Abstract
The family of leptospiral immunoglobulin-like (lig) genes comprises ligA, ligB and ligC. This study used PCR to demonstrate the presence of lig genes among serovars from a collection of leptospiral strains and clinical isolates. Whilst ligA and ligC appeared to be present in a limited number of pathogenic serovars, the ligB gene was distributed ubiquitously among all pathogenic strains. None of the lig genes were detected among intermediate or saprophytic Leptospira species. It was also shown that, similar to the previously characterized secY gene, a short specific PCR fragment of ligB could be used to correctly identify pathogenic Leptospira species. These findings demonstrate that ligB is widely present among pathogenic strains and may be useful for their reliable identification and classification.
INTRODUCTION
Leptospirosis is a re-emerging zoonotic disease caused by Leptospira species, which are transmitted to humans through direct or indirect contact with contaminated urine from a reservoir host, usually rats or other rodents (Faine et al., 1999). DNA–DNA hybridization studies have identified 19 Leptospira species to date (Yasuda et al., 1987; Brenner et al., 1999; Levett, 2001; Levett et al., 2006; Matthias et al., 2008; Slack et al., 2008). Among these, Leptospira interrogans, Leptospira borgpetersenii, Leptospira santarosai, Leptospira noguchii, Leptospira weilii, Leptospira kirschneri and Leptospira alexanderi are considered to be the main agents of leptospirosis (Levett et al., 2006). Serological methods have identified >300 serovars of which more than 200 are considered pathogenic (Faine et al., 1999; Levett, 2001; Bharti et al., 2003).
The lig genes, ligA, ligB and ligC, encode virulence determinants in pathogenic strains (Palaniappan et al., 2002; Matsunaga et al., 2003; Choy et al., 2007; Lin & Chang, 2007). The Lig proteins were identified as markers for the early diagnosis of leptospirosis (Croda et al., 2007; Srimanote et al., 2008) and as potential vaccine candidates (Koizumi & Watanabe, 2004; Palaniappan et al., 2006; Silva et al., 2007; Faisal et al., 2008; Yan et al., 2009). Previously, we determined that the lig genes are highly conserved (70–99 % identity) in virulent pathogenic Leptospira isolates (McBride et al., 2009). The ligB gene was present in all isolates, whilst ligA was limited to L. interrogans and L. kirschneri strains and ligC was a pseudogene in several isolates.
Molecular tools employed for the classification of Leptospira species include PFGE (Herrmann et al., 1992; Galloway & Levett, 2008), RFLP (Brown & Levett, 1997; Barocchi et al., 2001), arbitrarily primed PCR (Perolat et al., 1994), fluorescent amplified fragment length polymorphism (Vijayachari et al., 2004) and variable number tandem repeats (Majed et al., 2005; Slack et al., 2005; Salaün et al., 2006). However, these techniques lack reproducibility or have low sensitivity or specificity (Levett et al., 2006). 16S rRNA gene sequencing has been used in phylogenetic analyses (Hookey et al., 1993) but these genes exhibit a low degree of polymorphism, limiting their usefulness in typing. A limitation of a previous investigation of lig genes was the small number of isolates studied (McBride et al., 2009). To this end, we proposed to determine the presence of lig genes in an expanded collection of strains using a PCR-based assay. In addition, we found that it was possible to type the pathogenic leptospires to the species level using the ligB sequence. We therefore investigated the possibility of using the ligB sequences from the PCR assay for the molecular characterization of pathogenic Leptospira isolates.
METHODS
Bacterial strains and culture conditions.
Reference and clinical strains belonging to 10 species and including 40 serovars were obtained from the collections maintained at the Gonçalo Moniz Research Centre, Salvador, Brazil, and the National Reference Centre for Leptospirosis at the Institut Pasteur, Paris, France. Clinical strains were isolated from both humans and animals and from diverse geographical regions, including Brazil, Russia, Croatia and Guadeloupe (Majed et al., 2005; Silva et al., 2008). All strains were cultured at 30 °C in liquid Ellinghausen–McCullough–Johnson–Harris modified Tween 80/bovine albumin medium (Ellinghausen & McCullough, 1965; Johnson & Harris, 1967). A microscopic agglutination test was carried out using a standard method for putative serogroup determination (Levett et al., 2003).
Oligonucleotide design.
Primers were designed using Vector NTI 10 software (Invitrogen). The lig gene sequences deposited in GenBank were aligned, conserved regions were identified and degenerate primers were designed. Fragments from each of the lig genes were amplified and sequenced using primers specific for ligA (PSAF: 5′-CKGAWCTTGTRACYTGGARKTCYTC-3′; PSAR: 5′-TTGTTAATGTTTTCATRTTAYGGC-3′), ligB (PSBF: 5′-ACWRVHVHRGYWDCCTGGTCYTCTTC-3′; PSBR: 5′-TARRHDGCYBTAATATYCGRWYYTCCTAA-3′) and ligC (PSCF: 5′-GAGAAATAYAATCTCCTTCTTCCGG-3′; PSCR: 5′-CCTRTTCGTGTTGGARGAATTCC-3′).
DNA manipulation.
Genomic DNA was extracted using a GFX Genomic Blood DNA Purification kit following the protocol for Gram-negative bacteria recommended by the manufacturer (GE Healthcare). PCR amplification was performed using Taq DNA polymerase (Invitrogen) and the following cycling conditions: one denaturing cycle at 94 °C for 2 min; 35 cycles of denaturing at 94 °C for 30 s, annealing at 54 °C for 30 s and elongation at 72 °C for 45 s; and a final elongation at 72 °C for 10 min. The amplified products were analysed by 1 % agarose gel electrophoresis.
Sequencing.
PCR products were purified using a GFX PCR DNA and Gel Band Purification kit according to the manufacturer's instructions (GE Healthcare). Sequencing was performed using a MegaBACE 500 DNA sequencer (GE Healthcare) and Dynamic ET Terminator technology. The assembled sequences were analysed by blast alignment (http://www.ncbi.nlm.nih.gov/BLAST) against the available lig gene sequences in GenBank. The lig sequences were aligned using AlignX software (Invitrogen).
Phylogenetic analysis.
The ligB gene sequences from 48 pathogenic strains (Table 1) were used to assemble a phylogenetic tree with the mega 4 software (Tamura et al., 2007). 16S rRNA gene sequences were obtained from GenBank (Table 1) and aligned as described. One thousand bootstrap replications were used to provide confidence in the nodes. The trees were constructed by the neighbour-joining method using the Jukes–Cantor model (Tamura et al., 2007). Synonymous/non-synonymous data were calculated using mega 4.1β software. rpoB sequences used for comparison were obtained from GenBank (accession nos DQ296129–DQ296147; La Scola et al., 2006).
Table 1.
Distribution of lig genes
GenBank accession numbers for the 16S rRNA gene are given in parentheses. nd, Not determined.
| Species | Serovar | Strain | 16S rRNA gene* | PCR | ||
|---|---|---|---|---|---|---|
| ligA | ligB | ligC | ||||
| Pathogens | ||||||
| L. interrogans | Australis | Ballico | + (FJ154556) | + | + | +† |
| Autumnalis | Akiyami A | + (FJ154543) | + | + | + | |
| Bataviae | Van Tienen | + (FJ154566) | + | + | + | |
| Bratislava | Jez Bratislava | + (FJ154547) | + | + | + | |
| Canicola | Hond Utrech IV | + (FJ154561) | + | + | +† | |
| Canicola | Kito | + | +‡ | +‡ | +‡ | |
| Canicola | Mex 1 | + | +‡ | +‡ | +†‡ | |
| Copenhageni | Fiocruz L1-130 | + (AY461869) | +‡ | +‡ | +‡ | |
| Copenhageni | M 20 | + (FJ154542) | + | + | + | |
| Hardjo-prajitno | Hardjoprajitno | + (FJ154553) | + | + | + | |
| Hebdomadis | Hebdomadis | + (FJ154551) | + | + | + | |
| Icterohaemorrhagiae | RGA§ | + (FJ154549) | + | + | + | |
| Kennewicki | LT 1026 | + (FJ154571) | + | + | + | |
| Lai | 56601 | (AY461870) | – | + | + | |
| Lai | Lai | + | + | + | + | |
| Manilae | LT 398 | + (FJ154545) | + | + | + | |
| Muenchen | Munchen C90 | + (FJ154565) | + | + | + | |
| Pomona | PO-06-047 | + | +‡ | +‡ | +‡ | |
| Pomona | Pomona | + (FJ154544) | + | + | + | |
| Wolffi | 3705 | + (FJ154558) | + | + | + | |
| L. kirschneri | Cynopteri | 3522 C§ | + (FJ154546) | +† | + | +† |
| Djatzi | HS 26 | + | +† | + | + | |
| Erinaceiauriti | Erinaceus auritus 670 | + (FJ154560) | +† | + | + | |
| Grippotyphosa | 2.002.297|| | + | nd | +† | nd | |
| Grippotyphosa | 2.002.306|| | + | nd | +† | nd | |
| Grippotyphosa | 2000.11.449|| | + | nd | +† | nd | |
| Grippotyphosa | RM52 | + (AY461877) | +‡ | +‡ | +‡ | |
| Kambale | Kambale | + (FJ154562) | + | + | + | |
| Mozdok | 5621 | + (FJ154559) | + | + | + | |
| Ramisi | Musa | + (FJ154573) | +† | + | +† | |
| L. borgpetersenii | nd | 2E02 | + | –†‡ | +†‡ | – |
| Ceylonica | Piyasena | + (FJ154596) | – | + | – | |
| Istrica | M 18|| | + | nd | +† | nd | |
| Javanica | Veldrat Batavia 46§ | + (FJ154600) | – | + | – | |
| nd | 2002.10.110|| | + | nd | +† | nd | |
| Mini | Sari | + (FJ154592) | – | +† | – | |
| Poi | Poi | + (FJ154597) | – | + | – | |
| Hardjo-bovis | L550 | (NC_008508) | – | + | – | |
| Hardjo-bovis | JB197 | (NC_008510) | – | + | – | |
| Tarassovi | Perepelitsin | + (FJ154595) | – | +† | – | |
| L. noguchii | Bataviae¶ | Cascata | + (EU349495) | –‡ | +‡ | –‡ |
| Orleans | LSU 2580 | + (FJ154588) | – | + | – | |
| Panama | CZ 214 K§ | + (FJ154582) | – | + | + | |
| L. weilii | Hebdomadis¶ | Ecochallenge | + (AY034037) | –‡ | +‡ | +‡ |
| Celledoni | Celledoni§ | + (FJ154580) | – | + | – | |
| Coxi | Cox | + | – | +† | +† | |
| Vughia | LT 89-68 | + (FJ154590) | – | + | + | |
| nd | 2007.025.92|| | + | nd | + | nd | |
| L. santarosai | Alexi | HS 616 | + (FJ154585) | – | + | – |
| Shermani | LT 821§ | +(AY631883) | nd | + | nd | |
| Trinidad | TRVL 34056 | + (FJ154598) | – | +† | – | |
| nd | 2008.010.55|| | + | nd | + | nd | |
| Intermediates | ||||||
| L. fainei | Hurstbridge | But 6§ | + (FJ154578) | – | – | – |
| L. inadai | Lyme | 10§ | + | nd | – | nd |
| Saprophytes | ||||||
| L. meyeri | Semaranga | Veldrat Semarang 173§ | + | – | – | – |
| L. biflexa | Semaranga | Patoc 1§ | + | –‡ | –‡ | –‡ |
*Internal PCR control (Postic et al., 2000). For entries without a ‘+’, the sequence from GenBank was used but they were not amplified as a control in the experiment.
†PCR products not sequenced.
‡Confirmed by Southern blot analysis.
§Type strain.
||Clinical isolate.
¶Serogroup.
Southern blotting.
A total of 3 μg genomic DNA was digested with 20 U BamHI (Invitrogen) and separated by agarose gel electrophoresis. DNA was transferred from the gel to a positively charged Hybond-N nylon membrane (GE Healthcare) with a vacuum blotter (Bio-Rad). Probes for each of the lig genes were based on pooled PCR products amplified using the primers described and labelled using an ECL Direct Nucleic Acid Labelling and Detection System (GE Healthcare). Pre-hybridization was carried out at 42 °C for 1 h in hybridization buffer supplemented with 0.5 M NaCl and 5 % blocking agent. Hybridization was carried out overnight at 42 °C in roller bottles. Following hybridization, the membrane was washed twice for 10 min at 55 °C in wash solution (0.4 % SDS, 0.5× SSC). Finally, the membrane was washed twice in 2× SSC for 5 min per wash at room temperature. After incubation with ECL detection reagents, hybridization products were detected by exposure of the membrane to Hyperfilm ECL X-ray film (GE Healthcare).
RESULTS AND DISCUSSION
Distribution of the lig genes in Leptospira species
In our previous study, pairwise alignment of the lig genes allowed the identification of highly conserved regions within the lig genes (interspecies identity ranged from 68 to 99 %; McBride et al., 2009). Based on these regions, primers were designed to successfully amplify lig gene fragments from the Leptospira strains described in this study (Table 1). For ligA, the primers spanned nt 3482–3692 at the C-terminus, the ligB primers spanned nt 2125–2504 within the non-identical region and for ligC the primers spanned nt 1487–1734. The expected sizes of the amplicons were 211 bp (ligA), 380 bp (ligB) and 248 bp (ligC). The PCR results indicated that ligB was conserved in the genome of 100 % (52/52) of the pathogenic strains tested (Table 1) (Ren et al., 2003; Nascimenta et al., 2004; Bulach et al., 2006). Notably, ligA was limited to L. interrogans and L. kirschneri strains and was found in only 26/44 isolates. As well as being present in certain L. interrogans and L. kirschneri strains, ligC was also detected in L. noguchii and L. weilii strains (31/44 strains in total).
To confirm the negative PCR results as true negatives, Southern blot analysis was carried out (Table 1). The hybridization results corroborated the PCR assay findings. These results support previous studies suggesting that the lig genes are only found in pathogenic strains and that of the three lig genes, only ligB is conserved in all pathogenic Leptospira strains (Matsunaga et al., 2003; McBride et al., 2009). The findings presented here add to the growing body of evidence suggesting that Lig proteins are essential virulence determinants in Leptospira species (Matsunaga et al., 2005; Choy et al., 2007; McBride et al., 2009). To ensure that the PCR products were not artefacts, a selection of amplicons (see Table 1) were sequenced and analysed with the lig gene sequences available in GenBank.
Sequence variability of the lig gene fragments
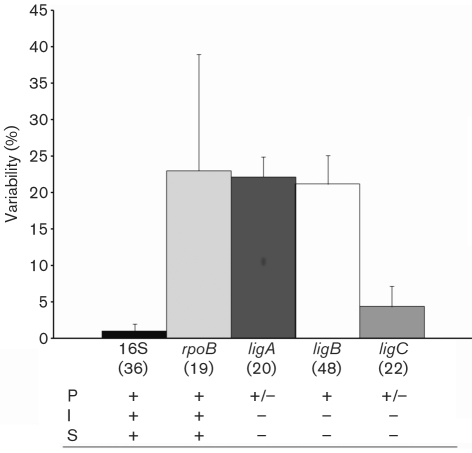
The ligB amplicons exhibited considerable DNA sequence polymorphism, due to indels, particularly at the 5′ and 3′ ends of the 380 bp fragment. Therefore, the ligB sequences were trimmed to remove these hypervariable regions as they did not appear to differ among strains of the same species, and a 214 bp region (nt 2236–2449, L. interrogans Fiocruz L1-130 strain) was identified that exhibited a high level of conservation. The hypervariable region of the 380 bp fragment was identified by comparison with the L. interrogans Fiocruz L1-130 ligB gene. The probability of recombination among the ligB hypervariable regions was confirmed (overall P <0.05) and the ratio between the non-synonymous and synonymous substitutions (dN/dS) was 2.41. This provided evidence for positive selection within this region. Of note, although we did not see any evidence of horizontal transfer within the 214 bp region of ligB, we cannot exclude the possibility that this may occur occasionally. The overall level of pairwise DNA sequence variability was determined to be 21.2±3.9 % (20.6±3.8 % at the amino acid level) for the ligB amplicon (Fig. 1). This DNA fragment demonstrated some interspecies polymorphism, but it was not significant (Fig. 1). The ligB gene is more variable than the previously evaluated ompL1 (15 %), lipL41 (9 %) and lipL32 (3 %) genes (Haake et al., 2004). The mean pairwise DNA sequence variability was 0.8±0.4, 3.7±1.5, 0, 1.2±0.9, 0.9±0.9 and 0 % among the L. interrogans, L. kirschneri, L. noguchii, L. borgpetersenii, L. santarosai and L. weilii strains, respectively (0.8±0.4, 3.7±1.5, 0, 1.7±1.2, 0.9±0.9 and 0 % at the amino acid level, respectively). Furthermore, 17 different ligB orthologues were identified among the 48 Leptospira strains that contained one or more base substitutions within the amplified region.
Fig. 1.
Comparison of the variability of the DNA sequences from the 16S rRNA, rpoB, ligA, ligB and ligC genes from Leptospira species. Results are shown as means±sd. The number of individual sequences used for the determination of sequence variability is indicated in parentheses. The presence (+) and absence (−) of each gene in pathogenic (P), intermediate (I) and saprophytic (S) strains is shown. The nucleotide positions used during the alignment analysis were: nt 75–1255 (16S rRNA gene), 1891–2462 (rpoB), 3482–3693 (ligA), 2236–2449 (ligB) and 1487–1734 (ligC).
The ligA amplicons demonstrated a mean pairwise variability of 21.5±2.4 % among L. interrogans strains and 0.8±0.8 % among L. kirschneri strains (25±3.9 and 0 % at the amino acid level, respectively). The overall mean pairwise DNA sequence variability of the ligA amplicons was 22.2±2.7 % (26.8±4.4 % at the amino acid level) (Fig. 1). The alignment of the ligA sequences revealed the presence of indels in some of the L. interrogans sequences corresponding to the loss of an amino acid codon. The ligC gene exhibited a mean pairwise variability of 1.9±1.7 and 0 % (1.9±1.8 and 0 % at the amino acid level, respectively) among the L. interrogans and L. kirschneri strains, respectively. The overall mean pairwise variability was 4.4±2.8 % (4.4±2.8 % at the amino acid level) (Fig. 1).
The lig genes encode an important family of outer-membrane proteins that are characterized by the presence of immunoglobulin-like domains (Palaniappan et al., 2002; Matsunaga et al., 2003) and are potential virulence determinants of Leptospira species (Choy et al., 2007; Lin & Chang, 2007). These proteins are surface-exposed and are upregulated within mammalian hosts (Matsunaga et al., 2005; Choy et al., 2007). Previous studies have demonstrated their usefulness as markers for diagnosis of leptospirosis (Palaniappan et al., 2004, 2005; Croda et al., 2007; Srimanote et al., 2008) and as potential vaccine candidates (Koizumi & Watanabe, 2004; Palaniappan et al., 2006; Silva et al., 2007; Faisal et al., 2008, 2009). More recently, their presence and conservation among virulent pathogenic strains of Leptospira species was confirmed (McBride et al., 2009). Of note, inactivation of ligB does not result in attenuation of virulence in animal models (Croda et al., 2008). This is probably due to functional redundancy of the Lig proteins, as LigA was expressed in the LigB-knockout strain. Both LigB and LigA can bind the same extracellular matrix and plasma proteins, suggesting that they both play a role during the colonization and dissemination stages of leptospirosis (Choy et al., 2007). In addition, we demonstrated that LigA was created from LigB in a gene duplication event. The N-terminal region of LigB and the LigA paralogue are essentially identical, further supporting the idea that LigA could replace LigB during pathogenesis (McBride et al., 2009). To clarify the role of the Lig proteins in virulence, a ligB/ligA double-knockout strain would be required. The findings of this study confirm the ubiquitous nature of LigB in pathogenic Leptospira species and that LigA and LigC are not present in all strains.
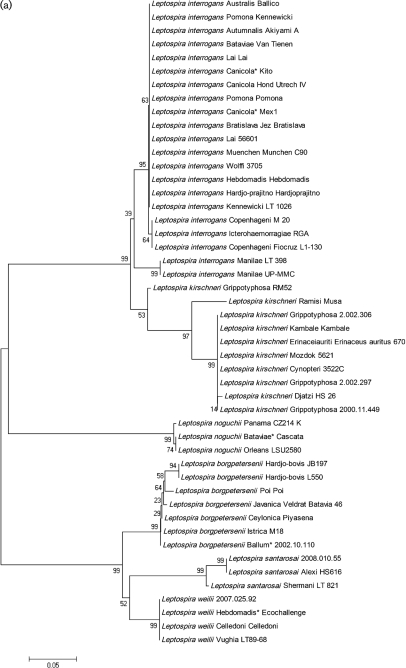
Phylogenetic analysis of ligB
The relatedness of the 48 ligB 214 bp DNA sequences is presented in Fig. 2(a). The Leptospira strains were resolved into two distinct clusters. The sequences from L. interrogans, L. kirschneri and L. noguchii grouped in one cluster, whilst those from L. borgpetersenii, L. santarosai and L. weilii formed the second cluster. The clustering pattern was similar to the phylogenetic tree based on the full-length ligB sequences (McBride et al., 2009). The individual Leptospira species were thus easily determined based on the ligB internal sequence.
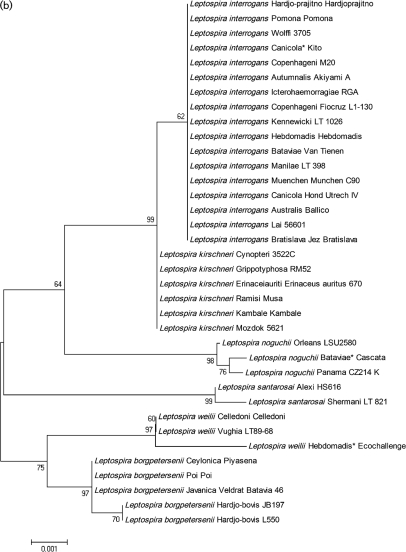
Fig. 2.
Unrooted phylogenetic trees were constructed from the ligB gene (214 bp) (a, this page) and the 16S gene (1181 bp) (b, opposite page). Bootstrap consensus values are indicated. Asterisks indicate serogroups rather than serovars.
The ligB amplicon is situated within a region of the ligB gene that was found to be phylogenetically clonal based on a multiple change-point model in the majority of strains (McBride et al., 2009). Of the two strains that showed evidence of rearrangements (L. interrogans and L. kirschneri), the amplicon was located outside these recombination hotspots. The results demonstrate that the internal ligB sequence can be used to discriminate Leptospira to the species level. Within each major cluster, there was evidence of further subclustering. For example, three out of five of the L. interrogans serogroup Icterohaemorrhagiae strains clustered together, including serovars Copenhageni and Icterohaemorrhagiae. Within the L. kirschneri and L. borgpetersenii cluster, various subclusters were identified but they did not correspond to the serogroups (Fig. 2a). However, there was insufficient discriminatory power to type the serovars beyond the species level. This is a similar situation to that reported for the 16S rRNA gene in Leptospira species (Morey et al., 2006). In addition, previous work by Victoria et al. (2008) demonstrated that the S10-spc-α locus is conserved within pathogenic Leptospira species, but less so among saprophytic and intermediate Leptospira species, indicating that it is a useful region for phylogenetic analysis. Sequence analysis of a short region (245 bp) of the secY gene that is normally used for PCR diagnosis found that it can be used to correctly identify Leptospira strains.
The number of synonymous substitutions within the ligB amplicons was equal or higher than the number of non-synonymous substitutions per site. The probability of the existence of recombination among the several ligB nucleotide sequences was not confirmed (overall P=1.00) and the dN/dS ratio was 0.34. This supports the hypothesis of sequence stability due to the absence of positive selection over this ligB locus. Rejection of the neutrality hypothesis (positive selection suggestive of recombination) in ligB was seen only in the L. borgpetersenii Poi and Veldrat Batavia 46 strains where dN/dS was 1.72 (P=0.04). However, this does not preclude the use of ligB for species typing, as both belong to the same species. The G+C content of the several ligB loci ranged from 37.8 mol% in L. kirschneri to 50 mol% in L. borgpetersenii (data not shown).
Phylogenetic analysis of the 16S rRNA gene sequences
The phylogenetic tree based on the available 16S rRNA gene sequences (Table 1) is presented in Fig. 2(b). The tree describes the relatedness for 36 sequences and the clustering pattern is similar to that described in previous studies (Haake et al., 2004; Levett et al., 2006). The strains clustered according to species: sequences from L. interrogans, L. kirschneri, L. noguchii and L. santarosai formed one cluster, whilst those from L. borgpetersenii and L. weilii formed a second cluster. The major difference between the predicted relatedness patterns is the clustering of the L. santarosai strains.
Traditionally, 16S rRNA gene sequences have been used for Leptospira species classification (Postic et al., 2000; Morey et al., 2006). However, this gene has few polymorphisms throughout its 1500 bp in Leptospira species (Janda & Abbott, 2007). Efforts to identify new markers for species differentiation have focused on the evaluation of partial rpoB (La Scola et al., 2006) and wzy (Wangroongsarb et al., 2007) polymerases, the gyrase subunit B (gyrB; Slack et al., 2006), the pre-protein translocase secY (Victoria et al., 2008) and the genes encoding the surface proteins LipL32, LipL41 and OmpL1 (Haake et al., 2004; Ahmed et al., 2006). The main advantage of selecting housekeeping genes for classification is the constant selection pressure over these genes in the genome. Indeed, secY PCR data alone were able to distinguish leptospires in three distinct lines of evolution, based on their pathogenic potential, and when associated with sequence-based data, conclusions regarding strain classification were possible: Leptospira meyeri strain ICF clustered with the pathogenic strains and Leptospira inadai strain H6 was in fact an L. interrogans strain (Victoria et al., 2008). However, as is the case for the 16S rRNA genes, this is associated with a low accumulation of polymorphisms and hence a lower resolution power in terms of strain differentiation. Genes such as rpoB and gyrB offer the advantage of being shorter and more polymorphic. Recently, La Scola et al. (2006) described three nucleotides that accounted for the differences between the L. kirschneri serovar Cynopteri and L. interrogans serovar Canicola 16S rRNA genes. In addition, Morey et al. (2006) reported that the difference between L. interrogans and L. kirschneri type strains was due to only two nucleotides. This is consistent with descriptions of the high degree of conservation of the 16S rRNA gene among other bacterial species (Janda & Abbott, 2007). rpoB was found to contain 51 polymorphisms over 600 bp when the Cynopteri and Canicola serovars were compared. In this study, the 214 bp ligB sequence contained 23 and 24 polymorphisms between the Cynopteri and Canicola serovars and the L. interrogans and L. kirschneri type strains, respectively.
The taxonomic analysis performed in this study demonstrated the discriminatory power of the ligB gene. We showed that ligB is a molecular marker that is able to differentiate serovars into their respective species (Fig. 2). Recently, we showed that some ligB genes contain mosaic sites, but they were located at the C-terminal end of the gene (McBride et al., 2009). Furthermore, some of the ligB domains were involved in the duplication events that led to the creation of ligA. In this study, we specifically chose a region outside the potential mosaic region that did not include the domains involved in the gene duplication events. In conclusion, the ligB molecular typing scheme demonstrates several major advantages: (i) the ability to differentiate strains to the species level, (ii) discrimination between pathogenic and non-pathogenic strains and (iii) the potential to be employed in multilocus sequence typing or multi-virulence-locus sequence typing analysis for identification of clonal derivation events during the seasonal epidemics and outbreaks associated with urban leptospirosis.
Acknowledgments
G. M. C. was supported by the CAPES Foundation, Brazilian Ministry of Education. This work was supported by Bio-Manguinhos, Oswaldo Cruz Foundation (09224-7 and PDTIS RVR05), the Brazilian National Research Council (grants 01.06.0298.00 3773/2005, 420067/2005, 554788/2006, 473006/2006-5), the National Institutes of Health (5R01 AI052473, 2D43 TW00919), and the Institut Pasteur.
Abbreviations
dN/dS, ratio of non-synonymous to synonymous substitutions
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the lig gene sequences of the Leptospira strains described in this study are EU938447–EU938521.
References
- Ahmed, N., Devi, S. M., Valverde, M. A., Vijayachari, P., Machang'u, R. S., Ellis, W. A. & Hartskeerl, R. A. (2006). Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann Clin Microbiol Antimicrob 5, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barocchi, M. A., Ko, A. I., Ferrer, S. R., Faria, M. T., Reis, M. G. & Riley, L. W. (2001). Identification of new repetitive element in Leptospira interrogans serovar Copenhageni and its application to PCR-based differentiation of Leptospira serogroups. J Clin Microbiol 39, 191–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti, A. R., Nally, J. E., Ricaldi, J. N., Matthias, M. A., Diaz, M. M., Lovett, M. A., Levett, P. N., Gilman, R. H., Willig, M. R. & other authors (2003). Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis 3, 757–771. [DOI] [PubMed] [Google Scholar]
- Brenner, D. J., Kaufmann, A. F., Sulzer, K. R., Steigerwalt, A. G., Rogers, F. C. & Weyant, R. S. (1999). Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int J Syst Bacteriol 49, 839–858. [DOI] [PubMed] [Google Scholar]
- Brown, P. D. & Levett, P. N. (1997). Differentiation of Leptospira species and serovars by PCR-restriction endonuclease analysis, arbitrarily primed PCR and low-stringency PCR. J Med Microbiol 46, 173–181. [DOI] [PubMed] [Google Scholar]
- Bulach, D. M., Zuerner, R. L., Wilson, P., Seemann, T., McGrath, A., Cullen, P. A., Davis, J., Johnson, M., Kuczek, E. & other authors (2006). Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc Natl Acad Sci U S A 103, 14560–14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy, H. A., Kelley, M. M., Chen, T. L., Moller, A. K., Matsunaga, J. & Haake, D. A. (2007). Physiological osmotic induction of Leptospira interrogans adhesion: LigA and LigB bind extracellular matrix proteins and fibrinogen. Infect Immun 75, 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croda, J., Ramos, J. G., Matsunaga, J., Queiroz, A., Homma, A., Riley, L. W., Haake, D. A., Reis, M. G. & Ko, A. I. (2007). Leptospira immunoglobulin-like proteins as a serodiagnostic marker for acute leptospirosis. J Clin Microbiol 45, 1528–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croda, J., Figueira, C. P., Wunder, E. A., Jr, Santos, C. S., Reis, M. G., Ko, A. I. & Picardeau, M. (2008). Targeted mutagenesis in pathogenic Leptospira species: disruption of the LigB gene does not affect virulence in animal models of leptospirosis. Infect Immun 76, 5826–5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghausen, H. C. & McCullough, W. G. (1965). Nutrition of Leptospira pomona and growth of 13 other serotypes: fractionation of oleic albumin complex and a medium of bovine albumin and polysorbate 80. Am J Vet Res 26, 45–51. [PubMed] [Google Scholar]
- Faine, S. B., Adler, B., Bolin, C. & Perolat, P. (1999). Leptospira and Leptospirosis. Melbourne, Australia: MediSci.
- Faisal, S. M., Yan, W., Chen, C. S., Palaniappan, R. U., McDonough, S. P. & Chang, Y. F. (2008). Evaluation of protective immunity of Leptospira immunoglobulin like protein A (LigA) DNA vaccine against challenge in hamsters. Vaccine 26, 277–287. [DOI] [PubMed] [Google Scholar]
- Faisal, S. M., Yan, W., McDonough, S. P. & Chang, Y. F. (2009). Leptospira immunoglobulin-like protein A variable region (LigAvar) incorporated in liposomes and PLGA microspheres produces a robust immune response correlating to protective immunity. Vaccine 27, 378–387. [DOI] [PubMed] [Google Scholar]
- Galloway, R. L. & Levett, P. N. (2008). Evaluation of a modified pulsed-field gel electrophoresis approach for the identification of Leptospira serovars. Am J Trop Med Hyg 78, 628–632. [PubMed] [Google Scholar]
- Haake, D. A., Suchard, M. A., Kelley, M. M., Dundoo, M., Alt, D. P. & Zuerner, R. L. (2004). Molecular evolution and mosaicism of leptospiral outer membrane proteins involves horizontal DNA transfer. J Bacteriol 186, 2818–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, J. L., Bellenger, E., Perolat, P., Baranton, G. & Girons, I. S. (1992). Pulsed field gel electrophoresis of NotI digests of leptospiral DNA: a new rapid method of serovar identification. J Clin Microbiol 30, 1696–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hookey, J. V., Bryden, J. & Gatehouse, L. (1993). The use of 16S rDNA sequence analysis to investigate the phylogeny of Leptospiraceae and related spirochaetes. J Gen Microbiol 139, 2585–2590. [DOI] [PubMed] [Google Scholar]
- Janda, J. M. & Abbott, S. L. (2007). 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 45, 2761–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R. C. & Harris, V. G. (1967). Differentiation of pathogenic and saprophytic leptospires. J Bacteriol 94, 27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi, N. & Watanabe, H. (2004). Leptospiral immunoglobulin-like proteins elicit protective immunity. Vaccine 22, 1545–1552. [DOI] [PubMed] [Google Scholar]
- La Scola, B., Bui, L. T. M., Baranton, G., Khamis, A. & Raoult, D. (2006). Partial rpoB gene sequencing for identification of Leptospira species. FEMS Microbiol Lett 263, 142–147. [DOI] [PubMed] [Google Scholar]
- Levett, P. N. (2001). Leptospirosis. Clin Microbiol Rev 14, 296–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levett, P. N. (2003). Usefulness of serologic analysis as a predictor of the infecting serovar in patients with severe leptospirosis. Clin Infect Dis 36, 447–452. [DOI] [PubMed] [Google Scholar]
- Levett, P. N., Morey, R. E., Galloway, R. L. & Steigerwalt, A. G. (2006). Leptospira broomii sp. nov., isolated from humans with leptospirosis. Int J Syst Evol Microbiol 56, 671–673. [DOI] [PubMed] [Google Scholar]
- Lin, Y. P. & Chang, Y. F. (2007). A domain of the Leptospira LigB contributes to high affinity binding of fibronectin. Biochem Biophys Res Commun 362, 443–448. [DOI] [PubMed] [Google Scholar]
- Majed, Z., Bellenger, E., Postic, D., Pourcel, C., Baranton, G. & Picardeau, M. (2005). Identification of variable-number tandem-repeat loci in Leptospira interrogans sensu stricto. J Clin Microbiol 43, 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga, J., Barocchi, M. A., Croda, J., Young, T. A., Sanchez, Y., Siqueira, I., Bolin, C. A., Reis, M. G., Riley, L. W. & other authors (2003). Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol Microbiol 49, 929–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga, J., Sanchez, Y., Xu, X. & Haake, D. A. (2005). Osmolarity, a key environmental signal controlling expression of leptospiral proteins LigA and LigB and the extracellular release of LigA. Infect Immun 73, 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthias, M. A., Ricaldi, J. N., Cespedes, M., Diaz, M. M., Galloway, R. L., Saito, M., Steigerwalt, A. G., Patra, K. P., Ore, C. V. & other authors (2008). Human leptospirosis caused by a new, antigenically unique Leptospira associated with a Rattus species reservoir in the Peruvian Amazon. PLoS Negl Trop Dis 2, e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride, A. J. A., Cerqueira, G. M., Suchard, M. A., Moreira, A. N., Zuerner, R. L., Reis, M. G., Haake, D. A., Ko, A. I. & Dellagostin, O. A. (2009). Genetic diversity of the leptospiral immunoglobulin-like (Lig) genes in pathogenic Leptospira spp. Infect Genet Evol 9, 196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey, R. E., Galloway, R. L., Bragg, S. L., Steigerwalt, A. G., Mayer, L. W. & Levett, P. N. (2006). Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing. J Clin Microbiol 44, 3510–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento, A. L., Ko, A. I., Martins, E. A., Monteiro-Vitorello, C. B., Ho, P. L., Haake, D. A., Verjovski-Almeida, S., Hartskeerl, R. A., Marques, M. V. & other authors (2004). Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J Bacteriol 186, 2164–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniappan, R. U., Chang, Y. F., Jusuf, S. S., Artiushin, S., Timoney, J. F., McDonough, S. P., Barr, S. C., Divers, T. J., Simpson, K. W. & other authors (2002). Cloning and molecular characterization of an immunogenic LigA protein of Leptospira interrogans. Infect Immun 70, 5924–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniappan, R. U., Chang, Y. F., Hassan, F., McDonough, S. P., Pough, M., Barr, S. C., Simpson, K. W., Mohammed, H. O., Shin, S. & other authors (2004). Expression of leptospiral immunoglobulin-like protein by Leptospira interrogans and evaluation of its diagnostic potential in a kinetic ELISA. J Med Microbiol 53, 975–984. [DOI] [PubMed] [Google Scholar]
- Palaniappan, R. U., Chang, Y. F., Chang, C. F., Pan, M. J., Yang, C. W., Harpending, P., McDonough, S. P., Dubovi, E., Divers, T. & other authors (2005). Evaluation of lig-based conventional and real time PCR for the detection of pathogenic leptospires. Mol Cell Probes 19, 111–117. [DOI] [PubMed] [Google Scholar]
- Palaniappan, R. U., McDonough, S. P., Divers, T. J., Chen, C. S., Pan, M. J., Matsumoto, M. & Chang, Y. F. (2006). Immunoprotection of recombinant leptospiral immunoglobulin-like protein A against Leptospira interrogans serovar Pomona infection. Infect Immun 74, 1745–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perolat, P., Merien, F., Ellis, W. A. & Baranton, G. (1994). Characterization of Leptospira isolates from serovars hardjo by ribotyping, arbitrarily primed PCR, and mapped restriction site polymorphisms. J Clin Microbiol 32, 1949–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic, D., Riquelme-Sertour, N., Merien, F., Perolat, P. & Baranton, G. (2000). Interest of partial 16S rDNA gene sequences to resolve heterogeneities between Leptospira collections: application to L. meyeri. Res Microbiol 151, 333–341. [DOI] [PubMed] [Google Scholar]
- Ren, S. X., Fu, G., Jiang, X. G., Zeng, R., Miao, Y. G., Xu, H., Zhang, Y. X., Xiong, H., Lu, G. & other authors (2003). Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422, 888–893. [DOI] [PubMed] [Google Scholar]
- Salaün, L., Mérien, F., Gurianova, S., Baranton, G. & Picardeau, M. (2006). Application of multilocus variable-number tandem-repeat analysis for molecular typing of the agent of leptospirosis. J Clin Microbiol 44, 3954–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, E. F., Medeiros, M. A., McBride, A. J., Matsunaga, J., Esteves, G. S., Ramos, J. G., Santos, C. S., Croda, J., Homma, A. & other authors (2007). The terminal portion of leptospiral immunoglobulin-like protein LigA confers protective immunity against lethal infection in the hamster model of leptospirosis. Vaccine 25, 6277–6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, E. F., Santos, C. S., Athanazio, D. A., Seyffert, N., Seixas, F. K., Cerqueira, G. M., Fagundes, M. Q., Brod, C. S., Reis, M. G. & other authors (2008). Characterization of virulence of Leptospira isolates in a hamster model. Vaccine 26, 3892–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack, A. T., Dohnt, M. F., Symonds, M. L. & Smythe, L. D. (2005). Development of a multiple-locus variable number of tandem repeat analysis (MLVA) for Leptospira interrogans and its application to Leptospira interrogans serovar Australis isolates from Far North Queensland, Australia. Ann Clin Microbiol Antimicrob 4, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack, A. T., Symonds, M. L., Dohnt, M. F. & Smythe, L. D. (2006). Identification of pathogenic Leptospira species by conventional or real-time PCR and sequencing of the DNA gyrase subunit B encoding gene. BMC Microbiol 6, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack, A. T., Kalambaheti, T., Symonds, M. L., Dohnt, M. F., Galloway, R. L., Steigerwalt, A. G., Chaicumpa, W., Bunyaraksyotin, G., Craig, S. & other authors (2008). Leptospira wolffii sp. nov., isolated from a human with suspected leptospirosis in Thailand. Int J Syst Evol Microbiol 58, 2305–2308. [DOI] [PubMed] [Google Scholar]
- Srimanote, P., Wongdeethai, N., Jieanampunkul, P., Samonkiert, S., Leepiyasakulchai, C., Kalambaheti, T. & Prachayasittikul, V. (2008). Recombinant LigA for leptospirosis diagnosis and ligA among the Leptospira spp. clinical isolates. J Microbiol Methods 72, 73–81. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Dudley, J., Nei, M. & Kumar, S. (2007). mega4: Molecular Evolutionary Genetics Analysis (mega) software version 4.0. Mol Biol Evol 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Victoria, B., Ahmed, A., Zurner, R. L., Ahmed, N., Bulach, D. M., Quinteiro, J. & Hartskeerl, R. A. (2008). Conservation of the S10-spc-α locus within otherwise highly plastic genomes provides phylogenetic insight into the genus Leptospira. PLoS One 3, e2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayachari, P., Ahmed, N., Sugunan, A. P., Ghousunnisa, S., Rao, K. R., Hasnain, S. E. & Sehgal, S. C. (2004). Use of fluorescent amplified fragment length polymorphism for molecular epidemiology of leptospirosis in India. J Clin Microbiol 42, 3575–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangroongsarb, P., Chanket, T., Gunlabun, K., Long, D. H., Satheanmethakul, P., Jetanadee, S., Thaipadungpanit, J., Wuthiekanun, V., Peacock, S. J. & other authors (2007). Molecular typing of Leptospira spp. based on putative O-antigen polymerase gene (wzy), the benefit over 16S rRNA gene sequence. FEMS Microbiol Lett 271, 170–179. [DOI] [PubMed] [Google Scholar]
- Yan, W., Faisal, S. M., McDonough, S. P., Divers, T. J., Barr, S. C., Chang, C. F., Pan, M. J. & Chang, Y. F. (2009). Immunogenicity and protective efficacy of recombinant Leptospira immunoglobulin-like protein B (rLigB) in a hamster challenge model. Microbes Infect 11, 230–237. [DOI] [PubMed] [Google Scholar]
- Yasuda, P. H., Steigerwalt, A. G., Sulzer, L. R., Kaufmann, A. F., Rogers, F. & Brenner, D. J. (1987). Deoxyribonucleic acid relatedness between serogroups and serovars in the family Leptospiraceae with proposals for seven new Leptospira species. Int J Syst Bacteriol 37, 407–415. [DOI] [PubMed] [Google Scholar]