Abstract
The genus Burkholderia includes many bacteria that cause serious human infections. As is the case with other Gram-negative bacteria, Burkholderia species produce LPS, which is an abundant component of the bacterial cell surface. Burkholderia cepacia complex (Bcc) bacteria (which include at least 17 separate species) produce LPS structures that are quite different. In an attempt to determine the degree of LPS epitope variation among Bcc species, a mAb was produced, designated 5D8, specific for the LPS of B. cepacia. Western blot analysis determined that mAb 5D8 was able to produce the classic ‘ladder pattern’ when used to probe B. cepacia and Burkholderia anthina lysates, although 5D8 did not produce this pattern with the other seven Bcc species tested. mAb 5D8 reacted with varying intensity to most but not all of the additional B. cepacia and B. anthina strains tested. Therefore, there seems to be significant epitope variation among Bcc LPS both between and within species. Additionally, mAb 5D8 reacted with a proteinase-K-sensitive 22 kDa antigen in all Bcc strains and also in a strain of Burkholderia pseudomallei.
INTRODUCTION
Bacteria in the genus Burkholderia are Gram-negative bacilli that include species with the potential to cause life-threatening human infections. People with cystic fibrosis (CF) are susceptible to infection by several Burkholderia species comprising the Burkholderia cepacia complex (Bcc) (LiPuma, 2005). ‘Cepacia syndrome’, which is characterized by necrotizing pneumonia and septicaemia resulting in rapid clinical deterioration, can develop in a small percentage of infected CF patients (Mahenthiralingam et al., 2005).
Although much remains unknown about the pathogenic mechanisms of Burkholderia, these species produce a number of putative virulence determinants. One such determinant is LPS, which is present in abundance on the outer membrane of Gram-negative bacteria. Bcc LPS has potent endotoxic activity and elicits significantly higher levels of pro-inflammatory cytokines such as tumour necrosis factor, interleukin-6 and interleukin-8 than LPS from Pseudomonas aeruginosa, another common CF pathogen (Shaw et al., 1995; Vinion-Dubiel & Goldberg, 2003).
The O antigen or O-specific polysaccharide, along with an oligosaccharide core and a lipid A molecule, are components of LPS (De Soyza et al., 2008). Bcc species produce many structurally distinct LPS O antigens (Burtnick et al., 2002; Vinion-Dubiel & Goldberg, 2003). In fact, 16 O-antigen serotypes have been identified among Bcc isolates recovered from CF patients (Nakamura et al., 1986; Soldatkina et al., 1989; Vinion-Dubiel & Goldberg, 2003). These serotypes do not correlate with Bcc species (Kenna et al., 2003; Rabkin et al., 1989). As a consequence, O-antigen serotype cannot be used to determine the specific Bcc species causing an infection. The O-antigen biosynthetic genes of Burkholderia stabilis (a member of the Bcc) have been mapped based on homology to other polysaccharide biosynthetic genes (Vinion-Dubiel & Goldberg, 2003). One gene, wbiI, was sequenced in a number of Bcc isolates and it was shown that the wbiI genotype did not correlate with Bcc species identity (Vinion-Dubiel et al., 2004).
In this study, we produced a mAb specific for B. cepacia LPS, designated 5D8. This mAb was used to analyse LPS epitope variability between and within different Bcc species. We determined that mAb 5D8 bound well to both B. cepacia and Burkholderia anthina but did not bind to the LPS of seven other Bcc species. mAb 5D8 bound to most but not all of the strains of B. cepacia and B. anthina tested. Interestingly, mAb 5D8 bound to a proteinase-K-sensitive 22 kDa antigen in all Bcc strains tested and in Burkholderia pseudomallei strain 1026b. Our data indicate that there is substantial epitope variation in the LPS between and within Bcc species.
METHODS
Bacterial strains.
The bacterial strains used in this study are listed in Table 1. Heat-inactivated B. pseudomallei (strain 1026b) was acquired from Dr Richard Bowen (Colorado State University). Burkholderia thailandensis and non-Burkholderia bacteria were purchased from the ATCC. Burkholderia mallei (inactivated by irradiation) was acquired from the Critical Reagents Program Antigen Repository (BEI Resources, VA, USA). All other Burkholderia species were selected from the Burkholderia cepacia Research Laboratory and Repository at the University of Michigan (MI, USA).
Table 1.
Bacteria used in this study
| Bacterium | Strain | mAb 5D8 LPS immunoreactivity* |
|---|---|---|
| B. cepacia | BTS13 | +++ |
| PC783 | +++ | |
| AU2543 | + | |
| AU2720 | + | |
| AU3398 | − | |
| AU4854 | +++ | |
| AU7573 | ++ | |
| AU8017 | + | |
| AU8316 | +++++ | |
| AU8858 | ++ | |
| AU8973 | − | |
| AU9148 | ++ | |
| AU10048 | ++ | |
| AU15126 | + | |
| B. anthina | AU1293 | ++++ |
| AU3904 | ++ | |
| AU5925 | +++ | |
| AU6369 | ++ | |
| AU12599 | +++ | |
| HI2738 | +++ | |
| HI3538 | − | |
| B. multivorans | HI2229 | − |
| B. cenocepacia | HI2718 | − |
| B. stabilis | HI2210 | − |
| B. vietnamiensis | PC259 | − |
| B. dolosa | AU0654 | − |
| B. ambifaria | HI2468 | − |
| B. pyrrocinia | BC11 | − |
| B. pseudomallei | 1026b | − |
| B. thailandensis | E264 | − |
| B. mallei | BEI resources | − |
| Pseudomonas aeruginosa | ATCC 27853 | − |
| Streptococcus pneumoniae | ATCC 10015 | − |
| Klebsiella pneumoniae | ATCC 13883 | − |
| Staphylococcus aureus | ATCC 25923 | − |
| Enterobacter cloacae | ATCC 23355 | − |
| Providencia stuartii | ATCC 33672 | − |
| Escherichia coli | ATCC 25922 | − |
*−, No reactivity; +, low reactivity; +++++, highest reactivity.
Burkholderia species (excluding B. pseudomallei and B. mallei) were grown on Luria–Bertani (LB) agar or Burkholderia cepacia selective agar plates (Hardy Diagnostics). LB broth was then inoculated with a single colony, followed by incubation at 32 °C for 24–48 h. B. pseudomallei strain 1026b was grown overnight at 37 °C in trypticase soy broth. The culture was pelleted by centrifugation and resuspended in sterile PBS. The procedure for heat-inactivation of B. pseudomallei (80 °C for 2 h) was adapted from a previous study (Steinmetz et al., 1995). Inactivated B. pseudomallei was plated on trypticase soy agar and no growth was observed after 48 h incubation at 37 °C. All non-Burkholderia bacteria were grown from lyophilized stocks in LB broth at 37 °C overnight.
Immunization of mice and production of LPS mAb.
Production of mAb 5D8 began with immunization of BALB/c mice with heat-inactivated B. cepacia (strain BTS13). An intraperitoneal injection of 2×108 bacteria was administered every 2 weeks for an 8-week period (Jones et al., 2002). An ELISA using heat-inactivated BTS13 in the solid phase was used to assess antibody levels to B. cepacia (Jones et al., 2002). Three days prior to the harvest of spleens, one final intraperitoneal immunization of 2×108 bacteria was administered. Hybridoma cells were produced as described previously (Kozel et al., 2004). Western blot analysis was carried out to identify hybridoma cell lines that were producing anti-LPS mAbs specific for B. cepacia. Hybridoma cell line 5D8 was grown in a CL 1000 culture flask (Integra Biosciences) and the mAb was purified by affinity chromatography over a protein A column.
Western blot analysis.
A standard Western blot procedure with semi-dry blotting was used in this study. Briefly, 8×106 bacterial cells were suspended in Laemmli sample buffer (Sigma) and incubated at 100 °C for 10 min. If required, 1 vol. proteinase K solution (3.3 mg in 1 ml Laemmli sample buffer) was added and the sample was incubated at 60 °C for 1 h. The samples were run on either a 10–20 % SDS gradient gel or a 10 % SDS gel followed by semi-dry transfer onto PVDF membrane. mAb 5D8 was used at a final concentration of 1 : 5000 (1 mg ml−1 stock). Horseradish-peroxidase-conjugated goat anti-mouse IgG (Southern Biotech) was used at a 1 : 10 000 dilution and the signal was detected with a chemiluminescent substrate (Pierce). Two silver staining procedures were performed to confirm LPS production in selected bacteria (Fomsgaard et al., 1990; Tsai & Frasch, 1982). Both of these procedures were used to treat the SDS gels prior to staining with a Pierce silver staining kit.
Production of B. cepacia polysaccharide extract.
The procedure for production of B. cepacia polysaccharide extract was adapted from Steinmetz et al. (1995). B. cepacia strain BTS13 was grown on mannitol-rich medium agar plates at 35 °C for 72 h. Bacteria were scraped from the plates and resuspended in 0.01 M PBS containing 1 % formaldehyde (37 %, w/w, stock). Cells were suspended in the PBS/formaldehyde solution for 48 h, followed by centrifugation of the cells at 2000 g for 30 min. The bacterial pellet was resuspended in PBS followed by the addition of 10 % (w/v) sodium acetate and 1 % (v/v) glacial acetic acid. The sample was stirred for 5 h followed by centrifugation at 15 000 g for 1 h to remove the cells. Next, 4 vols 95 % ethanol was added to the supernatant and the sample was placed at −20 °C for 3 h. The precipitate was centrifuged at 4 °C for 30 min at 3000 g followed by resuspension in 80 % ethanol. The precipitate was collected by centrifugation and washed in 95 % ethanol. The precipitate was then dissolved in PBS containing 0.01 M MgCl2. Undissolved particles were removed by centrifugation at 10 000 g for 30 min. RNase A (100 μg ml−1) and DNase I (100 μg ml−1) were added to the solution for 2.5 h at 60 °C, followed by the addition of proteinase K (100 μg ml−1) for 30 min at 60 °C. Enzymes were inactivated at 80 °C for 30 min. The solution was centrifuged at 20 000 g for 30 min at 4 °C. A 10 % (w/v) solution of sodium acetate containing 1 % acetic acid (v/v) was added to the supernatant. The sample was then stirred for 1 h followed by a 30 min centrifugation at 10 000 g. Polysaccharides were precipitated from the solution by the addition of 4 vols 95 % ethanol and the sample was then placed at −20 °C for 3 h. The precipitate was dissolved in water. A phenol/sulfuric acid test (DuBois et al., 1956) for carbohydrate was used to determine the final concentration, which was 1.2 mg ml−1.
Analysis of mAb binding.
An ELISA was performed to evaluate the binding of mAb 5D8 to B. cepacia polysaccharide extract. The procedure was exactly the same as an ELISA used to detect IgG antibodies specific to cryptococcal glucuronoxylomannan (Brandt et al., 2003). In place of glucuronoxylomannan, purified B. cepacia polysaccharide extract (4 μg ml−1) was used in the solid phase of the ELISA and mAb 5D8 was serially diluted across the 96-well plate at a starting concentration of 125 μg ml−1 (fluid phase).
Immunofluorescence analysis.
A suspension of bacteria was smeared onto a microscope slide and allowed to dry. Cells were fixed with methanol and the slide was washed by dipping repeatedly in water. mAb was applied directly to the slide and incubated for 90 min at room temperature. After rinsing by dipping in fresh water, mAb binding was detected using an Alexa Fluor 555-labelled goat anti-mouse IgG (Invitrogen) diluted 1 : 1000 in PBS and incubated for 1 h at room temperature. The slide was then rinsed in fresh water, allowed to dry and mounted under a coverslip with Vectashield (Vector Laboratories). Fluorescence was evaluated using a Nikon Eclipse E800 epifluorescence microscope fitted with a Nikon C1 confocal microscopy attachment (Nikon Instruments).
RESULTS
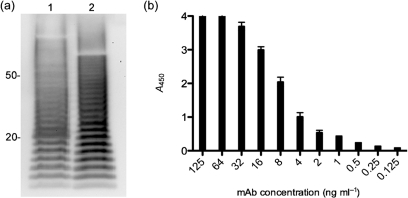
In order to generate mAbs specific to Bcc bacteria, heat-killed B. cepacia strain BTS13 was used to immunize BALB/c mice. mAb 5D8 was generated and used to probe a whole-cell lysate of BTS13 by Western blotting (Fig. 1a). The mAb reacted with multiple bands forming a ladder pattern; similar ladder patterns are seen when purified B. cepacia LPS is separated on polyacrylamide gels (Evans et al., 1999; Leone et al., 2006; Shaw et al., 1995). Next, BTS13 lysates were treated with proteinase K, which did not disrupt the ladder reaction, indicating that mAb 5D8 binds to a polysaccharide. mAb 5D8 was determined to be of the IgG2b subclass, as only anti-IgG2b subclass-specific horseradish-peroxidase-conjugated antibodies bound to the mAb by ELISA (data not shown). Analysis of mAb binding was performed by ELISA (Fig. 1b). Purified BTS13 polysaccharide extract (4 μg ml−1) was used in the solid phase and serially diluted mAb 5D8 was used in the fluid phase. A concentration of 2 ng 5D8 ml−1 was the minimum that could be used to detect BTS13 LPS in the polysaccharide extract. In addition, mAb 5D8 bound to purified antigen in a dose-dependent fashion. A mouse IgG2b mAb specific for the capsular polypeptide of Bacillus anthracis was substituted for mAb 5D8; binding above background levels was not detected (data not shown).
Fig. 1.
Western blot and binding analysis of mAb 5D8. (a) Western blot analysis of B. cepacia (strain BTS13) lysate probed with mAb 5D8. BTS13 cell lysate is shown in lane 1 and proteinase-K-treated BTS13 lysate in lane 2. (b) Analysis of mAb 5D8 binding was carried out by ELISA with purified BTS13 polysaccharide extract in the solid phase (1 μg ml−1) and serially diluted mAb 5D8 in the fluid phase. Data are shown as means±sd for three replicates.
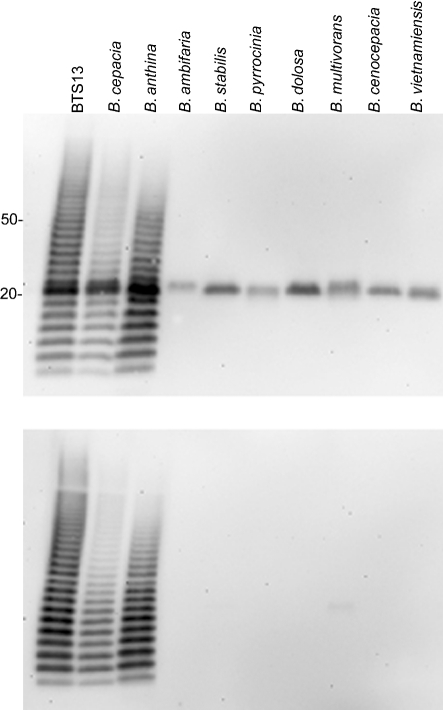
To determine variation in expression of the LPS epitope, we probed bacterial lysates from a panel of Bcc species with mAb 5D8. One representative strain of each of the nine Bcc species was selected from the Bcc experimental strain panel (Coenye et al., 2003; Mahenthiralingam et al., 2000). As shown by Western blotting, B. cepacia strain PC783 and B. anthina strain AU1293 were reactive with mAb 5D8 (Fig. 2). mAb 5D8 reacted with a 22 kDa antigen from the other seven Bcc species. The two B. cepacia strains and the B. anthina strain (Fig. 2, top panel) also expressed a similar 22 kDa antigen that reacted with mAb 5D8. Upon proteinase K treatment of the bacterial lysates, the 22 kDa antigen was no longer reactive by Western blotting (Fig. 2, lower panel).
Fig. 2.
Western blot analysis of Bcc bacterial lysates with mAb 5D8. B. cepacia strain PC783 and B. anthina strain AU1293 are shown; other specific strain designations are listed in Table 1. The lysates shown in the bottom panel were treated with proteinase K. The lanes contain the same bacteria in both immunoblots. B. cepacia strain BTS13 was used as a positive control.
We next wanted to determine whether lysates of B. pseudomallei or B. thailandensis were reactive with mAb 5D8. Western blot analysis determined that a ladder pattern was not produced, but B. pseudomallei did express a 22 kDa antigen that was reactive with mAb 5D8 (Fig. 3). However, B. thailandensis, which is genetically similar to B. pseudomallei (Brett et al., 1998), did not react with mAb 5D8. We also tested a variety of other bacteria outside the genus Burkholderia by Western blotting (Table 1); all were unreactive (data not shown).
Fig. 3.
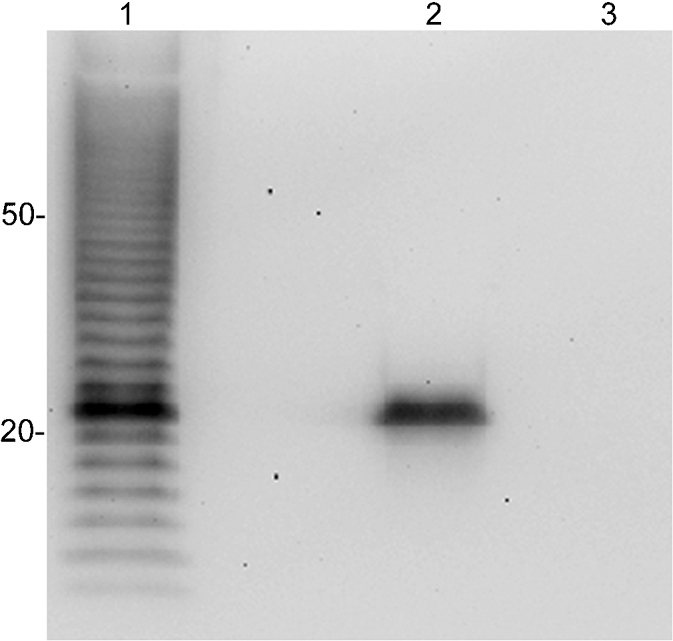
Western blot analysis of B. pseudomallei and B. thailandensis lysates. mAb 5D8 was used to probe B. cepacia strain BTS13 (lane 1), B. pseudomallei (lane 2) and B. thailandensis (lane 3).
Twelve additional strains of B. cepacia and six additional strains of B. anthina were tested by Western blotting to determine whether there was strain-to-strain variation in the epitope recognized by mAb 5D8. These data are summarized in Table 1. Six of the B. cepacia strains produced an obvious ladder pattern, four strains produced a faint ladder pattern and two strains produced no ladder pattern. The same 22 kDa band was present in all 12 of the strains (data not shown). Five of the six additional B. anthina strains also produced a ladder pattern when probed with mAb 5D8. Although difficult to observe when a strong ladder pattern was present, mAb 5D8 was reactive with the 22 kDa antigen of all B. anthina strains tested (data not shown). Two silver staining procedures (Fomsgaard et al., 1990; Tsai & Frasch, 1982) confirmed that LPS was actually produced by the strains that were unreactive with mAb 5D8 (data not shown).
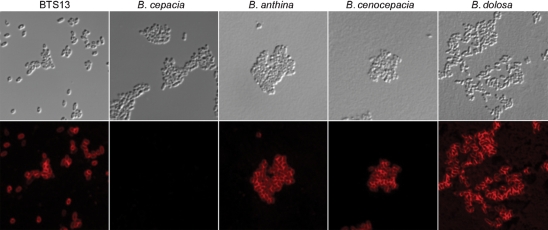
An immunofluorescence assay (IFA) was then performed on the nine representative Bcc strains. As expected, mAb 5D8 bound to the exterior of heat-killed B. cepacia BTS13 (Fig. 4). Interestingly, although B. cepacia strain PC783 produced a ladder reaction by Western blotting (Fig. 2), it did not react with mAb 5D8 by IFA. This was repeated multiple times with varying concentrations of mAb 5D8. Burkholderia dolosa, Burkholderia cenocepacia and B. anthina strain AU1293 (the only B. anthina strain tested) were also reactive by IFA. Notably, mAb 5D8 was reactive with the 22 kDa antigen (but not LPS) of B. cenocepacia and B. dolosa by Western blotting (Fig. 2). The other Bcc species (Burkholderia ambifaria, B. stabilis, Burkholderia pyrrocinia, Burkholderia multivorans and Burkholderia vietnamiensis) that expressed a reactive 22 kDa antigen by Western blotting were not positive by IFA (data not shown). The production of a ladder-type reaction or reactivity of the 22 kDa antigen by Western blotting did not correlate with a positive IFA in some Burkholderia strains. IgG2b Bacillus anthracis mAb F26G3 was substituted for mAb 5D8 as a negative control and no reactivity was evident (data not shown).
Fig. 4.
Immunofluorescence assay showing binding of mAb 5D8. The top panels show differential interference contrast microscopy and the lower panels show binding of mAb 5D8 using Alexa Fluor 555-labelled secondary antibody. B. cepacia strain PC783 and B. anthina AU1293 are shown; other specific strain designations are listed in Table 1.
DISCUSSION
B. cepacia strain BTS13, previously isolated from a CF patient (Lagatolla et al., 2002), was used to immunize BALB/c mice to produce mAbs specific for polysaccharide antigens on the surface of the bacterium. Originally, this strain was chosen because it produces an exopolysaccharide identical to one produced by B. pseudomallei (Cescutti et al., 2003). The goal was to develop a mAb using BTS13 that would bind to the exopolysaccharide produced by B. pseudomallei. However, the first mAb that was isolated (5D8) was specific for the LPS of BTS13 (Fig. 1) and we have shown that 5D8 does not bind to the LPS of B. pseudomallei (Fig. 3).
Bcc bacteria are phenotypically similar but genetically distinct, and it is clear that the LPS structures that are produced by each species (and within a species) may be variable. mAb 5D8 was only able to bind to the B. cepacia and B. anthina strains in our study. We also noted significant strain-to-strain variation in binding reactivity. The epitope recognized by mAb 5D8 seemed to be more conserved or more readily available in B. anthina strains than in B. cepacia strains, even though the mAb was produced by immunization with a B. cepacia strain. Seven Bcc species did not bind to mAb 5D8; however, it is quite possible that if more strains were tested from this negative group, reactive strains would be identified. Importantly, P. aeruginosa, a common lung pathogen in CF patients that can form mixed biofilms with Bcc species (Tomlin et al., 2001), was negative.
Silver staining was carried out on the B. cepacia and B. anthina strains that did not produce a ladder pattern by Western blotting when probed with mAb 5D8. This was done to determine whether the unreactive strains were actually producing polysaccharides consistent with LPS. Two silver staining procedures were needed (Fomsgaard et al., 1990; Tsai & Frasch, 1982) to visualize an LPS ladder from the unreactive strains (data not shown). The fact that different strains required separate protocols for silver staining is additional evidence for the variability of LPS in Bcc.
mAb 5D8 was reactive with a 22 kDa antigen in all Bcc species tested by Western blotting. However, this did not correlate well with binding by IFA. mAb 5D8 was not reactive with an LPS ladder from B. pseudomallei, but it was reactive with what is believed to be the same 22 kDa antigen by Western blotting.
The 22 kDa antigen may be masked on the exterior of some Bcc bacteria yielding a negative IFA result, whereas on B. cenocepacia and B. dolosa the epitope is accessible to 5D8. This may be the case for B. pseudomallei: a negative signal by IFA (data not shown), yet a positive 22 kDa band by Western blotting (Fig. 3).
Bcc species produce a variety of LPS molecules, which has resulted in the identification of at least 16 different LPS serotypes in strains recovered from CF patients (Knirel' Iu et al., 1988a; Nakamura et al., 1986; Vinion-Dubiel & Goldberg, 2003). Binding of LPS-specific bacteriophages (that can use some Bcc bacteria as a host) and O-antigen serotype do not correlate with Bcc species identity (Kenna et al., 2003). Also, no correlation exists between the RFLP pattern of the wbiI gene (which is part of the O-antigen biosynthetic locus) and Bcc species identity (Vinion-Dubiel et al., 2004). Many O-antigen structures from Bcc have been determined and one can speculate about which structure mAb 5D8 may bind to. A recent study has determined the structure of the O-antigen from a strain of B. anthina as [→3)α-l-Rhap-(1→2)α-l-Rhap-(1→2)-α-d-Galp-(1→]n (Carillo et al., 2009). This structure is similar to an O-antigen that was identified in a B. cepacia clinical isolate: [→3)α-d-Rhap-(1→3)α-d-Rhap-(1→4)-α-d-Galp-(1→]n (Cerantola et al., 1999; Faure et al., 2007). However, it is not clear in the literature whether the aforementioned B. cepacia clinical isolate is actually a B. cepacia strain. Also, an additional O-antigen from a B. cepacia strain varies greatly; the structure was determined to be a disaccharide repeat of a glucopyranose followed by a mannopyranose (Cox & Wilkinson, 1990; Knirel' Iu et al., 1988b). Additional studies will need to be completed utilizing strains that produce known O-antigen structures to determine which O antigen(s) react with mAb 5D8.
To our knowledge, this is the first description of a Bcc-specific mAb. There are some potential uses for mAb 5D8. As the mAb binds to a 22 kDa antigen in all Bcc species tested, it may be helpful in diagnosing Bcc infections in CF patients. Also, a Bcc murine model of pulmonary infection has been developed in which certain Bcc species persist in the lungs of infected mice (Chu et al., 2002; Conway et al., 2004). Targeting the 5D8 epitope by active or passive immunization might facilitate clearance of a normally persistent infection.
Acknowledgments
This study was supported by Public Health Service grant (Pacific Southwest Regional Center of Excellence for Biodefence and Emerging Infectious Diseases) no. AI065359 from the National Institute of Allergy and Infectious Diseases. mAb 5D8 is freely available to the research community by contacting the corresponding author.
Abbreviations
Bcc, Burkholderia cepacia complex
CF, cystic fibrosis
IFA, immunofluorescence assay
References
- Brandt, S., Thorkildson, P. & Kozel, T. R. (2003). Monoclonal antibodies reactive with immunorecessive epitopes of glucuronoxylomannan, the major capsular polysaccharide of Cryptococcus neoformans. Clin Diagn Lab Immunol 10, 903–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett, P. J., DeShazer, D. & Woods, D. E. (1998). Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int J Syst Bacteriol 48, 317–320. [DOI] [PubMed] [Google Scholar]
- Burtnick, M. N., Brett, P. J. & Woods, D. E. (2002). Molecular and physical characterization of Burkholderia mallei O antigens. J Bacteriol 184, 849–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carillo, S., Silipo, A., Perino, V., Lanzetta, R., Parrilli, M. & Molinaro, A. (2009). The structure of the O-specific polysaccharide from the lipopolysaccharide of Burkholderia anthina. Carbohydr Res 344, 1697–1700. [DOI] [PubMed] [Google Scholar]
- Cerantola, S., Lemassu-Jacquier, A. & Montrozier, H. (1999). Structural elucidation of a novel exopolysaccharide produced by a mucoid clinical isolate of Burkholderia cepacia. Characterization of a trisubstituted glucuronic acid residue in a heptasaccharide repeating unit. Eur J Biochem 260, 373–383. [DOI] [PubMed] [Google Scholar]
- Cescutti, P., Impallomeni, G., Garozzo, D., Sturiale, L., Herasimenka, Y., Lagatolla, C. & Rizzo, R. (2003). Exopolysaccharides produced by a clinical strain of Burkholderia cepacia isolated from a cystic fibrosis patient. Carbohydr Res 338, 2687–2695. [DOI] [PubMed] [Google Scholar]
- Chu, K. K., Davidson, D. J., Halsey, T. K., Chung, J. W. & Speert, D. P. (2002). Differential persistence among genomovars of the Burkholderia cepacia complex in a murine model of pulmonary infection. Infect Immun 70, 2715–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenye, T., Vandamme, P., LiPuma, J. J., Govan, J. R. & Mahenthiralingam, E. (2003). Updated version of the Burkholderia cepacia complex experimental strain panel. J Clin Microbiol 41, 2797–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway, B. A., Chu, K. K., Bylund, J., Altman, E. & Speert, D. P. (2004). Production of exopolysaccharide by Burkholderia cenocepacia results in altered cell-surface interactions and altered bacterial clearance in mice. J Infect Dis 190, 957–966. [DOI] [PubMed] [Google Scholar]
- Cox, A. D. & Wilkinson, S. G. (1990). Structure of the O-specific polymer for Pseudomonas cepacia serogroup O7. Carbohydr Res 198, 153–156. [DOI] [PubMed] [Google Scholar]
- De Soyza, A., Silipo, A., Lanzetta, R., Govan, J. R. & Molinaro, A. (2008). Chemical and biological features of Burkholderia cepacia complex lipopolysaccharides. Innate Immun 14, 127–144. [DOI] [PubMed] [Google Scholar]
- DuBois, M., Gilles, K. A., Hamilton, J. K., Rebers, P. A. & Smith, F. (1956). Colorimetric method for determination of sugars and related substances. Anal Chem 28, 350–356. [Google Scholar]
- Evans, E., Poxton, I. R. & Govan, J. R. (1999). Lipopolysaccharide chemotypes of Burkholderia cepacia. J Med Microbiol 48, 825–832. [DOI] [PubMed] [Google Scholar]
- Faure, R., Shiao, T. C., Lagnoux, D., Giguere, D. & Roy, R. (2007). En route to a carbohydrate-based vaccine against Burkholderia cepacia. Org Biomol Chem 5, 2704–2708. [DOI] [PubMed] [Google Scholar]
- Fomsgaard, A., Freudenberg, M. A. & Galanos, C. (1990). Modification of the silver staining technique to detect lipopolysaccharide in polyacrylamide gels. J Clin Microbiol 28, 2627–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, S. M., Ellis, J. F., Russell, P., Griffin, K. F. & Oyston, P. C. (2002). Passive protection against Burkholderia pseudomallei infection in mice by monoclonal antibodies against capsular polysaccharide, lipopolysaccharide or proteins. J Med Microbiol 51, 1055–1062. [DOI] [PubMed] [Google Scholar]
- Kenna, D. T., Barcus, V. A., Langley, R. J., Vandamme, P. & Govan, J. R. (2003). Lack of correlation between O-serotype, bacteriophage susceptibility and genomovar status in the Burkholderia cepacia complex. FEMS Immunol Med Microbiol 35, 87–92. [DOI] [PubMed] [Google Scholar]
- Knirel' Iu, A., Shashkov, A. S., Soldatkina, M. A., Paramonov, N. A. & Zakharova, I. (1988a). Antigenic polysaccharides of bacteria. 32. The structure of O-specific polysaccharide chains of Pseudomonas cepacia serotype B and E lipopolysaccharides containing d-fucose. Bioorg Khim 14, 1208–1213. (in Russian) [PubMed] [Google Scholar]
- Knirel' Iu, A., Soldatkina, M. A., Shashkov, A. S., Tanamar, N. V. & Paramonov, N. A. (1988b). Antigenic polysaccharides of bacteria. 34. Structure of O-specific polysaccharide chains of lipopolysaccharides from Pseudomonas cepacia strains IMV 4207 (Serotype A) and IMV 598/2. Bioorg Khim 14, 1678–1683. (in Russian) [PubMed] [Google Scholar]
- Kozel, T. R., Murphy, W. J., Brandt, S., Blazar, B. R., Lovchik, J. A., Thorkildson, P., Percival, A. & Lyons, C. R. (2004). mAbs to Bacillus anthracis capsular antigen for immunoprotection in anthrax and detection of antigenemia. Proc Natl Acad Sci U S A 101, 5042–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagatolla, C., Skerlavaj, S., Dolzani, L., Tonin, E. A., Monti Bragadin, C., Bosco, M., Rizzo, R., Giglio, L. & Cescutti, P. (2002). Microbiological characterisation of Burkholderia cepacia isolates from cystic fibrosis patients: investigation of the exopolysaccharides produced. FEMS Microbiol Lett 209, 99–106. [DOI] [PubMed] [Google Scholar]
- Leone, S., Molinaro, A., Gerber, I. B., Dubery, I. A., Lanzetta, R. & Parrilli, M. (2006). The O-chain structure from the LPS of the endophytic bacterium Burkholderia cepacia strain ASP B 2D. Carbohydr Res 341, 2954–2958. [DOI] [PubMed] [Google Scholar]
- LiPuma, J. J. (2005). Update on the Burkholderia cepacia complex. Curr Opin Pulm Med 11, 528–533. [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam, E., Coenye, T., Chung, J. W., Speert, D. P., Govan, J. R., Taylor, P. & Vandamme, P. (2000). Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J Clin Microbiol 38, 910–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam, E., Urban, T. A. & Goldberg, J. B. (2005). The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol 3, 144–156. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., Hyodo, S., Chonan, E., Shigeta, S. & Yabuuchi, E. (1986). Serological classification of Pseudomonas cepacia by somatic antigen. J Clin Microbiol 24, 152–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabkin, C. S., Jarvis, W. R., Anderson, R. L., Govan, J., Klinger, J., LiPuma, J., Martone, W. J., Monteil, H., Richard, C. & other authors (1989). Pseudomonas cepacia typing systems: collaborative study to assess their potential in epidemiologic investigations. Rev Infect Dis 11, 600–607. [DOI] [PubMed] [Google Scholar]
- Shaw, D., Poxton, I. R. & Govan, J. R. (1995). Biological activity of Burkholderia (Pseudomonas) cepacia lipopolysaccharide. FEMS Immunol Med Microbiol 11, 99–106. [DOI] [PubMed] [Google Scholar]
- Soldatkina, A., Knirel' Iu, A., Tanatar, N. V. & Zakharova, I. (1989). Immunologic and structural studies of lipopolysaccharides from Pseudomonas cepacia. Mikrobiol Zh 51, 32–38. [PubMed] [Google Scholar]
- Steinmetz, I., Rohde, M. & Brenneke, B. (1995). Purification and characterization of an exopolysaccharide of Burkholderia (Pseudomonas) pseudomallei. Infect Immun 63, 3959–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlin, K. L., Coll, O. P. & Ceri, H. (2001). Interspecies biofilms of Pseudomonas aeruginosa and Burkholderia cepacia. Can J Microbiol 47, 949–954. [PubMed] [Google Scholar]
- Tsai, C. M. & Frasch, C. E. (1982). A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem 119, 115–119. [DOI] [PubMed] [Google Scholar]
- Vinion-Dubiel, A. D. & Goldberg, J. B. (2003). Lipopolysaccharide of Burkholderia cepacia complex. J Endotoxin Res 9, 201–213. [DOI] [PubMed] [Google Scholar]
- Vinion-Dubiel, A. D., Spilker, T., Dean, C. R., Monteil, H., LiPuma, J. J. & Goldberg, J. B. (2004). Correlation of wbiI genotype, serotype, and isolate source within species of the Burkholderia cepacia complex. J Clin Microbiol 42, 4121–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]