Abstract
Mycobacterium avium is an opportunistic pathogen associated with pulmonary disease in non-AIDS patients and disseminated infection in patients with AIDS. The chief route of infection is by colonization and invasion of the mucosa of the gastrointestinal tract, but infection through the respiratory route also occurs. After crossing the mucosa, M. avium infects and replicates within tissue macrophages. To identify M. avium genes required for survival in vivo, a library of signature-tagged transposon mutants was constructed and screened for clones attenuated in mice. Thirty-two clones were found to be attenuated for their virulence, from which eleven were sequenced and tested further. All the mutants studied grew similarly in vitro to the wild-type MAC104. Ten mutants were tested individually in mice, confirming the attenuated phenotype. MAV_2450, a polyketide synthase homologue to Mycobacterium tuberculosis pks12, was identified. STM5 and STM10 genes (encoding two hypothetical proteins MAV_4292 and MAV_4012) were associated with susceptibility to oxidative products. Mutants MAV_2450, MAV_4292, MAV_0385 and MAV_4264 live in macrophage vacuoles with acidic pH (below 6.9). Mutants MAV_4292, MAV_0385 and MAV_4264 were susceptible to nitric oxide in vitro. The study of individual mutants can potentially lead to new knowledge about M. avium pathogenic mechanisms.
INTRODUCTION
Mycobacterium avium is an opportunistic pathogenic bacterium that infects many host cell types, although the mononuclear phagocyte is the primary bacterial target. The bacterium is taken up by macrophages and lives inside a cytoplasmic vacuole, which does not acidify (Inderlied et al., 1993; Sturgill-Koszycki et al., 1994). The M. avium vacuole also does not follow the normal course of maturation, such as excluding proteins like Rab7 (Via et al., 1997), which has a role in the docking of the late phagosome to lysosome. It is clear that M. avium, although not as virulent as Mycobacterium tuberculosis, is capable of surviving within host macrophages by subverting the phagocytic cell functions (Bermudez et al., 2004; Crowle et al., 1991). To do so, the bacterium has an array of virulence factors, a few of them already described (Honer Zu Bentrup et al., 1999; Krzywinska et al., 2005; Li et al., 2005), but the majority are still unknown. It has also been shown that the virulence of M. avium, both in macrophages as well as in mice, depends on the bacterium morphotype. Smooth, rough, transparent or opaque, represent the bacterial morphotypes encountered. Although the smooth transparent morphotype is associated with virulence in mouse, the opaque and even the rough (without glycopeptidolipid) morphotypes have been isolated from patients with AIDS (Falkinham, 1996; Inderlied et al., 1993).
Several studies have identified M. avium genes expressed within the macrophages. Hou and colleagues reported on M. avium genes upregulated in macrophages using selective capture of transcribed sequences (Hou et al., 2002). More recently, Danelishvili et al. (2004) identified M. avium genes expressed in macrophages by differential fluorescence induction, using a green fluorescent protein promoter. Still other virulent genes were identified by screening of a transposon library, such as the PPE-encoding gene described by Li et al. (2005).
Despite the knowledge acquired regarding M. avium genes associated with the pathogenesis of infection, very little is known about their function. In addition, the genes important for bacterial survival in vivo are mostly unknown. In mice, following intravenous infection (via the tail vein), a large portion of M. avium organisms are located in the spleen and liver (Bermudez et al., 1999). Oral infection is also associated with disseminated disease (Bermudez et al., 1992). Bacteria cross the intestinal mucosa through enterocytes, and many of them survive in a latency-like state in mesenteric lymph nodes (Petrofsky & Bermudez, 2005; Sangari et al., 2000). Dissemination to the spleen, liver and bone marrow still often occurs (Petrofsky & Bermudez, 2005). Once in the liver and spleen, M. avium is found within macrophages and Kupffer cells, respectively (L. E. Bermudez & M. Petrofsky, unpublished results). The spread of the infection and systemic dissemination requires escape from macrophages and infection of, presumably, another macrophage (Bermudez et al., 2004). A recent study demonstrated that M. tuberculosis dissemination does seem to occur secondary to transport within phagocytic cells (Clay et al., 2007).
Once within macrophages, M. avium can resist reactive oxygen intermediates (ROIs) and nitric oxide (NO) (Appelberg & Orme, 1993; Bermudez & Young, 1989). Differently from M. tuberculosis, the inactivation of induced NO has no effect on M. avium survival in mice (Cooper et al., 2000; Gomes et al., 1999). Therefore, thus far no mechanism of killing of intracellular M. avium has been identified, although it is known that macrophage activation by tumour necrosis factor alpha and gamma interferon (IFN-γ) have important roles in M. avium killing (Appelberg & Orme, 1993; Bermudez & Young, 1989).
Signature-tagged mutagenesis (STM) has been used by a number of groups to identify bacterial genes required for the survival and replication in the host (Camacho et al., 1999; Cox et al., 1999; Hensel et al., 1995). It allows for large pools of bacterial clones to be screened simultaneously. Therefore, to get new insight on M. avium pathogenesis, we created and screened STM mutants in mice. We identified several genes that, when inactivated, resulted in attenuation of M. avium in vivo, and initial studies have suggested potential mechanisms of action.
METHODS
Bacterial strains and growth conditions.
Escherichia coli XL1-blue was used for the cloning experiments. It was grown at 37 °C on Luria–Bertani broth supplemented with 50 μg kanamycin ml−1. The M. avium strain 104, isolated from the blood of an AIDS patient and shown to be virulent in mice, as well as the mutants, were grown on Middlebrook 7H9 broth or 7H11 agar (Difco) supplemented with 0.2 % glycerol, 0.05 % Tween 80 and oleic acid, albumin glucose and catalase, as reported previously (Bermudez et al., 1992, 2004).
Construction of plasmid pYJTags.
The transposon Tn5367 derived from IS1096 was extracted from pYUB285 on a HindIII and EcoRI fragment, and cloned into the HindIII and EcoRI restriction sites of the E. coli vector pUC19 to create pYJL1. The mycobacteria temperature-sensitive origin of replication was amplified from pYUB285 by using primers with HindIII restriction site, and was cloned into pYJL1. The resulting plasmid was named pYJL2. Double-stranded DNA tags were produced by PCR amplification using a variable oligonucleotide pool RTCG as template DNA that was 5′-AAACTAACTAGTTACAACCTCAAGCTT-(NK)20-AAGCTTGGTTAGATGACTAGTATTAAA-3′. The primers used to amplify the tags were pTag1 (5′-AAACTAACGCGTTACAACCTC-3′), pTag2 (5′-TTTAATACGCGTCATTCTAACC-3′), pTag3 (5′-TAGTTACAACCTCAAGCTT-3′) and pTag4 (5′-TAGTCATTCTAACCAAGCTT-3′). For the cloning, the tags were amplified using the RTCG template, and pTag1 and pTag2 as primers. The PCR was performed using an AmpliTaq kit (Perkins-Elmer) with a 50 μl reaction containing 200 pg RTCG, pTag1 and pTag2 (100 pg), 1.5 M MgCl2 and 250 μM each of dATP, dGTP, dCTP dTTP, 1×Taq buffer and 5 U AmpliTaq. The cycle conditions used were the same as described by Hensel et al. (1995). PCR products were purified using a PCR purification kit (Qiagen), digested with MluI, dephosphorylated and ligated into the vector pYJL2. The ligation mixture was used to transform E. coli, and the transformants were selected on Luria–Bertani agar plate containing 50 μg kanamycin ml−1. The plasmids were analysed by restriction enzyme digestion to verify that tags had been incorporated into pYJL2. The resulting plasmid with a mixture of tags was named pYJTags.
Construction of the M. avium mutant library.
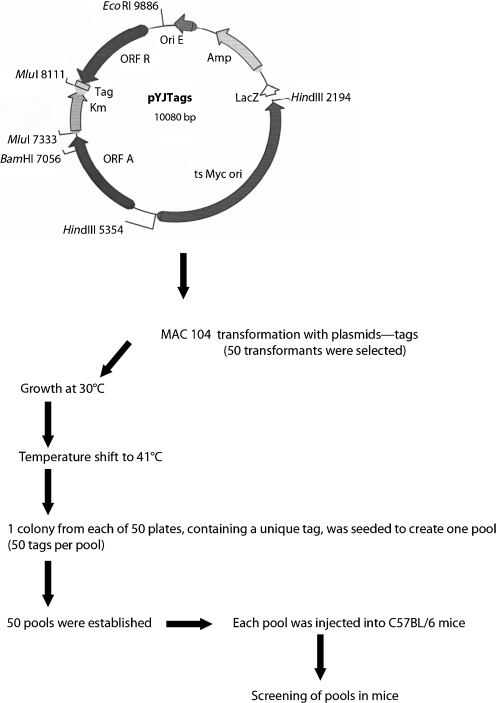
A pool of 5000 E. coli transformants was used to prepare plasmid pYJTags DNA (Fig. 1). These plasmids were electroporated into M. avium 104 strain, as described previously (Dam et al., 2006; Li et al., 2005), and transformants were selected on 7H11 medium containing 200 μg kanamycin ml−1. A total of 50 transformants containing pYJTags were obtained and used to produce 50 different mutant banks. A library of 2064 mutants picked randomly from 50 banks was organized and grown in microtitre plates containing 7H9 medium supplemented with 400 μg kanamycin ml−1 at 30 °C. After shifting the temperature to 41 °C for 3 days, the suspension was plated onto 7H11 agar plates with 400 μg kanamycin ml−1. Hundreds of mutants were obtained from each well. One colony was picked up from each of 50 individual plates to form 48 pools of 50 clones, each containing 50 different tags. A series of membranes containing the 50 different tags was produced by amplifying the different tags using pTag3 and pTag4 primers, and transferring them onto nylon N+ membrane using a manifold apparatus.
Fig. 1.
Schematic representation of the approach used. The pYJTags was created from pUC19 with the insertion of a temperature-sensitive (ts) Myc ori. It allowed for the integration of the transposon once the environmental temperature was elevated to 41 °C.
Labelling of input and output probe, and hybridization.
For amplification of the output probes from the spleens of the infected mice, homogenized spleen was plated onto solid medium supplemented with 400 μg kanamycin ml−1. After 7 to 10 days, mutant pools were recovered from the plates as output probes. For amplifying tags from the input of M. avium mutants, the pool containing 50 different tags was grown in 7H9 broth with 400 μg kanamycin ml−1, and bacterial cells were lysed using a high temperature incubation protocol (100 °C for 10 min and 4 °C for 1 min, 4 cycles). A 20 μl aliquot of supernatant was used for the 50 μl PCR; the PCR product was purified with a PCR purification kit (Qiagen), and labelled with non-radioactive digoxin with a DIG-Chem-Link labelling and detection set (Roche). The probe was digested with HindIII and purified to remove the non-specific arms consisting of the tags. Prehybridization, hybridization and washes were carried out at 68 °C using the DIG Wash and Block Buffer set hybridization kit from Roche. CDP-Star chemiluminescent substrate for alkaline phosphatase was used for visualization. X-ray films (Kodak) were exposed for 1 to 10 min at room temperature, depending on the signal strength.
Mapping of transposon insertion and sequence analysis.
To identify the transposon insertion site, chromosomal DNA from the mutant isolates was prepared. The interrupted unknown sequence was amplified using non-specific PCR as described previously (Danelishvili et al., 2004). The product was purified and directly sequenced using an ABI 373 DNA sequencer with a Taq dideoxy terminator cycle sequencing kit (Perkin-Elmer Cetus). Only sequences containing the transposon were considered. The sequences were mapped on the M. avium genome sequence [The Institute for Genomic Research (TIGR) database] and M. tuberculosis H37Rv genome sequence using the blast program.
Complementation of genes.
To address the importance of the genes inactivated on M. avium virulence, complementation of the genes MAV_2450, MAV_4292, MAV_1346, MAV_4012 and MAC_4264 was carried out using a recently published strategy (Li et al., 2005). Specific primers were designed and used to amplify the four genes from genomic DNA using PCR as described previously (Li et al., 2005). The amplification product was then cloned in pMV261-Apr plasmid and used to transform the mutants STM1 (STM1 comp), STM5 (STM5 comp), STM8 (STM8 comp), STM10 (STM10 comp) and STM11 (STM11 comp). Clones containing the plasmid were selected in presence of 200 μg apramycin ml−1. Selected clones were confirmed for the presence of the plasmid.
Mice infection studies.
In order to screen clones for attenuated mutants, 0.2 ml (1×108 c.f.u.) each input pool were injected intravenously into C57BL/6 mice (Jackson Laboratories). Mice were observed, and 3 weeks post-infection mice were sacrificed by cervical dislocation, spleens removed aseptically and homogenized. Series dilutions of spleen homogenates were plated onto 7H11 with kanamycin at 400 μg ml−1. The colonies were collected and processed, as described above, to make the output probe tags.
To prove that the strains identified in the initial screen were, in fact, attenuated mutants, mice were injected intravenously with 3×107 individual bacterial mutant. Twelve mice were used for each of the time points. In two independent experiments (with a total of 48 animals per time point), organs were removed 1 day, 1 week, 3 weeks and 5 weeks post-infection, homogenized, and serially diluted and plated onto 7H11 plates, as previously reported (Bermudez et al., 1992, 1997). The number of c.f.u. was determined and compared with the number of colonies in the spleen of mice infected with the wild-type (WT) bacterium.
Macrophages and specimen preparation.
Human monocyte-derived macrophages were isolated and prepared as previously described (Bermudez et al., 1997, 2004). Cells were adjusted to 105 cells ml−1 and seeded for 18 to 24 h on sterile, Formvar-coated London finder-gold grids (Electron Microscopy Sciences) in RPMI 1640 supplemented with 10 % fetal bovine serum (Sigma Chemicals). The monolayers were infected with M. avium strains for 1 h with an approximate ratio of 10 bacteria per macrophage, as reported previously (Bermudez et al., 1997, 2004). After infection, specimens were fixed in 1 % paraformaldehyde, pH 7.2, for 30 min. The grids were then washed and chambers were observed for identification of bacteria or phagosomes, as previously reported (Wagner et al., 2005). In other studies, RAW 246.7 macrophage monolayers (5×105 cells), cultured in presence of Dulbecco's modified Eagle's medium supplemented with 10 % heat inactivated fetal bovine serum, were stimulated with 100 mg recombinant mouse IFN-γ ml−1 and subsequently infected with different M. avium strains for 1 h (m.o.i. of 10), and washed to remove extracellular bacteria (Bermudez et al., 1992; Li et al., 2005). Depending on the experiment, 2 mg bovine superoxide dismutase (SOD) ml−1 (Sigma Chemicals) or N-methylarginine (Sigma) at 100 μM concentration was added concomitantly with the bacteria. These concentrations of inhibitors are known to suppress O2 and NO, respectively, in infected macrophages (Bermudez, 1993). Prior studies have shown that neither SOD nor N-methyl-arginine has any effect on monocyte and macrophage phagocytosis (data not shown). Monolayers were kept for 24 h and then lysed as previously described (Bermudez et al., 1992; Li et al., 2005). The macrophage lysate was serially diluted and plated onto 7H10 agar plates. Plates were incubated at 37 °C for 10 days and the number of colonies quantified (Bermudez et al., 1992; Li et al., 2005).
In some experiments, intracellular bacteria were allowed to grow inside macrophages for 4 days before lysing. Macrophage viability was monitored during the experiments to evidence differential detachment of macrophages from monolayers (Bermudez et al., 1992; Li et al., 2005).
Vacuole pH.
Measure of the vacuole pH was performed as previously described (Li et al., 2005). Briefly, to label bacteria, mid-exponential growth phase M. avium, WT and mutants, were washed in PBS and resuspended in PBS with 0.05 % Tween 80. Bacteria were labelled with fluorescent probes NHS-CF and NHS-Rho (10 μg each). Measurements were made in a fluorimeter (CytoFluor II; Biosearch). The filters used consisted of an excited band pass at 465–505 nm and emission at 505–555 nm. Calculation of phagosome pH for each well was carried out according to the standard calibration curve based on CF/Rho ratio versus pH, according to the method previously reported (Li et al., 2005).
Susceptibility to NO.
The NO donor NOR-3 was used to determine the effects of reactive nitrogen substances on M. avium strains. NOR-3 was dissolved in 15 mM Tris/HCl, pH 7.4, and was quantified spectrophotometrically. The NOR-3 (NO donor) was added to the culture of M. avium, WT or mutants, for 60 min. Afterwards, bacteria were quantified by plating the diluted suspension to 7H10 agar medium.
Statistical analysis.
Results of experimental studies in mice were analysed by Student's t-test. Each number represented the total number of mice divided into two different experiments. Experiments in vitro were analysed using ANOVA. The P values <0.05 were considered significant.
RESULTS AND DISCUSSION
M. avium is an intracellular opportunistic pathogen that infects and survives within host macrophages. M. avium is acquired from the environment, and its ability to cross the mucosal barrier and infect submucosal macrophages appears to be key for the establishment of infection (Bermudez et al., 1997; Inderlied et al., 1993; Sangari et al., 2000). Several studies have identified virulence determinants in vitro that allow the bacterium to enter (Dam et al., 2006; Miltner et al., 2005) and survive within macrophages (Honer Zu Bentrup et al., 1999; Hou et al., 2002; Krzywinska et al., 2005; Li et al., 2005). Although some other studies have determined the expression of M. avium genes in mononuclear phagosomes in vitro (Danelishvili et al., 2004; Hou et al., 2002) and in vivo (Li et al., 2005), very little has been performed in an attempt to identify virulence genes important for the survival in the host using an in vivo model.
Identification of attenuated mutants
To identify M. avium mutants attenuated in vivo, we partially screened an STM library in mice (2064 clones). Thirty-two clones with significant attenuation in vivo, in comparison to the WT bacterium, were identified by comparing the hybridization of the pool of DNA from bacteria in the spleen after 3 or 5 weeks of infection with the pool of DNA of the bacterial inoculum before administration (Fig. 1). Eleven clones were sequenced and further tested individually. The clones STM1 and STM7 had the transposon inserted in the same gene. The inactivated genes were sequences as described in Methods and elsewhere (Danelishvili et al., 2004; Li et al., 2005). The genes inactivated in the sequenced mutants are listed in Table 1. To confirm the phenotype of the isolated mutants, we infected C57BL/6 mice intravenously with the individual mutants (except for STM7 and STM9), and the number of bacteria in the spleen (compared with the WT MAC 104) was determined at 24 h, 1 week, 3 weeks and 5 weeks. STM7, as for STM1, had the pks12 gene inactivated, and STM9 had a transposon in a chaperonin and was considered less relevant than other mutants. As shown in Table 2, some mutants were attenuated at the first week of infection (STM5, STM6 and STM10), some were attenuated after 3 weeks of infection (STM1, STM3, STM4, STM8 and STM11) and only one clone showed attenuation in mice only after 5 weeks of infection (STM2).
Table 1.
Genes inactivated in the identified mutants
Attenuated mutants were identified by screening an STM library. The mutants were tested individually for attenuation.
| Clone | M. avium gene | Function |
|---|---|---|
| STM1/STM7 | MAV_2450 | Polyketide synthase 12 |
| STM2 | MAV_3321 | ABC transporter, ATP-binding subunit |
| STM3 | MAV_3105 | Polyketide synthase 11 |
| STM4 | MAV_2723 | PAP A2 condensation domain |
| STM5 | MAV_4292 | Hypothetical protein |
| STM6 | MAV_0385 | N-Acetylmuramyl-l-alanine amidase |
| STM8 | MAV_1346 | PE family protein |
| STM9 | MAV_4366 | Chaperonin GroES |
| STM10 | MAV_4012 | Conserved hypothetical protein |
| STM11 | MAV_4264 | Hypothetical protein, homology with bacterial regulatory protein TetR domain |
Table 2.
Comparison of the number of organisms in the WT strain MAC 104 and the other STM clones at several time points following infection of mice
C57BL/6 mice were infected intravenously and their spleens were harvested at 24 h, and at weeks 1, 3 and 5, for quantification of the bacterial load.
| Clone | No. of c.f.u. per organ | |||
|---|---|---|---|---|
| 24 h | 1 week | 3 weeks | 5 weeks | |
| STM1 | 6.7±0.3×106 | 6.7±0.4×106 | 6.1±0.4×105* | 5.2±0.3×104* |
| STM2 | 6.1±0.3×106 | 6.5±0.4×106 | 6.0±0.2×106 | 4.9±0.2×103* |
| STM3 | 5.5±0.5×106 | 2.3±0.2×106 | 1.2±0.1×106* | 8.6±0.4×105* |
| STM4 | 5.6±0.4×106 | 3.2±0.3×106 | 2.0±0.2×106* | 1.4±0.3×106* |
| STM5 | 3.8±0.2×106 | 1.1±0.2×106* | 1.5±0.1×106 | 9.4±0.7×105* |
| STM6 | 2.3±0.9×106 | 1.1±0.7×106* | 8.3±0.6×105* | 8.1±0.5×105* |
| STM8 | 1.6±0.2×106 | 1.8±0.6×106* | 1.8±0.4×106 | 1.9±0.3×105* |
| STM10 | 3.2±0.4×106 | 1.1±0.3×106* | 2.0±0.4×106 | 7.3±0.3×104* |
| STM11 | 5.1±0.3×106 | 2.7±0.4×106 | 8.5±0.3×105* | 5.3±0.6×104* |
| WT (MAC 104) | 1.9±0.2×106 | 3.8±0.2×106 | 1.3±0.2×107 | 4.6±0.4×107 |
*P<0.05 compared with the WT MAC 104 at the same time point.
In vitro growth
To determine whether or not the identified mutants had impaired growth ability in vitro, growth curves were established in 7H9 broth. STM1, STM7, STM5–6 and STM8–11 were grown in parallel to M. avium 104, and the growth curve determined over 10 days. Growth of all mutant strains was similar to the WT bacterium.
In the current study, we identified a number of M. avium clones that were attenuated in vivo by using an STM system. The attenuation of the individual mutants was then confirmed by challenge experiments, and some of the genes were complemented, leading to the recovery of the virulent phenotype. Some of the genes associated with attenuation in vivo were those encoding the polyketide synthases 11 and 12. Recent studies showed that genes involved in the synthesis and transport of polyketides are required for M. tuberculosis growth and virulence in mice (Appelberg & Orme, 1993; Hahn et al., 2005; Jain & Cox, 2005; Wagner et al., 2005). In addition, the work by Hou et al. (2002) demonstrated that the polyketide synthase 11 encoding gene was upregulated upon uptake of M. avium by macrophages. Polyketides are involved in cell-wall-lipid synthesis and the integrity of the outer surface of mycobacteria. They participate in the synthesis of dimycocerosyl phthiocerol, which has been suggested as playing a role in the permeability of the cell wall (Camacho et al., 2001). In M. tuberculosis, 24 polyketide synthases have been annotated (Cole et al., 1998); while in M. avium, there are at least 22 polyketide synthases, all with significant homology to the ones in M. tuberculosis (L. E. Bermudez & Y. Li, unpublished observation). Polyketides were also associated with modulation of the host immune response to infection by impacting on cytokine production by infected macrophages (Converse et al., 2003). Polyketides are transported to the surface of M. tuberculosis by a unique transport mechanism involving several genes of the mmpL family (Camacho et al., 1999; Converse et al., 2003; Domenech et al., 2004). In M. avium, the role of mmpL genes has not yet been defined, but infections of macrophages have been associated with the expression of the MmpL transport proteins (Danelishvili et al., 2004; Hou et al., 2002), and it is likely to be somewhat similar to the role in M. tuberculosis.
The identification of an ABC transporter subunit as virulence factor points to the importance of secreted proteins. ABC transporters are two-protein complexes, one protein involved in transport and the other providing energy for the process. One recent ABC transporter linked with virulence in mycobacteria is involved in the transport of oxidoreductase associated with the invasion of epithelial cells (Patel et al., 2006). Other ABC transporters have been identified in screening for mycobacterial virulence genes using several methods, but the functions have not been characterized.
Our screen also identified a PE protein (mycobacterial protein rich in proline and glutamic acid), GroES, a polyketide synthase-associated protein and many proteins of unknown function, some of them not present in M. tuberculosis but with homology to Mycobacterium paratuberculosis. PE, PPE and PE-PGRS are families of proteins that have been linked with virulence of Mycobacterium marinum and M. tuberculosis (Dubnau et al., 2002; Ramakrishnan et al., 2000). Some PE proteins have more recently been described as exported proteins, with expression on the surface of the bacterium (Delogu & Brennan, 2001) and are expressed differently in host tissues (Delogu et al., 2006). In fact, PE–PGRS proteins apparently have different patterns of expression, as evidenced by studies with M. tuberculosis, M. marinum and M. avium (Dheenadhayalan et al., 2006b; Li et al., 2005; Ramakrishnan et al., 2000). Initially, PE, PPE and PE–PGRS groups of proteins were suggested as being involved in the interaction with the host immune system, and being important antigenic determinants of M. tuberculosis (Cole et al., 1998). Although these families of proteins may be important to the immune response of the host, it is increasingly evident that several of them are involved in the pathogenesis of the infection, as shown by the requirement for survival in granulomas (Ramakrishnan et al., 2000) and within macrophages (Li et al., 2005). Our current work identified another PE protein in M. avium important for full virulence in vivo. Its function is unknown at this point, but recent work demonstrates that PE–PGRS 33 of M. tuberculosis, when expressed in Mycobacterium smegmatis, triggers necrosis of macrophages (Dheenadhayalan et al., 2006a). Chaperonins, such as GroEL, have recently been associated with mycobacterial virulence (Ojha et al., 2005). GroEL1 has been linked with the ability of M. smegmatis to form biofilms (Ojha et al., 2005) by modulating the synthesis of mycolates. The role in cell-wall synthesis may impact on the ability of the bacterium to interact with the host. In the case of GroES, the association with virulence has not been established yet, but the fact that the inactivation of the genes is associated with attenuation early in the infection in vivo, indicates that GroES might have other, still unknown, function(s). The different functions of stress-related proteins are still not completely known.
Inactivation of the genes encoding the polyketide synthases 11 and 12 was associated with lack of growth from the beginning of the infection, but major attenuation was only observed after 3 weeks following challenge. Those results agree with what has been described for clones with the inactivation of pks12 in M. tuberculosis (Sirakova et al., 2003). In contrast, the inactivation of the PE-encoding gene was associated with significant bacterial attenuation 5 weeks after infection, indicating that this protein may have function important for survival during the host adaptive response leading to the activation of macrophages or with the spreading phase of the infection, as recently suggested (Dheenadhayalan et al., 2006a). Polyketide synthases 11 and 12, as proteins associated with surface moieties, could potentially impact on the ability of the bacterium to interact with the environment.
Attenuation in macrophages
To examine whether the STM mutants were attenuated in macrophages, RAW 246.7 mouse macrophages were infected and the growth was followed for 4 days. It was observed that all mutants but STM3 and STM4 had no significant attenuation in macrophages in vitro. It suggests that the attenuation of STM3 and STM4 in vivo is probably not related with the innate immune response (Supplementary Table S1 available with the online journal). It was also determined that attenuation was due to increased susceptibility to superoxide anion or nitric oxide. STM5 and STM10 showed increased sensitivity to ROIs (Table 3), while STM10 and STM11 were susceptible to nitric oxide (Table 4). The concentrations of inhibitors used in these assays are knows to suppress O2 and NO in infected macrophages.
Table 3.
Susceptibility of M. avium attenuated clones to superoxide anion
| M. avium strain | No. of c.f.u. (ml macrophage lysate)−1* | ||
|---|---|---|---|
| 1 h | 24 h | ||
| −SOD | +SOD | ||
| WT (MAC 104) | 2.1±0.3×105 | 3.3±0.5×105 | 2.1±0.4×105 |
| STM1 | 3.6±0.4×105 | 1.1±0.3×105 | 1.5±0.2×105 |
| STM2 | 2.7±0.3×105 | 9.0±0.2×104 | 8.6±0.3×104 |
| STM3 | 5.1±0.5×105 | 3.0±0.2×105 | 2.6±0.4×105 |
| STM4 | 2.3±0.4×105 | 2.0±0.5×105 | 1.8±0.3×105 |
| STM5 | 3.0±0.5×105 | 8.2±0.4×104 | 2.0±0.5×105† |
| STM6 | 3.9±0.2×105 | 1.9±0.3×105 | 1.6±0.4×105 |
| STM8 | 3.6±0.5×105 | 8.1±0.2×104 | 7.6±0.4×104 |
| STM10 | 3.7±0.3×105 | 8.5±0.4×104 | 1.0±0.3×105† |
| STM11 | 4.7±0.3×105 | 8.1±0.6×104 | 8.5±0.2×104 |
*RAW 246.7 macrophage monolayers were infected with M. avium for 1 h and extracellular bacteria were removed by washing. Some wells were incubated with 2 mg SOD ml−1, as reported in Methods. Monolayers were lysed at 24 h and viable intracellular bacteria quantified. The experiment was repeated three times.
†P<0.05 compared with the number of c.f.u. (ml macrophage lysate)−1 at 24 h without adding SOD.
Table 4.
Susceptibility of M. avium clones to the effect of nitric oxide
| M. avium strain | No. of c.f.u. (ml macrophage lysate)−1* | ||
|---|---|---|---|
| 1 h | 24 h | ||
| −N-MA | +N-MA | ||
| WT (MAC 104) | 6.4±0.3×105 | 8.3±0.3×105 | 8.7±0.5×105 |
| STM1 | 2.0±0.4×105 | 8.1±0.3×104 | 8.5±0.6×104 |
| STM2 | 4.4±0.2×105 | 7.6±0.3×104 | 8.1±0.2×104 |
| STM3 | 5.2±0.5×105 | 4.1±0.3×105 | 3.8±0.4×105 |
| STM4 | 2.6±0.2×105 | 1.0±0.3×105 | 9.8±0.2×105 |
| STM5 | 3.4±0.2×105 | 9.8±0.3×104 | 1.0±0.4×105 |
| STM6 | 3.9±0.5×105 | 2.8±0.4×105 | 2.7±0.6×105 |
| STM8 | 3.8±0.2×105 | 8.3±0.3×104 | 7.6±0.5×104 |
| STM10 | 4.6±0.3×105 | 4.1±0.5×104 | 1.6±0.3×105† |
| STM11 | 2.5±0.5×105 | 7.8±0.4×104 | 1.2±0.6×105† |
*RAW 246.7 macrophage monolayers were infected for 1 h with M. avium, washed, and all of the wells were treated with IFN-γ and with 100 μM N-methylarginine. The monolayers were lysed after 24 h and the intracellular bacteria quantified. The experiments were repeated three times.
†P<0.05 compared with the number of bacteria at 24 h without N-methylarginine.
Susceptibility to NO
To examine whether STM strains are susceptible to NO in vitro, we incubated the strains STM1, 5, 10 and 11 in the presence of NO donor and verified the viability afterward. As shown in Supplementary Table S2 (available with the online journal), the mutants STM5 and STM10 were susceptible to NO at a concentration of 1 μM.
We identified a number of mutations in M. avium genes of unknown function that are associated with attenuation in vivo. Some of the genes inactivated, such as those homologous to MAV_4292 and MAV_4012, lead to increased susceptibility to SOD and NO (MAV_4012 and MAV_4264), raising interesting questions about the mechanisms of bacterial resistance to these toxic products of phagocytic cells. Production of superoxide anion is ineffective as a mechanism of killing of the majority of the M. avium strains (Bermudez & Young, 1989; Gunzel et al., 2006; Sarmento & Appelberg, 1996). Likewise, the role of NO, in contrast to M. tuberculosis, has not been established (Chan et al., 1992; Gomes et al., 1999). In fact, it seems that the inability of macrophages to produce nitric oxide leads to increased control of the infection (Appelberg & Orme, 1993; Bermudez, 1993; Gomes et al., 1999). MAV4012 is a transcription regulator, and its inactivation was associated with increased susceptibility to ROIs and NO. Because MAV_4012 is a transcription regulator, one might suggest that the expression of this gene regulates the expression of a number of membrane-associated genes. When tested individually for susceptibility against NO, STM10, STM11 and, to a lesser extent, STM5 were susceptible. The reason for the discrepancy between the results obtained in macrophages and regarding STM5 susceptibility in vitro to NO is unknown at this point. MAV_4292 is a gene encoding a protein of unknown function. Of importance is the finding that other proteins, but not SOD or correlated protein, were identified associated with resistance to superoxide anion and reactive nitrogen intermediates. Future studies will address these specific questions.
Vacuole pH
We determined the pH of the vacuole of STM mutants and compared it to the WT pH. As shown in Table 5, while the M. avium 104 vacuole pH was 6.4 at 15 min and 6.9 at 24 h, STM1, STM5, STM10 and STM11 had significant acidification in the macrophage vacuole. The vacuole of STM6, although significantly more acidic than the WT bacterium, was less acidic than the vacuole of the other STM mutants.
Table 5.
Vacuole pH of RAW 264.7 macrophages
| Strain | pH | |
|---|---|---|
| 15 min | 24 h | |
| WT (MAC 104) | 6.4 | 6.9 |
| STM1 | 5.3* | 5.6* |
| STM5 | 5.5* | 5.7* |
| STM6 | 5.7* | 6.5* |
| STM10 | 5.2* | 5.1* |
| STM11 | 4.8* | 4.5* |
*P<0.05 compared with the WT MAC 104. Experiments were repeated three times.
The pH in the vacuole of the STM mutants was acidic, in contrast to the pH of the vacuole of the WT bacterium. Except for one strain, in which the gene inactivated is a regulator, all other mutants had significantly less decrease in the pH of the vacuole compared with M. avium 104. The interpretation of the findings suggests that MAV_4264 regulates a series of genes that participate in the bacterial inhibition of phagosome acidification.
Complementation of phenotype
Complementation of STM1, STM5, STM8, STM10 and STM11 mutants was accomplished and the complemented clones were then evaluated for attenuation in RAW 246.7 macrophages. The results, shown in Table 6, indicate that the complementation of STM1, STM5, STM8 and STM10 recovered the virulent phenotype, with similar results to WT, while the attempt to complement STM11 was only partially successful (Table 6).
Table 6.
Ability of complemented strains STM1 comp, STM5 comp, STM8 comp, STM10 comp and STM11 comp to replicate in murine macrophages
Assays were repeated three times. The numbers represent means±sd.
| Strain | No. of c.f.u. (ml macrophage lysate)−1 | |
|---|---|---|
| 1 h | 4 days | |
| WT MAC 104 | 2.3±0.3×105 | 7.1±0.2×105 |
| SMT1 | 2.7±0.4×105 | 7.0±0.4×104* |
| STM1 comp | 2.1±0.5×105 | 6.7±0.4×105† |
| STM5 | 3.8±0.4×105 | 1.6±0.3×104* |
| STM5 comp | 2.5±0.6×105 | 5.9±0.5×105† |
| STM8 | 5.0±0.4×105 | 6.7±0.3×104* |
| STM8 comp | 4.1±0.3×105 | 6.9±0.4×105† |
| STM10 | 2.8±0.2×105 | 9.8±0.6×103* |
| STM10 comp | 3.4±0.4×105 | 4.1±0.5×105† |
| STM11 | 3.0±0.3×105 | 6.1±0.3×103* |
| STM11 comp | 4.4±0.5×105 | 9.5±0.4×104† |
*P<0.05 compared with the WT MAC 104 at day 4.
†P<0.05 compared with the mutated strain at day 4.
Conclusions
In summary, by screening an STM library, we have identified a number of virulence genes in M. avium required for the survival of the bacteria in macrophages and in the experimental host. Several of the genes appear to be directly associated with the resistance to macrophage mechanisms of killing.
Supplementary Material
Acknowledgments
We are indebted to Denny Weber for editing the manuscript, Mary Petrofsky and Bernadette Stang for helping with the mouse experiments, and Julie Early for technical help. This work was supported by the grants AI-43199 and AI-40710 from the National Institute of Allergy and Infectious Diseases.
Abbreviations
IFN-γ, gamma interferon
ROI, reactive oxygen intermediate
SOD, superoxide dismutase
STM, signature-tagged mutagenesis
WT, wild-type
Footnotes
Tables of growth and susceptibility data for the wild-type and mutant isolates are available as supplementary data with the online version of this paper.
References
- Appelberg, R. & Orme, I. M. (1993). Effector mechanisms involved in cytokine-mediated bacteriostasis of Mycobacterium avium infections in murine macrophages. Immunology 80, 352–359. [PMC free article] [PubMed] [Google Scholar]
- Bermudez, L. E. (1993). Differential mechanisms of intracellular killing of Mycobacterium avium and Listeria monocytogenes by activated human and murine macrophages. The role of nitric oxide. Clin Exp Immunol 91, 277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez, L. E. & Young, L. S. (1989). Oxidative and non-oxidative intracellular killing of Mycobacterium avium complex. Microb Pathog 7, 289–298. [DOI] [PubMed] [Google Scholar]
- Bermudez, L. E., Petrofsky, M., Kolonoski, P. & Young, L. S. (1992). An animal model of Mycobacterium avium complex disseminated infection after colonization of the intestinal tract. J Infect Dis 165, 75–79. [DOI] [PubMed] [Google Scholar]
- Bermudez, L. E., Parker, A. & Goodman, J. R. (1997). Growth within macrophages increases the efficiency of Mycobacterium avium in invading other macrophages by a complement receptor-independent pathway. Infect Immun 65, 1916–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez, L. E., Kolonoski, P., Wu, M., Aralar, P. A., Inderlied, C. B. & Young, L. S. (1999). Mefloquine is active in vitro and in vivo against Mycobacterium avium complex. Antimicrob Agents Chemother 43, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez, L. E., Petrofsky, M. & Sangari, F. (2004). Intracellular phenotype of Mycobacterium avium enters macrophages primarily by a macropinocytosis-like mechanism and survives in a compartment that differs from that with extracellular phenotype. Cell Biol Int 28, 411–419. [DOI] [PubMed] [Google Scholar]
- Camacho, L. R., Ensergueix, D., Perez, E., Gicquel, B. & Guilhot, C. (1999). Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol Microbiol 34, 257–267. [DOI] [PubMed] [Google Scholar]
- Camacho, L. R., Constant, P., Raynaud, C., Laneelle, M. A., Triccas, J. A., Gicquel, B., Daffe, M. & Guilhot, C. (2001). Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J Biol Chem 276, 19845–19854. [DOI] [PubMed] [Google Scholar]
- Chan, J., Xing, Y., Magliozzo, R. S. & Bloom, B. R. (1992). Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J Exp Med 175, 1111–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay, H., Davis, J. M., Beery, D., Huttenlocher, A., Lyons, S. E. & Ramakrishnan, L. (2007). Dichotomous role of the macrophage in early Mycobacterium marinum infection of the Zebrafish. Cell Host Microbe 2, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S. & other authors (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. [DOI] [PubMed] [Google Scholar]
- Converse, S. E., Mougous, J. D., Leavell, M. D., Leary, J. A., Bertozzi, C. R. & Cox, J. S. (2003). MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc Natl Acad Sci U S A 100, 6121–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, A. M., Pearl, J. E., Brooks, J. V., Ehlers, S. & Orme, I. M. (2000). Expression of the nitric oxide synthase 2 gene is not essential for early control of Mycobacterium tuberculosis in the murine lung. Infect Immun 68, 6879–6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J. S., Chen, B., McNeil, M. & Jacobs, W. R., Jr (1999). Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402, 79–83. [DOI] [PubMed] [Google Scholar]
- Crowle, A. J., Dahl, R., Ross, E. & May, M. H. (1991). Evidence that vesicles containing living, virulent Mycobacterium tuberculosis or Mycobacterium avium in cultured human macrophages are not acidic. Infect Immun 59, 1823–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam, T., Danelishvili, L., Wu, M. & Bermudez, L. E. (2006). The fadD2 gene is required for efficient Mycobacterium avium invasion of mucosal epithelial cells. J Infect Dis 193, 1135–1142. [DOI] [PubMed] [Google Scholar]
- Danelishvili, L., Poort, M. J. & Bermudez, L. E. (2004). Identification of Mycobacterium avium genes up-regulated in cultured macrophages and in mice. FEMS Microbiol Lett 239, 41–49. [DOI] [PubMed] [Google Scholar]
- Delogu, G. & Brennan, M. J. (2001). Comparative immune response to PE and PE_PGRS antigens of Mycobacterium tuberculosis. Infect Immun 69, 5606–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delogu, G., Sanguinetti, M., Pusceddu, C., Bua, A., Brennan, M. J., Zanetti, S. & Fadda, G. (2006). PE_PGRS proteins are differentially expressed by Mycobacterium tuberculosis in host tissues. Microbes Infect 8, 2061–2067. [DOI] [PubMed] [Google Scholar]
- Dheenadhayalan, V., Delogu, G. & Brennan, M. J. (2006a). Expression of the PE_PGRS 33 protein in Mycobacterium smegmatis triggers necrosis in macrophages and enhanced mycobacterial survival. Microbes Infect 8, 262–272. [DOI] [PubMed] [Google Scholar]
- Dheenadhayalan, V., Delogu, G., Sanguinetti, M., Fadda, G. & Brennan, M. J. (2006b). Variable expression patterns of Mycobacterium tuberculosis PE_PGRS genes: evidence that PE_PGRS16 and PE_PGRS26 are inversely regulated in vivo. J Bacteriol 188, 3721–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech, P., Reed, M. B., Dowd, C. S., Manca, C., Kaplan, G. & Barry, C. E., III (2004). The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J Biol Chem 279, 21257–21265. [DOI] [PubMed] [Google Scholar]
- Dubnau, E., Fontan, P., Manganelli, R., Soares-Appel, S. & Smith, I. (2002). Mycobacterium tuberculosis genes induced during infection of human macrophages. Infect Immun 70, 2787–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkinham, J. O., III (1996). Epidemiology of infection by nontuberculous mycobacteria. Clin Microbiol Rev 9, 177–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, M. S., Florido, M., Pais, T. F. & Appelberg, R. (1999). Improved clearance of Mycobacterium avium upon disruption of the inducible nitric oxide synthase gene. J Immunol 162, 6734–6739. [PubMed] [Google Scholar]
- Gunzel, D., Kucharski, L. M., Kehres, D. G., Romero, M. F. & Maguire, M. E. (2006). The MgtC virulence factor of Salmonella enterica serovar Typhimurium activates Na+,K+-ATPase. J Bacteriol 188, 5586–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, M. Y., Raman, S., Anaya, M. & Husson, R. N. (2005). The Mycobacterium tuberculosis extracytoplasmic-function sigma factor SigL regulates polyketide synthases and secreted or membrane proteins and is required for virulence. J Bacteriol 187, 7062–7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel, M., Shea, J. E., Gleeson, C., Jones, M. D., Dalton, E. & Holden, D. W. (1995). Simultaneous identification of bacterial virulence genes by negative selection. Science 269, 400–403. [DOI] [PubMed] [Google Scholar]
- Honer Zu Bentrup, K., Miczak, A., Swenson, D. L. & Russell, D. G. (1999). Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis. J Bacteriol 181, 7161–7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, J. Y., Graham, J. E. & Clark-Curtiss, J. E. (2002). Mycobacterium avium genes expressed during growth in human macrophages detected by selective capture of transcribed sequences (SCOTS). Infect Immun 70, 3714–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inderlied, C. B., Kemper, C. A. & Bermudez, L. E. (1993). The Mycobacterium avium complex. Clin Microbiol Rev 6, 266–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, M. & Cox, J. S. (2005). Interaction between polyketide synthase and transporter suggests coupled synthesis and export of virulence lipid in M. tuberculosis. PLoS Pathog 1, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinska, E., Bhatnagar, S., Sweet, L., Chatterjee, D. & Schorey, J. S. (2005). Mycobacterium avium 104 deleted of the methyltransferase D gene by allelic replacement lacks serotype-specific glycopeptidolipids and shows attenuated virulence in mice. Mol Microbiol 56, 1262–1273. [DOI] [PubMed] [Google Scholar]
- Li, Y., Miltner, E., Wu, M., Petrofsky, M. & Bermudez, L. E. (2005). A Mycobacterium avium PPE gene is associated with the ability of the bacterium to grow in macrophages and virulence in mice. Cell Microbiol 7, 539–548. [DOI] [PubMed] [Google Scholar]
- Miltner, E., Daroogheh, K., Mehta, P. K., Cirillo, S. L., Cirillo, J. D. & Bermudez, L. E. (2005). Identification of Mycobacterium avium genes that affect invasion of the intestinal epithelium. Infect Immun 73, 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha, A., Anand, M., Bhatt, A., Kremer, L., Jacobs, W. R., Jr & Hatfull, G. F. (2005). GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123, 861–873. [DOI] [PubMed] [Google Scholar]
- Patel, D., Danelishvili, L., Yamazaki, Y., Alonso, M., Paustian, M. L., Bannantine, J. P., Meunier-Goddik, L. & Bermudez, L. E. (2006). The ability of Mycobacterium avium subsp. paratuberculosis to enter bovine epithelial cells is influenced by preexposure to a hyperosmolar environment and intracellular passage in bovine mammary epithelial cells. Infect Immun 74, 2849–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrofsky, M. & Bermudez, L. E. (2005). CD4+ T cells but not CD8+ or γδ+ lymphocytes are required for host protection against Mycobacterium avium infection and dissemination through the intestinal route. Infect Immun 73, 2621–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan, L., Federspiel, N. A. & Falkow, S. (2000). Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288, 1436–1439. [DOI] [PubMed] [Google Scholar]
- Sangari, F. J., Goodman, J. & Bermudez, L. E. (2000). Mycobacterium avium enters intestinal epithelial cells through the apical membrane, but not by the basolateral surface, activates small GTPase Rho and, once within epithelial cells, expresses an invasive phenotype. Cell Microbiol 2, 561–568. [DOI] [PubMed] [Google Scholar]
- Sarmento, A. & Appelberg, R. (1996). Involvement of reactive oxygen intermediates in tumor necrosis factor α-dependent bacteriostasis of Mycobacterium avium. Infect Immun 64, 3224–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirakova, T. D., Dubey, V. S., Kim, H. J., Cynamon, M. H. & Kolattukudy, P. E. (2003). The largest open reading frame (pks12) in the Mycobacterium tuberculosis genome is involved in pathogenesis and dimycocerosyl phthiocerol synthesis. Infect Immun 71, 3794–3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill-Koszycki, S., Schlesinger, P. H., Chakraborty, P., Haddix, P. L., Collins, H. L., Fok, A. K., Allen, R. D., Gluck, S. L., Heuser, J. & Russell, D. G. (1994). Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 263, 678–681. [DOI] [PubMed] [Google Scholar]
- Via, L. E., Deretic, D., Ulmer, R. J., Hibler, N. S., Huber, L. A. & Deretic, V. (1997). Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J Biol Chem 272, 13326–13331. [DOI] [PubMed] [Google Scholar]
- Wagner, D., Maser, J., Lai, B., Cai, Z., Barry, C. E., III, Honer Zu Bentrup, K., Russell, D. G. & Bermudez, L. E. (2005). Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J Immunol 174, 1491–1500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.