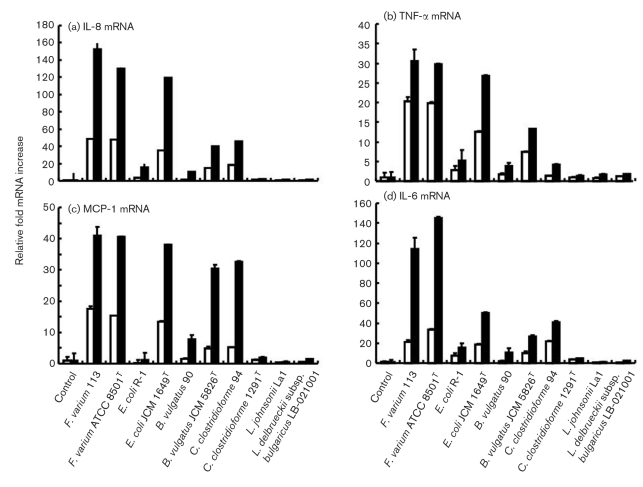
Induction of cytokine mRNAs in SW-480 cells infected with commensal bacteria and probiotics. Dotted bars, the attachment assay; black bars, the invasion assay. (a) IL-8 mRNA. In the attachment assay, the IL-8 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T and B. vulgatus JCM 5826T were significantly higher than those with E. coli R-1, C. clostridioforme 94 and JCM 1291T, B. vulgatus 90 and probiotics (P <0.05). In the invasion assay, the IL-8 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T, C. clostridioforme 94 and B. vulgatus JCM 5826T were significantly higher than those with B. vulgatus 90, C. clostridioforme JCM 1291T and probiotics (P <0.0001). The IL-8 mRNA levels with F. varium ATCC 8501T and 113, E. coli JCM 1649T, C. clostridioforme 94 and B. vulgatus JCM 5826T in the invasion assay were significantly higher than those in the attachment assay (P <0.0001). (b) TNF-α mRNA. In the attachment assay, the TNF-α mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T and B. vulgatus JCM 5826T were significantly higher than those with E. coli R-1, C. clostridioforme 94 and JCM 1291T, B. vulgatus 90 and probiotics (P <0.0001). In the invasion assay, the TNF-α mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T and B. vulgatus JCM 5826T were significantly higher than those with C. clostridioforme JCM 1291T and probiotics (P <0.0001). (c) MCP-1 mRNA. In the attachment assay, the MCP-1 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T and B. vulgatus JCM 5826T were significantly higher than those with E. coli R-1, C. clostridioforme 94 and JCM 1291T, B. vulgatus 90 and probiotics (P <0.0001). In the invasion assay, the MCP-1 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T, C. clostridioforme 94 and B. vulgatus JCM 5826T were significantly higher than those with C. clostridioforme JCM 1291T, E. coli R-1 and probiotics (P <0.0001). (d) IL-6 mRNA. In the attachment assay, the IL-6 mRNA levels after incubation with F. varium ATCC 8501T and 113 and E. coli JCM 1649T were significantly higher than those with probiotics (P=0.023–<0.0001). In the invasion assay, the IL-6 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T, C. clostridioforme 94 and B. vulgatus JCM 5826T were significantly higher than those with B. vulgatus 90, C. clostridioforme JCM 1291T and probiotics (P=0.0081–<0.0001).
Fig. 5.
Induction of cytokine mRNAs in HT-29 cells infected with commensal bacteria and probiotics. White bars, the attachment assay; black bars, the invasion assay. (a) IL-8 mRNA. In the attachment assay, the IL-8 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T and B. vulgatus JCM 5826T were significantly higher than those with E. coli R-1, C. clostridioforme 94 and JCM 1291T, B. vulgatus 90 and probiotics (P <0.01). In the invasion assay, the IL-8 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T, C. clostridioforme 94 and B. vulgatus JCM 5826T were significantly higher than those with B. vulgatus 90, C. clostridioforme JCM 1291T and probiotics (P <0.0001). The IL-8 mRNA levels with F. varium ATCC 8501T and 113, E. coli JCM 1649T, C. clostridioforme 94 and B. vulgatus JCM 5826T in the invasion assay were significantly higher than those in the attachment assay (P <0.0001). (b) TNF-α mRNA. In the attachment assay, the TNF-α mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T and B. vulgatus JCM 5826T were significantly higher than those with E. coli R-1, C. clostridioforme 94 and JCM 1291T, B. vulgatus 90 and probiotics (P <0.0001). In the invasion assay, the TNF-α mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T and B. vulgatus JCM 5826T were significantly higher than those with C. clostridioforme JCM 1291T and probiotics (P <0.0001). (c) MCP-1 mRNA. In the attachment assay, the MCP-1 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T and B. vulgatus JCM 5826T were significantly higher than those with E. coli R-1, C. clostridioforme 94 and JCM 1291T, B. vulgatus 90 and probiotics (P <0.0001). In the invasion assay, the MCP-1 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T, C. clostridioforme 94 and B. vulgatus JCM 5826T were significantly higher than those with C. clostridioforme JCM 1291T, E. coli R-1 and probiotics (P <0.0001). (d) IL-6 mRNA. In the attachment assay, the IL-6 mRNA levels after incubation with F. varium ATCC 8501T and 113 and E. coli JCM 1649T were significantly higher than those with probiotics (P=0.023–<0.0001). In the invasion assay, the IL-6 mRNA levels after incubation with F. varium ATCC 8501T and 113, E. coli JCM 1649T, C. clostridioforme 94 and B. vulgatus JCM 5826T were significantly higher than those with B. vulgatus 90, C. clostridioforme JCM 1291T and probiotics (P <0.0001).
The bacteria tested, with the exception of C. clostridioforme JCM 1291T, L. johnsonii La1 and/or L. delbrueckii subsp. bulgaricus LB-021001 strains in the attachment assay, and C. clostridioforme JCM 1291T in the invasion assay, stimulated TNF-α mRNA expression by SW-480 and HT-29 cells (Figs 4b, 5b). In the invasion assays, TNF-α mRNA expression was stimulated by F. varium ATCC 8501T (SW-480, 115.2±4.2; HT-29, 29.7±5.9), F. varium 113 (34.8±1.2; 30.6±1.3), E. coli JCM 1649T (36.6±2.5; 26.8±2.7) and B. vulgatus JCM 5826T (14.1±0.9; 13.3±0.7).
F. varium ATCC 8501T and 113, E. coli JCM 1649T, C. clostridioforme 94 and JCM 1291T, and B. vulgatus JCM 5826T strains in the attachment assay, as well as B. vulgatus 90, C. clostridioforme 94 and E. coli R-1 tested in the invasion assay significantly stimulated MCP-1 mRNA expression in SW-480 and HT-29 cells (Figs 4c, 5c). In the invasion assay, F. varium 113 (SW-480, 42.6±3.1; HT-29, 41.1±2.3), F. varium ATCC 8501T (142.1±3.1; 40.6±2.7), E. coli JCM 1649T (39.6±3.1; 38.0±2.3), B. vulgatus JCM 5826T (33.7±2.6; 30.4±1.5) and C. clostridioforme 94 (35.6±0.6; 32.6±1.2) strains stimulated marked MCP-1 mRNA expression.
F. varium ATCC 8501T and 113, E. coli JCM 1649T and R-1, and B. vulgatus JCM 5826T strains in the attachment assay, as well as the bacteria tested with the exception of L. johnsonii La1 in the invasion assay, significantly stimulated IL-6 mRNA expression in SW-480 and HT-29 cells (Figs 4d, 5d). In the invasion assays, IL-6 mRNA expression was stimulated higher by F. varium ATCC 8501T (SW-480, 447.9±13.4; HT-29, 145.4±11.5), F. varium 113 (115.1±2.8; 113.8±2.2), E. coli JCM 1649T (52.1±11.9; 50.4±4.3), C. clostridioforme 94 (42.4±1.0; 40.9±1.4) and B. vulgatus JCM 5826T (27.0±7.4; 26.5±4.3). IL-18 mRNA expression was not detected in either bacterially stimulated or unstimulated SW-480 cells. This is in line with heterogeneous expression among cell lines being different from that among normal colonic mucosa cells, which usually display IL-18 mRNA and protein in relation to IFN-γ and Fas-L-dependent cytotoxicity (Pagès et al., 1999; Paulukat et al., 2001). Noninvasive bacteria, i.e. C. clostridioforme JCM 1291T, L. johnsonii La1 and L. delbrueckii subsp. bulgaricus LB-021001, slightly stimulated IL-6 mRNA expression.
Thus we found that certain commensal bacteria, which can invade epithelial cells, stimulated IL-8 and TNF-α secretion and correspondingly increased expression of IL-8, TNF-α, MCP-1 and IL-6 mRNA in the invaded epithelial cells. However, there was no enhancement of IL-8 or TNF-α secretion and minimal expression of IL-8, TNF-α, MCP-1 and IL-6 mRNA with noninvasive commensal bacteria and probiotics. Expression of IL-8 and TNF-α mRNAs correlated somewhat with cytokine secretion, indicating definite cytokine production in response to bacterial invasion. However, MCP-1 and IL-6 were below their detection limits, despite considerable mRNA expression being demonstrated by real-time RT-PCR. This discrepancy may be due to the relatively high detection limits for MCP-1 and IL-6. These results suggest that commensal bacteria can both induce and maintain colonic inflammation.
In previous studies, we have shown that bacterial rods and cocci, including F. varium, adhered to and invaded the intestinal epithelia and invaded the mucosa propria in patients with UC but not in those with Crohn's disease (Ohkusa et al., 1993, 2002). In this study, we used the type strain and a clinical isolate of F. varium; the two strains have almost the same virulence factors, such as enhancement of inflammatory cytokine secretion and invasion of epithelial cells.
Immunoreactive cytokine expression in inflamed mucosal epithelia in patients with UC
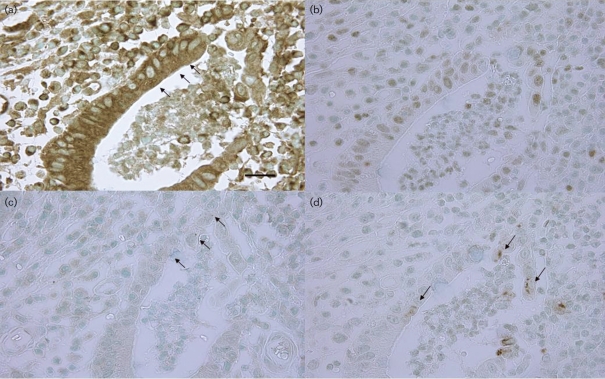
To confirm the results described above by in vitro co-culture experiments, we attempted to visualize the immunoreactive expression of TNF-α, IL-8, NF-κB p65 and phosphor-NF-κB p65 in inflamed crypt epithelia with active UC (cryptitis and crypt abscesses) and in rectal epithelia from patients with active UC, using an immunohistochemical method, and results were compared among rectal mucosae of normal controls, inactive UC specimens and active UC specimens. Cytoplasmic granular expression of TNF-α was extensive in epithelia of half of the crypts with cryptitis and 68 % of those with crypt abscesses, while no expression was detected in crypts of normal control or inactive UC samples. Similarly, inflamed epithelium expressed IL-8 in the cytoplasm in active UC specimens but not in crypts of normal control and inactive UC specimens. NF-κB p65 was ubiquitously expressed in the cytoplasm of rectal epithelial cells. However, crypts with epithelia showing nuclear expression were frequently found in active UC specimens, whereas such crypts were rarely seen in normal control and inactive UC samples. Activated phosphor-NF-κB p65 was also frequently found in nuclei of inflamed epithelia in active UC samples, but not in those of normal control and inactive UC samples (Fig. 6, Table 2).
Fig. 6.
Representative photographs of immunohistochemical expression of TNF-α, IL-8, NF-κB p65 and phosphor-NF-κB p65 in semiserial sections of inflamed epithelium featuring cryptitis or crypt abscesses. (a) TNF-α expression in a crypt abscess. Note positive granules in the epithelial cytoplasm (arrows) (bar, 25 μm). (b) IL-8 expression in a crypt abscess. Note the positive reaction in the epithelial cytoplasm. (c) NF-κB p65 expression in epithelial nuclei in a crypt abscess (arrows). (d) Phosphor-NF-κB p65 expression in epithelial nuclei in a crypt abscess (arrows).
Table 2.
Immunoreactive cytokine expression in mucosal epithelium (positive crypts/crypts examined)
Crypts contained at least two positive epithelia.
| Rectal mucosa* | TNF-α | IL-8 | NF-κB p65 in nuclei | Phosphor-NF-κB p65 in nuclei |
|---|---|---|---|---|
| Normal control | 0/20 (0.0 %) | 0/20 (0.0 %) | 1/20 (5.0 %) | 0/20 (0.0 %) |
| UC, inactive | 0/24 (0.0 %) | 0/24 (0.0 %) | 5/24 (20.8 %) | 3/24 (12.5 %) |
| UC, active | ||||
| Cryptitis | 6/12 (50.0 %)† | 10/12 (83.3 %)‡ | 12/12 (100.0 %)‡ | 12/12 (83.3 %)‡ |
| Crypt abscess | 17/25 (68.0 %)‡ | 21/25 (84.0 %)‡ | 25/25 (100.0 %)‡ | 19/25 (76.0 %)‡ |
*More than 10 cases examined.
†P <0.0005 as compared to normal control or inactive UC samples.
‡P <0.0001 as compared to normal control or inactive UC samples.
Although epithelia from crypts of normal control or inactive UC samples were not positive for either TNF-α or IL-8, those of active UC samples were positive for both, supporting our proposed pathway. Furthermore, nuclear expression of NF-κB p65 and phosphor-NF-κB p65 was also demonstrated in most of these inflamed crypts, indicating that NF-κB activation is due to stimulation of inflammatory cytokine production and release. In our previous study, F. varium invasion was lower in inactive UC (Ohkusa et al., 2002), and bacterial invasion may spontaneously decrease without eradication therapy in inactive UC. Therefore, epithelia from crypts in inactive UC samples may not be positive for either TNF-α or IL-8.
Previous studies by other investigators have shown that although human intestinal epithelial cell lines constitutively express IL-8 and TGF-β1, other cytokines such as TNF-α are expressed only after challenge with invasive strains of enteropathogenic bacteria (Eckmann et al., 1993a, b). Moreover, expression of TNF-α and IL-8 genes is not altered by incubation with noninvasive E. coli or Enterococcus faecium (Jung et al., 1995). Adherence is a common characteristic shared by many pathogens since it is a crucial step for establishing an infection (Mann & Petri, 1995). However, we found that noninvasive commensal bacteria and probiotics, despite having the ability to adhere to cells, could not stimulate IL-8 secretion. Therefore, bacterial invasion appears to be a critical factor for the enhancement of secretion and expression of proinflammatory cytokines in colonic epithelial cells.
We conclude that there are harmful commensal bacteria in the colon that can invade the colonic epithelia, induce epithelial production of proinflammatory cytokines, and activate intracellular inflammatory signalling. Therefore, F. varium isolated from the colonic mucosa of patients with UC may be among the causative agents of active colitis. Recently, Andoh et al. (2007) performed terminal restriction fragment length polymorphism analysis of the faecal microbiota from UC patients and reported that terminal restriction fragments derived from Fusobacterium and other unclassified bacteria were predominantly detected in the active UC patients but not in the inactive UC patients and the healthy individuals. These findings have implications for antibiotic therapy against harmful commensal bacteria, aimed at preventing colonic inflammation (Nomura et al., 2005; Ohkusa et al., 2005).
Acknowledgments
This work was supported in part by Grants in Aid for Scientific Research (15590688, 18659102) from the Japanese Ministry of Education, Culture, Sports, Science and Technology, NIH grants AI09268, GM54666, DE08293 and DE08240, and by grants from the Uehara Memorial Foundation. We thank Malcolm Moore, PhD, for revision of the scientific English language.
Abbreviations
IL, interleukin
SEM, scanning electron microscopy
TEM, transmission electron microscopy
UC, ulcerative colitis
References
- Aihara, M., Tsuchimoto, D., Takizawa, H., Azuma, A., Wakebe, H., Ohmoto, Y., Imagawa, K., Kikuchi, M., Mukaida, N. & Matsuhima, K. (1997). Mechanisms involved in Helicobacter pylori-induced interleukin-8 production by a gastric cancer cell line, MKN45. Infect Immun 65, 3218–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andoh, A., Sakata, S., Koizumi, Y., Mitsuyama, K., Fujiyama, Y. & Benno, Y. (2007). Terminal restriction fragment length polymorphism analysis of the diversity of fecal microbiota in patients with ulcerative colitis. Inflamm Bowel Dis 13, 955–962. [DOI] [PubMed] [Google Scholar]
- Burke, D. A. & Axon, A. T. (1988). Adhesive Escherichia coli in inflammatory bowel disease and infective diarrhoea. BMJ 297, 102–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann, L., Kagnoff, M. F. & Fierer, J. (1993a). Epithelial cells secrete the chemokine interleukin-8 in response to bacterial entry. Infect Immun 61, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann, L., Jung, H. C., Schurer-Maly, C., Panja, A., Morzycka-Wroblewska, E. & Kagnoff, M. F. (1993b). Differential cytokine expression by human intestinal epithelial cell lines: regulated expression of interleukin 8. Gastroenterology 105, 1689–1697. [DOI] [PubMed] [Google Scholar]
- Guarner, F., Casellas, F., Borruel, N., Antolin, M., Videla, S., Vilaseca, J. & Malagelada, J. R. (2002). Role of microecology in chronic inflammatory bowel diseases. Eur J Clin Nutr 56 (Suppl. 4), S34–S38. [DOI] [PubMed] [Google Scholar]
- Han, Y. W. & Miller, V. L. (1997). Reevaluation of the virulence phenotype of the inv yadA double mutants of Yersinia pseudotuberculosis. Infect Immun 65, 327–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino, K., Takeuchi, O., Kawai, T., Sanjo, H., Ogawa, T., Takeda, Y., Takeda, K. & Akira, S. (1999). Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol 162, 3749–3752. [PubMed] [Google Scholar]
- Jung, H. C., Eckmann, L., Yang, S. K., Panja, A., Fierer, J., Morzycka-Wroblewska, E. & Kagnoff, M. F. (1995). A distinct array of proinflammatory cytokines is expressed in human colon epithelial cells in response to bacterial invasion. J Clin Invest 95, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhara, T., Iigo, M., Itoh, T., Ushida, Y., Sekine, K., Terada, N., Okamura, H. & Tsuda, H. (2000). Orally administered lactoferrin exerts an antimetastatic effect and enhances production of IL-18 in the intestinal epithelium. Nutr Cancer 38, 192–199. [DOI] [PubMed] [Google Scholar]
- Mann, B. J. & Petri, W. A., Jr (1995). The role of microbial adherence factors in gastrointestinal disease. In Infections of the Gastrointestinal Tract, pp. 99–105. Edited by M. J. Blaser, P. D. Smith, J. I. Ravdin, H. B. Greenberg & R. L. Guerrant. New York: Raven Press.
- Matsuda, H., Fujiyama, Y., Andoh, A., Ushijima, T., Kajinami, T. & Bamba, T. (2000). Characterization of antibody responses against rectal mucosa-associated bacterial flora in patients with ulcerative colitis. J Gastroenterol Hepatol 15, 61–68. [DOI] [PubMed] [Google Scholar]
- Mikami, T., Yoshida, T., Akino, F., Motoori, T., Yajima, M. & Okayasu, I. (2003). Apoptosis regulation differs between ulcerative colitis-associated and sporadic colonic tumors: association with surviving and Bcl-2. Am J Clin Pathol 119, 723–730. [DOI] [PubMed] [Google Scholar]
- Mitchell, T. J., Whittaker, S. J. & John, S. (2003). Dysregulated expression of COOH-terminally truncated Stat5 and loss of IL2-inducible Stat5-dependent gene expression in Sezary syndrome. Cancer Res 63, 9048–9054. [PubMed] [Google Scholar]
- Mitsuhashi, J., Mikami, T., Saigenji, K. & Okayasu, I. (2005). Significant correlation of morphological remodeling in ulcerative colitis with disease duration and between elevated p53 and p21WAF1 expression in rectal mucosa and neoplastic development. Pathol Int 55, 113–121. [DOI] [PubMed] [Google Scholar]
- Mylonaki, M., Rayment, N. B., Rampton, D. S., Hudspith, B. N. & Brostoff, J. (2005). Molecular characterization of rectal mucosa-associated bacterial flora in inflammatory bowel disease. Inflamm Bowel Dis 11, 481–487. [DOI] [PubMed] [Google Scholar]
- Nomura, T., Ohkusa, T., Okayasu, I., Yoshida, T., Sakamoto, M., Hayashi, H., Benno, Y., Hirai, S., Hojo, M. & other authors (2005). Mucosa-associated bacteria in ulcerative colitis before and after antibiotic combination therapy. Aliment Pharmacol Ther 21, 1017–1027. [DOI] [PubMed] [Google Scholar]
- Ohkusa, T., Okayasu, I., Tokoi, S. & Ozeki, Y. (1993). Bacterial invasion into the colonic mucosa in ulcerative colitis. J Gastroenterol Hepatol 8, 116–118. [DOI] [PubMed] [Google Scholar]
- Ohkusa, T., Sato, N., Ogihara, T., Morita, K., Ogawa, M. & Okayasu, I. (2002). Fusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibody. J Gastroenterol Hepatol 17, 849–853. [DOI] [PubMed] [Google Scholar]
- Ohkusa, T., Okayasu, I., Ogihara, T., Morita, K., Ogawa, M. & Sato, N. (2003). Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut 52, 79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkusa, T., Nomura, T. & Sato, N. (2004). The role of bacterial infection in the pathogenesis of inflammatory bowel disease. Intern Med 43, 534–539. [DOI] [PubMed] [Google Scholar]
- Ohkusa, T., Nomura, T., Terai, T., Miwa, H., Kobayashi, O., Hojo, M., Takei, Y., Ogihara, T., Hirai, S. & other authors (2005). Effectiveness of antibiotic combination therapy in patients with active ulcerative colitis: a randomized, controlled pilot trial with long-term follow up. Scand J Gastroenterol 40, 1334–1342. [DOI] [PubMed] [Google Scholar]
- Pagès, F., Berger, A., Henglein, B., Piqueras, B., Danel, C., Zinzindohoue, F., Thiounn, N., Cugnenc, P. H. & Fridman, W. H. (1999). Modulation of interleukin-18 expression in human colon carcinoma: consequences for tumor immune surveillance. Int J Cancer 84, 326–330. [DOI] [PubMed] [Google Scholar]
- Paulukat, J., Bosmann, M., Nold, M., Garkisch, S., Kämpfer, H., Frank, S., Raedle, J., Zeuzem, S., Pfeilschifter, J. & Mühl, H. (2001). Expression and release of IL-18 binding protein in response to IFN-gamma. J Immunol 167, 7038–7043. [DOI] [PubMed] [Google Scholar]
- Sadlack, B., Merz, H., Chorley, H., Schimple, A., Feller, A. C. & Horak, I. (1993). Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell 75, 253–261. [DOI] [PubMed] [Google Scholar]
- Sartor, R. B. (2004). Microbial influences in inflammatory bowel disease: role in pathogenesis and clinical implications. In Kirsner's Inflammatory Bowel Disease, 6th edn, pp. 138–140. Edited by R. B. Sartor & W. J. Sandborn. Edinburgh: Saunders.
- Sellon, R. K., Tonkonogy, S., Schultz, M., Dieleman, L. A., Grenther, W., Balish, E., Rennick, D. M. & Sartor, B. B. (1998). Resident enteric flora are necessary for development of spontaneous colitis and immune system activation in IL-10-deficient mice. Infect Immun 66, 5224–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, M., Okada, M., Yao, T., Ueki, M., Arima, S. & Okumura, M. (1992). An index of disease activity in patients with ulcerative colitis. Am J Gastroenterol 87, 971–977. [PubMed] [Google Scholar]
- Singer, M. & Sansonetti, P. J. (2004). IL-8 is a key chemokine regulating neutrophil recruitment in a new mouse model of Shigella-induced colitis. J Immunol 173, 4197–4206. [DOI] [PubMed] [Google Scholar]
- Suzuki, M., Hisamatsu, T. & Podolsky, D. K. (2003). Gamma interferon augments the intracellular pathway for lipopolysaccharide (LPS) recognition in human intestinal epithelial cells through coordinated up-regulation of LPS uptake and expression of the intracellular Toll-like receptor 4-MD-2 complex. Infect Immun 71, 3503–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski, A., Ladhoff, A., Pernthaler, A., Swidsinski, S., Loening-Baucke, V., Ortner, M., Weber, J., Hoffmann, U., Schreiber, S. & other authors (2002). Mucosal flora in inflammatory bowel disease. Gastroenterology 122, 44–54. [DOI] [PubMed] [Google Scholar]
- Vitiello, M., D'Isanto, M., Galdiero, M., Raieta, K., Tortora, A., Rotondo, P., Peluso, L. & Galdiero, M. (2004). Interleukin-8 production by THP-1 cells stimulated by Salmonella enterica serovar Typhimurium porins is mediated by AP-1, NF-kappaB and MAPK pathways. Cytokine 27, 15–24. [DOI] [PubMed] [Google Scholar]
- Weinstein, D. L., O'Neill, B. L. & Metcalf, E. S. (1997). Salmonella typhi stimulation of human intestinal epithelial cells induces secretion of epithelial cell-derived interleukin-6. Infect Immun 65, 395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, T., Haga, S., Numata, Y., Yamashita, K., Mikami, T., Ogawa, T., Ohkusa, T. & Okayasu, I. (2006). Disruption of p53-p53R2 DNA repair system in ulcerative colitis contributes to colon tumorigenesis. Int J Cancer 118, 1395–1403. [DOI] [PubMed] [Google Scholar]