Abstract
Using constructs that encode the individual West Nile virus (WNV) NS3helicase (NS3hel) and NS3hel linked to the hydrophilic, N-terminal 1–50 sequence of NS4A, we demonstrated that the presence of NS4A allows NS3hel to conserve energy in the course of oligonucleotide substrate unwinding. Using NS4A mutants, we also determined that the C-terminal acidic EELPD/E motif of NS4A, which appears to be functionally similar to the acidic EFDEMEE motif of hepatitis C virus (HCV) NS4A, is essential for regulating the ATPase activity of NS3hel. We concluded that, similar to HCV NS4A, NS4A of WNV acts as a cofactor for NS3hel and allows helicase to sustain the unwinding rate of the viral RNA under conditions of ATP deficiency.
After flavivirus entry into the host cell, its approximately 11 kb genome is uncoated and serves as a template for the translation of a single C-prM-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5 polyprotein precursor (Chambers et al., 1990; Sampath & Padmanabhan, 2009). The polyprotein is inserted into the endoplasmic reticulum membrane and processed by host and viral proteinases into three structural proteins (C, prM and E) and seven non-structural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) (Padmanabhan et al., 2006). The structural proteins are components of mature virus particles. The NS proteins are expressed only in the infected host cell and are not packaged into mature particles. Similar to other flaviviruses, the full-length NS3 peptide sequence of West Nile virus (WNV) represents a multifunctional protein in which the N-terminal 184 residues encode serine proteinase (NS3pro) and the C-terminal 440 residues code for an RNA triphosphatase, an NTPase and an RNA helicase (NS3hel) (Benarroch et al., 2004; Borowski et al., 2001). NS3hel is a member of the DEAH/D-box family within the helicase superfamily 2 (Gorbalenya et al., 1989; Li et al., 1999; Luking et al., 1998; Luo et al., 2008). NS3hel is required for unwinding of RNA during replication. The ATPase activity of NS3hel does not require the binding of the enzyme to its nucleotide substrate (Chernov et al., 2008). As a result, high concentrations of ATP (in the mM range) are necessary to support the unwinding activity of NS3hel in vitro.
The presence of a cofactor, NS2B, is necessary for NS3pro to exhibit its proteolytic activity (Aleshin et al., 2007; Erbel et al., 2006). Although evidence suggests that the small, hydrophobic NS4A protein (approx. 16 kDa) is required for viral replication, and that NS4A may serve as a central ‘organizer’ of the replication complex of flaviviruses (Lindenbach & Rice, 1999; Paredes & Blight, 2008; Umareddy et al., 2006), its precise functional role is not characterized sufficiently. NS4A associates with membranes via four internal hydrophobic regions. Its C-terminal region (frequently designated 2K fragment) serves as a signal sequence for the translocation of the adjacent NS4B into the endoplasmic reticulum lumen. In the lumen, the 2K fragment is removed from the N terminus of NS4B by the host signallase. Proteolytic removal of the 2K peptide induces membrane alterations (Miller et al., 2007; Roosendaal et al., 2006). The N terminus of NS4A is generated in the cytoplasm by the viral NS2B–NS3pro, suggesting that both the NS3hel domain and the hydrophilic N-terminal portion of NS4A are exposed to the cytoplasm. The hydrophilic sequence of NS4A is homologous among flaviviruses (Fig. 1a). Functionally, WNV NS4A is likely to be similar to NS4A of hepatitis C virus (HCV) (Paredes & Blight, 2008; Thompson et al., 2009). Several groups have repeatedly observed NS3–NS4A as a transient intermediate in polyprotein processing in both yellow fever and dengue flaviviruses (Cahour et al., 1992; Preugschat & Strauss, 1991; Preugschat et al., 1990; Zhang et al., 1992; Zhang & Padmanabhan, 1993).
Fig. 1.
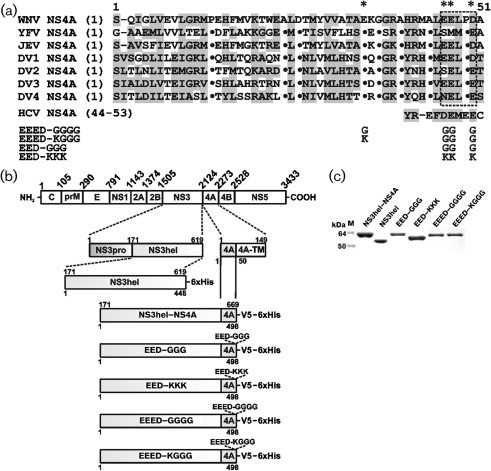
(a) Sequence alignment of the 1–50 N-terminal cytoplasmic portion of the flaviviral NS4A. Virus abbreviations: WNV, West Nile; YFV, yellow fever; JEV, Japanese encephalitis; and DV1–4, dengue virus types 1–4. Homologous residues are shaded grey. Dots indicate identical residues. The negatively charged EELPD motif is in a frame. The acidic sequence motif (44–53) of HCV NS4A is shown below the sequence alignment of flaviviral NS4A. Asterisks show the modified residues in the EED–GGG, EED–KKK, EEED–GGGG and EEED–KGGG mutants. (b) The structure of the WNV polyprotein precursor and the recombinant NS3hel–NS4A constructs. (c) SDS gel electrophoresis of the purified recombinant constructs (Coomassie-stained). The individual WNV NS3hel domain was directly linked with the N-terminal 1–50 sequence of WNV NS4A. NS3hel was tagged with a 6× His tag and other constructs were tagged with both V5 and 6× His tags. M, Molecular mass markers.
Based on these data, we hypothesized that, similar to NS2B (the cofactor with NS3pro), NS4A is a cofactor that is essential for the optimal performance of NS3hel. To support our hypothesis, we studied the unwinding and ATPase activities of the individual NS3hel domain and the wild-type and mutant WNV NS3hel–NS4A constructs (Chernov et al., 2008). For these purposes, we constructed the soluble WNV NS3hel–NS4A chimera and the individual NS3hel 1676–2123 domain lacking the NS3pro sequence. The constructs (Fig. 1b) were amplified by PCR using the WNV strain NY99 cDNA template (Hayes et al., 2005) and the specific direct and reverse primers (Supplementary Table S1, available in JGV Online). The constructs were cloned in the pET101/D-TOPO vector (Invitrogen) and their authenticity was verified by DNA sequencing.
In the chimera construct, the hydrophilic 1–50 aa N-terminal part of the natural membrane-tethered 2124–2273 NS4A sequence was directly linked to the 1676–2123 sequence of the WNV polyprotein precursor which encodes the NS3hel domain. The N- and C-terminal sequences of NS4A represent transmembrane segments and, therefore, they were not included in our constructs. To facilitate the isolation and analysis of the recombinant proteins from Escherichia coli cells, the constructs were C-terminally tagged with a V5 tag followed by a 6×His tag. We also constructed mutants in which the conserved negatively charged Glu and Asp residues of NS4A were replaced with Gly and Lys. As a result, we obtained NS4A mutants called EED–GGG, EED–KKK, EEED–GGGG and EEED–KGGG. The structure of WNV constructs and the relative positions of the mutations are shown in Fig. 1(b).
Competent E. coli BL21 CodonPlus (DE3)-RIPL cells (Stratagene) were transformed with the resulting recombinant pET101/D-TOPO plasmids. Transformed cells were grown in 2 l LB broth at 37 °C to reach A600=0.6. The protein expression was induced for an additional 6 h at 37 °C using 1 mM isopropyl β-D-thiogalactoside. The majority of the synthesized constructs appeared in a soluble form. The collected cells were resuspended in 40 ml 20 mM Tris/HCl, pH 7.8 (buffer A), supplemented with 1 M NaCl, the complete proteinase inhibitor cocktail (Roche) and 1 mg lysozyme ml−1, and disrupted by sonication. Cell debris was removed by centrifugation. The NS3hel–NS4A and mutant constructs were purified from the supernatant using a HiTrap Co2+-chelating Sepharose FastFlow column (1.6×2.5 cm; GE Healthcare). The 6×His-tagged recombinant constructs were eluted using a 0–500 mM gradient of imidazole concentrations. The fractions were analysed using SDS gel electrophoresis and also by Western blotting with a 6×His antibody. These procedures yielded a high purity individual NS3hel construct and the wild-type and mutant NS3hel–NS4A chimeras (Fig. 1c).
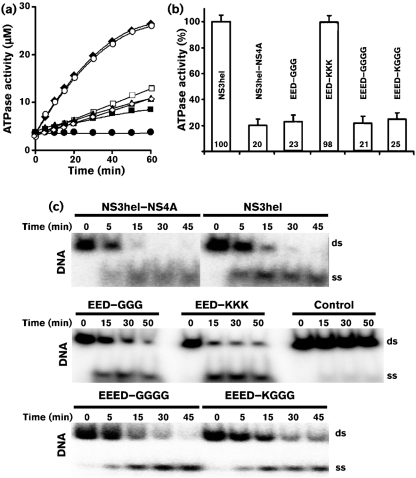
The ATPase activity of the constructs was measured at 37 °C for 5–60 min in wells of a 96-well plate using the NTPase assay colorimetric system (Innova Biosciences). The unwinding activity of NS3hel requires the presence of ATP (5 mM) in in vitro reactions (Chernov et al., 2008). In turn, the ATPase activity does not require the binding of NS3hel to a nucleotide substrate. As a result, the reactions did not include oligonucleotide substrates. The 100 μl reactions were measured in triplicate and contained the protein samples (0.04 nM) and 1 mM ATP in 50 mM Tris/HCl, pH 7.5, supplemented with 5 mM MgCl2. The resulting absorbance was measured (at 620 nm) on a fluorescence reader. The rate of ATP hydrolysis was quantified using a standard curve. A direct comparison demonstrated that NS3hel–NS4A had a fivefold reduced ATPase activity compared with the individual NS3hel (Fig. 2a, b). Analysis of the mutants with the EED–GGG, EED–KKK, EEED–GGGG and EEED–KGGG substitutions in the NS4A sequence showed that only the EED–KKK mutation abolished the ATP-saving function of NS4A, while the level of ATPase activity in the other mutants was as high as that of the wild-type NS3hel–NS4A construct. These data suggest that the negatively charged EELPD/E motif in the C-terminal portion of the NS4A hydrophilic region is essential for the role that NS4A plays in the regulation of the ATPase activity of NS3hel. A similar acidic motif (EFDEMEE) exists in the C-terminal portion of the hydrophilic region of HCV NS4A (Fig. 1a). Experimental evidence directly suggests that, in HCV, this acidic motif is also essential for the helicase cofactor function of NS4A (Beran et al., 2009).
Fig. 2.
(a) The ATPase activity of the WNV NS3hel (⧫) and NS3hel–NS4A constructs (▴, EED–GGG; ○, EED–KKK; ◊, EEED–GGGG; □, EEED–KGGG; ▪, NS3hel–NS4A; •, no enzyme). The reactions were performed using 1 mM ATP. (b) The ATPase activity (at 30 min) of the constructs relative to the individual NS3hel (set as 100 %). Error bars indicate sem. (c) The DNA unwinding activity of the WNV NS3hel and NS3hel–NS4A constructs. Control, no enzyme.
To determine whether mutations affected the unwinding activity of the helicase, we used the HPLC-purified double-stranded DNA substrates with a 3′-single-stranded terminus, formed by annealing an 18 bp sequence (5′-GCCTCGCTGCCGTCGCCA-3′; D1) to a 32P-labelled 38 bp oligonucleotide (5′-TGGCGACGGCAGCGAGGCTTTTTTTTTTTTTTTTTTTT-3′; D2). D1 (50 pmol) was 5′-labelled using T4 polynucleotide kinase and [γ-32P]-ATP (Perkin Elmer). The labelled product was separated from the free label using a micro bio-spin 6 column (Bio-Rad). To generate duplex DNA, labelled D1 (50 pmol) was mixed with D2 (100 pmol) in buffer A, containing 150 mM NaCl and 0.1 mM EDTA. The samples were boiled for 1 min and then cooled to 20 °C.
The unwinding activity of the constructs was measured at 37 °C in 200 μl reactions containing buffer A supplemented with 25 mM NaCl, 3 mM MgCl2, 2 mM DTT, 20 μg BSA ml−1, 5 mM ATP, 5 nM DNA oligonucleotide preformed duplex and protein sample (500 nM). Aliquots of 20 μl were withdrawn from the reactions between 5 and 60 min after the start of the reaction. The reactions were stopped by adding 6 μl 50 mM EDTA to the aliquots. Single-stranded oligonucleotides were separated from the duplex using non-denaturing 10 % polyacrylamide gel. Gels were visualized by autoradiography using an FLA-5100 imaging system and the obtained images were digitized using Multi Gauge software (Fujifilm). In agreement with our earlier studies (Chernov et al., 2008), the individual NS3hel domain demonstrated a high unwinding activity (Fig. 2c). The unwinding activity of the wild-type and mutant NS3hel–NS4A constructs was similar to that of the individual NS3hel domain. These findings suggested that NS4A had no significant effect on the rate with which NS3hel unwound the oligonucleotide duplex. NS4A, however, allowed the NS3hel–NS4A constructs to accomplish the substrate unwinding by consuming lower quantities of ATP when compared with NS3hel alone. Our results directly complement the very recent findings of Beran et al. (2009) who determined that HCV NS4A enhanced the ability of NS3hel to bind RNA in the presence of ATP, thereby acting as a cofactor for helicase activity in HCV. As a combined result of these studies, it now becomes clear that in the flaviviridae, including WNV and HCV, the hydrophilic NS4A sequence functions as an NS3hel cofactor.
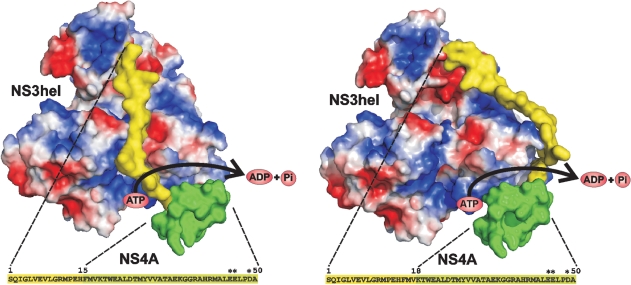
Because the C-terminal part of NS4A spans the membrane, only the hydrophilic, 50 nt N-terminal portion of NS4A is flexible and available for interacting with NS3hel. To estimate if the length of this hydrophilic portion is sufficient for directly accessing the ATP-binding pocket of the helicase domain, we modelled the NS3hel–NS4A complex. The crystal structure of Kunjin NS3 helicase (PDB 2QEQ) (Mastrangelo et al., 2007) was used to model NS3hel. Because the crystal structure of 1–50 NS4A is not yet determined, we used the secondary structure prediction programs SSpro v 4.5 and SSpro8 (Cheng et al., 2005) for estimating the folding of NS4A. Both programs predicted that NS4A included an extended 1–18 loop followed by a two-helical bundle. This bundle includes the mutant EED–KKK residues. Fig. 3, which shows the modelling results, suggests that the two-helical bundle of NS4A can access the ATPase site of NS3hel and explains why the EED–KKK mutations (which are close to the ATPase site) inactivated both the interaction of NS4A with NS3hel and the ATP-conserving function of NS4A.
Fig. 3.
Two alternative predicted structures of the WNV NS3hel–NS4A construct. The C-terminal 19–50 part of NS4A (green) directly interacts with the ATP-binding region of NS3hel, while the N-terminal 1–18 loop portion is perpendicular to the RNA-binding groove of NS3hel. Mutant positions which inactivated the ATP-conserving function of NS4A are indicated by asterisks. The 19–50 portion of NS4A is modelled as a typical two-helical bundle of identical size and represents the actual structure of NS4A.
Overall, we concluded that, in vivo, NS4A plays an important role of a cofactor in virus replication and that NS4A allows the flaviviral NS3hel to sustain the unwinding rate of the viral RNA under the conditions of ATP deficiency in the host cell. Our data support the recent data of others (Beran et al., 2009) and provide a biochemical rationale for the important role that NS4A plays in replication of the flaviviridae viruses (Lindenbach & Rice, 1999; Roosendaal et al., 2006; Sampath & Padmanabhan, 2009).
Supplementary Material
Acknowledgments
The study was supported by NIH grants RR020843 and AI055789.
Footnotes
A supplementary table of primer sequences is available with the online version of this paper.
References
- Aleshin, A. E., Shiryaev, S. A., Strongin, A. Y. & Liddington, R. C. (2007). Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci 16, 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benarroch, D., Selisko, B., Locatelli, G. A., Maga, G., Romette, J. L. & Canard, B. (2004). The RNA helicase, nucleotide 5′-triphosphatase, and RNA 5′-triphosphatase activities of Dengue virus protein NS3 are Mg2+-dependent and require a functional Walker B motif in the helicase catalytic core. Virology 328, 208–218. [DOI] [PubMed] [Google Scholar]
- Beran, R. K., Lindenbach, B. D. & Pyle, A. M. (2009). The NS4A protein of hepatitis C virus promotes RNA-coupled ATP hydrolysis by the NS3 helicase. J Virol 83, 3268–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowski, P., Niebuhr, A., Mueller, O., Bretner, M., Felczak, K., Kulikowski, T. & Schmitz, H. (2001). Purification and characterization of West Nile virus nucleoside triphosphatase (NTPase)/helicase: evidence for dissociation of the NTPase and helicase activities of the enzyme. J Virol 75, 3220–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahour, A., Falgout, B. & Lai, C. J. (1992). Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B–NS3, whereas NS4A/NS4B may be processed by a cellular protease. J Virol 66, 1535–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers, T. J., Hahn, C. S., Galler, R. & Rice, C. M. (1990). Flavivirus genome organization, expression, and replication. Annu Rev Microbiol 44, 649–688. [DOI] [PubMed] [Google Scholar]
- Cheng, J., Randall, A. Z., Sweredoski, M. J. & Baldi, P. (2005). SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res 33, W72–W76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernov, A. V., Shiryaev, S. A., Aleshin, A. E., Ratnikov, B. I., Smith, J. W., Liddington, R. C. & Strongin, A. Y. (2008). The two-component NS2B–NS3 proteinase represses DNA unwinding activity of the West Nile virus NS3 helicase. J Biol Chem 283, 17270–17278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbel, P., Schiering, N., D'Arcy, A., Renatus, M., Kroemer, M., Lim, S. P., Yin, Z., Keller, T. H., Vasudevan, S. G. & Hommel, U. (2006). Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol 13, 372–373. [DOI] [PubMed] [Google Scholar]
- Gorbalenya, A. E., Koonin, E. V., Donchenko, A. P. & Blinov, V. M. (1989). Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res 17, 4713–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, E. B., Komar, N., Nasci, R. S., Montgomery, S. P., O'Leary, D. R. & Campbell, G. L. (2005). Epidemiology and transmission dynamics of West Nile virus disease. Emerg Infect Dis 11, 1167–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Clum, S., You, S., Ebner, K. E. & Padmanabhan, R. (1999). The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J Virol 73, 3108–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach, B. D. & Rice, C. M. (1999). Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol 73, 4611–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luking, A., Stahl, U. & Schmidt, U. (1998). The protein family of RNA helicases. Crit Rev Biochem Mol Biol 33, 259–296. [DOI] [PubMed] [Google Scholar]
- Luo, D., Xu, T., Watson, R. P., Scherer-Becker, D., Sampath, A., Jahnke, W., Yeong, S. S., Wang, C. H., Lim, S. P. & other authors (2008). Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J 27, 3209–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo, E., Milani, M., Bollati, M., Selisko, B., Peyrane, F., Pandini, V., Sorrentino, G., Canard, B., Konarev, P. V. & other authors (2007). Crystal structure and activity of Kunjin virus NS3 helicase; protease and helicase domain assembly in the full length NS3 protein. J Mol Biol 372, 444–455. [DOI] [PubMed] [Google Scholar]
- Miller, S., Kastner, S., Krijnse-Locker, J., Buhler, S. & Bartenschlager, R. (2007). The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem 282, 8873–8882. [DOI] [PubMed] [Google Scholar]
- Padmanabhan, R., Mueller, N., Reichert, E., Yon, C., Teramoto, T., Kono, Y., Takhampunya, R., Ubol, S., Pattabiraman, N. & other authors (2006). Multiple enzyme activities of flavivirus proteins. Novartis Found Symp 277, 74–84. [DOI] [PubMed] [Google Scholar]
- Paredes, A. M. & Blight, K. J. (2008). A genetic interaction between hepatitis C virus NS4B and NS3 is important for RNA replication. J Virol 82, 10671–10683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preugschat, F. & Strauss, J. H. (1991). Processing of nonstructural proteins NS4A and NS4B of dengue 2 virus in vitro and in vivo. Virology 185, 689–697. [DOI] [PubMed] [Google Scholar]
- Preugschat, F., Yao, C. W. & Strauss, J. H. (1990). In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J Virol 64, 4364–4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosendaal, J., Westaway, E. G., Khromykh, A. & Mackenzie, J. M. (2006). Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J Virol 80, 4623–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath, A. & Padmanabhan, R. (2009). Molecular targets for flavivirus drug discovery. Antiviral Res 81, 6–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, A. A., Zou, A., Yan, J., Duggal, R., Hao, W., Molina, D., Cronin, C. N. & Wells, P. A. (2009). Biochemical characterization of recombinant hepatitis C virus nonstructural protein 4B: evidence for ATP/GTP hydrolysis and adenylate kinase activity. Biochemistry 48, 906–916. [DOI] [PubMed] [Google Scholar]
- Umareddy, I., Chao, A., Sampath, A., Gu, F. & Vasudevan, S. G. (2006). Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J Gen Virol 87, 2605–2614. [DOI] [PubMed] [Google Scholar]
- Zhang, L. & Padmanabhan, R. (1993). Role of protein conformation in the processing of dengue virus type 2 nonstructural polyprotein precursor. Gene 129, 197–205. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Mohan, P. M. & Padmanabhan, R. (1992). Processing and localization of Dengue virus type 2 polyprotein precursor NS3-NS4A-NS4B-NS5. J Virol 66, 7549–7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.