Abstract
The alphavirus non-structural protein 3 (nsP3) has a conserved N-terminal macro domain and a variable highly phosphorylated C-terminal domain. nsP3 forms complexes with cellular proteins, but its role in virus replication is poorly understood and protein interaction domains have not been defined. As the N-terminal macro domain can bind poly(ADP-ribose) (PAR), and PAR polymerase-1 (PARP-1) is activated and autoribosylated during Sindbis virus (SINV) infection, it was hypothesized that PARP-1 and nsP3 may interact. Co-immunoprecipitation studies showed that PARP-1 interacted with nsP3 during SINV infection of NSC34 neuronal cells and was most abundantly present in replication complexes that contained plus- and minus-strand SINV RNAs 10–14 h after infection, prior to PARP-1 activation or automodification with PAR. Treatment with an inhibitor of PARP enzymic activity did not affect the interaction between nsP3 and PARP-1 or SINV replication. Co-expression of individual domains of nsP3 with PARP-1 showed that nsP3 interacted with PARP-1 through the C-terminal domain, not the N-terminal macro domain, and that phosphorylation was not required. It was concluded that PARP-1 interacts with the C-terminal domain of nsP3, is present in replication complexes during virus amplification and may play a role in regulating virus RNA synthesis in neuronal cells.
INTRODUCTION
Sindbis virus (SINV) is the prototype of the genus Alphavirus, family Togaviridae, and has a positive-strand RNA genome. In humans, alphaviruses can cause encephalitis, and SINV causes arthritis and a rash (Calisher, 1994; Griffin, 2007; Laine et al., 2004). In mice, SINV causes encephalomyelitis, and neurons are the main target cells for infection in the central nervous system. SINV non-structural proteins (nsPs) are translated as polyproteins (P123 and P1234) from the 5′ two-thirds of the genome and are cleaved in a regulated fashion into four individual nsPs (nsP1–4) (de Groot et al., 1990). Within replication complexes, the polyproteins and individual nsPs have different functions in genomic and subgenomic plus- and minus-strand synthesis (Lemm et al., 1994, 1998; Shirako & Strauss, 1994). nsP1 plays a role in capping virus RNAs (Mi et al., 1989; Scheidel et al., 1987), nsP2 is a multifunctional protein with helicase and 5′ triphosphatase activities and is the protease that cleaves the non-structural polyprotein (Gomez de Cedrón et al., 1999; Ding & Schlesinger, 1989; Hardy & Strauss, 1989; Vasiljeva et al., 2000) and nsP4 is the RNA-dependent RNA polymerase and terminal adenyltransferase (Kamer & Argos, 1984; Tomar et al., 2006).
The function of nsP3 is least understood. The protein has a conserved N-terminal macro domain, an intermediate linker domain and a variable highly phosphorylated C-terminal domain. nsP3 associates with cellular membranes (Peranen & Kaariainen, 1991) and plays a role in regulating RNA synthesis (De et al., 1996; LaStarza et al., 1994b; Wang et al., 1994), but its exact function is unknown. nsP3 interacts with a variety of cellular proteins that are likely to play essential roles in the formation or function of virus replication complexes (Cristea et al., 2006; Despres et al., 1995; Frolova et al., 2006; Gorchakov et al., 2008), and a primary function for nsP3 may be to recruit the required cellular proteins to sites of SINV replication. However, the role of the different domains of nsP3 and their post-translational modifications in recruitment of cellular proteins are unknown.
The C-terminal domain of nsP3 is highly variable, both in sequence and length, among alphaviruses and is heavily phosphorylated at serine/threonine (Li et al., 1990; Peranen et al., 1988; Vihinen & Saarinen, 2000). This domain is not essential for SINV replication, but plays a host-cell-dependent role in SINV minus-strand RNA synthesis (De et al., 2003; LaStarza et al., 1994a). Phosphorylation of this domain is mediated by unidentified host-cell kinases (Li et al., 1990; Peranen, 1991). Complete elimination of the phosphorylation sites from Semliki Forest virus nsP3 decreases the rate of RNA synthesis and attenuates the neurovirulence of this virus for mice (Vihinen et al., 2001). Otherwise, the function of this domain and the role of phosphorylation in regulating this function are unclear.
Unlike the C terminus, the N-terminal 150 aa of nsP3 are well conserved among alphaviruses and this region was first recognized as a domain of unknown function (X domain) (Gorbalenya et al., 1991; Koonin et al., 1992). The X domain was based on homology to the non-histone region of histone macroH2A, later identified as a macro domain (Pehrson & Fried, 1992). Macro domains are ancient and widely distributed throughout all eukaryotic organisms, bacteria and archaea (Pehrson & Fuji, 1998). Macro domains are also found in nsPs of some positive-strand RNA viruses including alphaviruses, coronaviruses, hepatitis E virus and rubella virus (Gorbalenya et al., 1991; Koonin et al., 1992; Liang et al., 2000). Many macro domains can bind various forms of ADP-ribose (Comstock & Denu, 2007; Egloff et al., 2006; Karras et al., 2005; Kustatscher et al., 2005). The alphavirus macro domain efficiently binds poly(ADP-ribose) (PAR), which is synthesized by PAR polymerases (PARPs) (Neuvonen & Ahola, 2009; Park & Griffin, 2009). PARP-1, the founding member of the PARP protein family, can attach 50–200 units of ADP-ribose to target proteins, including itself, upon activation of its enzymic activity.
As PARP-1 associates with macroH2A, the protein with the best-studied macro domain (Nusinow et al., 2007), the macro domain of nsP3 binds PAR (Neuvonen & Ahola, 2009; Park & Griffin, 2009) and PARP-1 is activated by SINV infection (Nargi-Aizenman et al., 2002; Ubol et al., 1996), we explored the possibility that PARP-1 is one of the cellular proteins that interacts with nsP3 in SINV-infected neuronal cells.
METHODS
Cell culture.
The murine neuronal NSC34 cell line was derived by fusion of mouse N18TB2 neuroblastoma cells with mouse motor neurons and was a gift from Neil Cashman (University of British Columbia, USA) (Cashman et al., 1992; Durham et al., 1993). NSC34 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10 % heat-inactivated fetal bovine serum (FBS), l-glutamine (2 mM), penicillin (100 U ml−1) and streptomycin (100 μg ml−1) (all from Gibco-BRL) at 37 °C in a 5 % CO2 incubator. Baby hamster kidney 21 (BHK21) and 293T cells were grown in DMEM/10 % FBS supplemented as above. BHK21 cells were used to grow virus stocks and to quantify virus by plaque assay. 293T cells were used for transfection experiments.
Virus infection and 1,5-dihydroxyisoquinoline (DHIQ) treatment.
The TE recombinant strain of SINV (Lustig et al., 1988) was used. Virus was diluted in DMEM/1 % FBS and cells were infected at an m.o.i. of 10 for 1 h at 37 °C. Cells were then washed three times with medium and the medium was replaced. To inhibit PARP enzymic activity, a competitive inhibitor, DHIQ (300 μM; Sigma) (Southan & Szabo, 2003), was added to the medium after infection.
Plasmids and transfection.
pcDNA3.1 SINV nsP3 was generated by PCR amplification of full-length SINV nsP3 (aa 1–549) from the TE plasmid, followed by insertion into pcDNA3.1 using a pcDNA 3.1 Directional TOPO Expression kit (Invitrogen). Haemagglutinin (HA)-tagged individual nsP3 domains were generated by PCR amplification of the macro domain (aa 2–152), middle region (aa 152–307) and C terminus (aa 307–549) from the TE plasmid, followed by insertion into pCMV-HA (Clontech). pRC3.1 human PARP was a gift from Valina Dawson (Johns Hopkins University School of Medicine, USA). 293T cells at 90 % confluency were transfected with plasmids using Lipofectamine 2000 (Invitrogen). Two days after transfection, 293T cells were lysed and used for immunoprecipitation.
Immunoprecipitation and immunoblot analysis.
Cells were lysed using radioimmune precipitation assay (RIPA) buffer [10 mM Tris/HCl (pH 7.5), 1 % NP-40, 0.1 % SDS, 0.1 % sodium deoxycholate, 150 mM NaCl, 1 mM EDTA] containing a protease inhibitor cocktail (Sigma) and Halt phosphatase inhibitor cocktail (Pierce). Cell lysates were centrifuged at 16 000 g for 30 min to remove cell debris and then used for immunoprecipitation or immunoblotting. For immunoprecipitation, 200 μg cell lysate protein was incubated with antibody to nsP3 (10 μg, polyclonal rabbit antibody raised against aa 212–230 of nsP3; Quality Controlled Biochemicals) or antibody to PARP-1 (diluted 1 : 100; Cell Signalling) in PBS/0.5 % Triton X-100 (NSC34 cell lysates) or RIPA buffer (293T cell lysates) overnight with gentle rocking at 4 °C. ImmunoPure Immobilized Protein G (10 μl settled gel; Pierce) was added to the mixture of cell lysate and antibody, followed by overnight incubation with gentle rocking at 4 °C. To immunoprecipitate HA-tagged proteins, anti-HA agarose conjugate (20 μl settled gel; Sigma) was incubated with cell lysates in RIPA buffer overnight at 4 °C. The gel was washed three times with RIPA buffer and bound proteins were eluted by heating to 90 °C with 2× SDS sample buffer for 5 min. Eluted proteins were analysed by PAGE and immunoblotting. Antibodies used for immunoblotting were against actin (Chemicon), apoptosis-inducing factor (AIF; Santa Cruz), PARP-1 (Alexis and Cell Signalling), PAR (BD Biosciences), nsP1 (Hardy & Strauss, 1989), nsP2 (polyclonal rabbit antibody raised against aa 113–130 of nsP2; Quality Controlled Biochemicals) and nsP3.
RNA co-immunoprecipitation and RNA detection.
Cells were lysed using lysis buffer [10 mM Tris/HCl (pH 7.4), 140 mM NaCl, 1 % NP-40, 0.4 % sodium deoxycholate] and immunoprecipitation was performed in lysis buffer as described above using antibodies to PARP-1 and enhanced green fluorescent protein (EGFP) as a control. RNA was eluted from the immunoprecipitate by boiling the gel in 300 μl elution buffer [10 mM Tris (pH 7.4), 2 mM EDTA, 0.5 % SDS] for 5 min, and eluted RNAs were purified using an RNeasy kit (Qiagen). To quantify virus plus- and minus-strand RNA, each cDNA was synthesized from RNA using a Transcriptor First Strand cDNA Synthesis kit (Roche Applied Science) with a SINV-specific primer (SINV8456F, 5′-CACGGCAATGTGTTTGCT-3′, for the cDNA synthesis of minus-strand RNA; SINV9899R, 5′-AGCATTGGCCGACCTAACGCAGCAC-3′, for the cDNA synthesis of plus-strand RNA). Real-time PCR was performed with the synthesized cDNA, primers SVE2F (5′-TGGGACGAAGCGGACGATAA-3′) and SVE2R (5′-CTGCTCCGCTTTGGTCGTAT-3′) and the Taqman probe 5′-FAM-CGCATACAGACTTCCGCCCAGT-TAMRA-3′ (Applied Biosystems) using a TaqMan PCR Core Reagent kit (Applied Biosystems). Real-time PCR was run and analysed with a 7500 Real-time PCR System (Applied Biosystems).
RESULTS
PARP-1 is present in SINV replication complexes
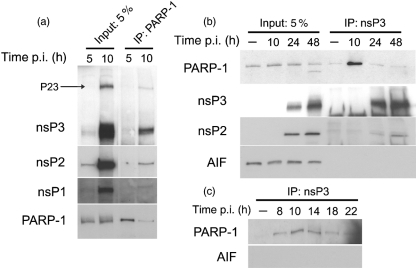
Upon activation, PARP-1 is automodified with PAR. As nsP3 contains a macro domain that can bind PAR in vitro (Egloff et al., 2006; Karras et al., 2005; Park & Griffin, 2009), we explored the possibility that nsP3 interacts with activated PARP-1. Co-immunoprecipitation assays were performed using SINV-infected NSC34 cell lysates collected at different times after infection. Antibody to PARP-1 co-immunoprecipitated nsP3, nsP2 and nsP1, as well as a protein with the migration characteristics of P23, at 10 h post-infection (p.i.) (Fig. 1a). In reciprocal immunoprecipitation reactions, antibody to nsP3 co-immunoprecipitated PARP-1 as well as nsP2, but not AIF (Fig. 1b). nsP3 was not visualized at 10 h in this panel because of interference by the antibody heavy chain band when the immunoprecipitation was carried out with the nsP3 antibody. To examine more closely the transient interaction between PARP-1 and nsP3, co-immunoprecipitation was performed at more frequent intervals in the first 24 h after infection (Fig. 1c). PARP-1 was most abundant in the immunoprecipitated protein complex at 10 and 14 h, and had declined by 18 h after infection.
Fig. 1.
PARP-1 interacts with SINV nsPs. NSC34 cells were infected with SINV (m.o.i.=10) or mock-infected. Cell lysates were incubated with antibody to PARP-1 (a) or nsP3 (b, c), and co-immunoprecipitated proteins were analysed by immunoblotting using antibodies to nsP3, nsP2, nsP1, PARP-1 and AIF. Input, 5 % of the total lysate used for the co-immunoprecipitation assay.
To determine whether the nsP3–PARP-1 complex contained virus RNA, co-immunoprecipitation assays from SINV-infected NSC34 cell lysates collected at 10 h after infection were probed for plus- and minus-strand RNA. Antibody to PARP-1 co-immunoprecipitated both plus- and minus-strand virus RNAs, whilst no RNA was detected in the absence of antibody or with control (EGFP) antibody (data not shown). These studies indicated that PARP-1 was present in the virus replication complexes.
PARP enzymic activity is not required for the interaction between PARP-1 and nsPs or for SINV replication
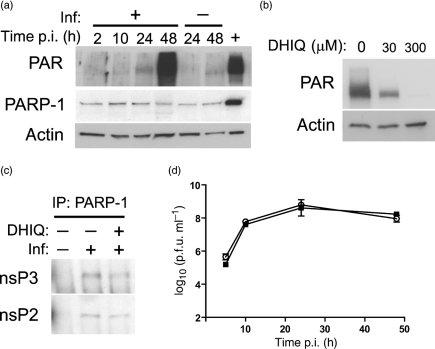
We hypothesized that the nsP3 macro domain binds automodified PARP-1 through PAR; therefore, we examined the activation of PARP and the synthesis of PAR after SINV infection (Fig. 2). PAR was detectable at 24 h after infection and reached a peak at 48 h (Fig. 2a). However, the interaction between PARP-1 and nsPs was stronger before the detection of PAR (Fig. 1), suggesting that PAR may not be important for the interaction. Therefore, we tested the effect of inhibition of PARP enzymic activity with DHIQ, a competitive inhibitor of PARP (Southan & Szabo, 2003). DHIQ completely inhibited PAR synthesis at 300 μM (Fig. 2b), but had no effect on co-immunoprecipitation of nsP3 and nsP2 with PARP-1 (Fig. 2c), indicating that the interaction is not mediated by PAR binding to the macro domain.
Fig. 2.
Inhibition of PARP enzymic activity does not affect the interaction between PARP-1 and nsPs or SINV replication. (a) Time course of PARP activation after SINV infection (Inf) of NSC34 cells. Cells were infected with SINV (m.o.i.=10) or mock-infected. Cell lysates were analysed by immunoblotting using antibodies to PAR, PARP-1 or actin. The positive control (+) was obtained from HeLa cells treated with hydrogen peroxide. (b) Effect of DHIQ on PAR synthesis in SINV-infected NSC34 cells. Cells were infected with SINV and treated with DHIQ, a PARP inhibitor, at 30 or 300 μM, or mock-treated. Cell lysates were collected 24 h after infection and analysed as in (a). (c) Effect of inhibition of PAR synthesis on the interaction between PARP-1 and nsPs in NSC34 cells. Cells were infected with SINV (m.o.i.=10) or mock-infected and treated with DHIQ (300 μM) or mock-treated. Cell lysates at 10 h p.i. were incubated with antibody to PARP-1, and co-immunoprecipitated proteins were analysed by immunoblotting using antibodies to nsP2 and nsP3. (d) Effect of inhibition of PAR synthesis on SINV replication in NSC34 cells. Cells were infected with SINV (m.o.i.=10) and treated with DHIQ (▪; 300 μM) or mock-treated (○). Virus released into the cell-culture medium was assessed by plaque assay. The means±sem of two independent experiments are presented. In each experiment, duplicate samples were assessed.
We also explored the possibility that PARP-1 enzymic activity is required for SINV replication, as PARP-1 exists in virus replication complexes and could function through PARylation of other proteins. SINV-infected NSC34 cells were treated with DHIQ or mock-treated, and virus replication was compared. Inhibition of PAR synthesis did not affect SINV replication (Fig. 2d).
Other SINV proteins are not required for the interaction between PARP-1 and nsP3
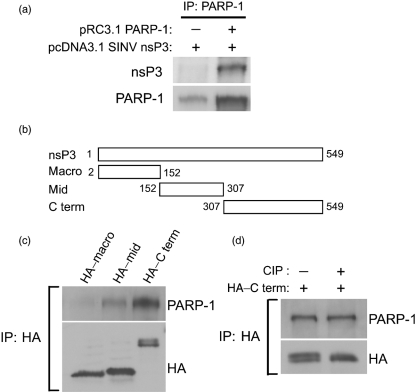
To determine whether nsP3 required other SINV proteins to interact with PARP-1, 293T cells were singly or doubly transfected with plasmids expressing PARP-1 and nsP3, and immunoprecipitation was performed using antibody to PARP-1 (Fig. 3a). Expressed PARP-1 interacted with expressed nsP3, indicating that nsP3 does not require other SINV proteins to bind PARP-1.
Fig. 3.
The nsP3 C terminus mediates PARP-1 interaction, but phosphorylation is not required. (a) Interaction between PARP-1 and nsP3 expressed in 293T cells. At 48 h post-transfection, cell lysates were incubated with antibody to PARP-1, and co-immunoprecipitated proteins were analysed by immunoblotting. (b) Construction of plasmids expressing the HA-tagged macro domain (Macro), middle region (Mid) and C terminus (C term) of nsP3. (c) Interaction between PARP-1 and different regions of nsP3. 293T cells were doubly transfected with plasmids expressing PARP-1 and the nsP3 macro domain, middle or C terminus. At 48 h post-transfection, cell lysates were incubated with antibody to HA, and co-immunoprecipitated proteins were analysed by immunoblotting. (d) Effect of eliminating phosphorylation on the interaction between PARP-1 and the nsP3 C terminus. 293T cells were doubly transfected with plasmids expressing PARP-1 and the HA-tagged nsP3 C terminus. At 48 h post-transfection, cell lysates were collected and treated with CIP or mock-treated. Immunoprecipitation was performed as in (b).
The nsP3 C terminus mediates the interaction with PARP-1 independent of phosphorylation
To determine which domain of nsP3 is important for the interaction between nsP3 and PARP-1, we generated plasmids expressing HA-tagged versions of the macro domain, the middle (linker) region and the C terminus of nsP3 (Fig. 3b). 293T cells were doubly transfected with plasmids expressing PARP-1 and the individual nsP3 domains (macro, middle and C-terminal). Immunoprecipitation was performed using antibody to HA (Fig. 3c). PARP-1 interacted strongly with the C-terminal domain of nsP3 but did not react with the macro domain.
As the nsP3 C terminus can be phosphorylated, we investigated whether phosphorylation modulated the interaction with PARP-1. Cell lysates from 293T cells doubly transfected with PARP-1 and HA-tagged nsP3 C terminus were treated with calf intestinal alkaline phosphatase (CIP) or mock-treated, and immunoprecipitated with antibody to HA (Fig. 3d). Treatment with CIP dephosphorylated the nsP3 C terminus, as evidenced by the disappearance of the upper band, but did not affect the established interaction with PARP-1, suggesting that the interaction is not dependent on nsP3 phosphorylation.
DISCUSSION
In this study, we showed that nsP3 binds and recruits PARP-1 to replication complexes containing SINV RNA early during infection. Although the nsP3 macro domain can bind PAR (Park & Griffin, 2009) and PARP is activated by SINV infection, the interaction between PARP-1 and SINV nsP3 was not mediated by PAR. nsP3 interacted with PARP-1 prior to PARP activation, and inhibition of PAR synthesis with DHIQ had no effect on the interaction or on SINV replication. Rather, binding of PARP-1 to nsP3 was through the C-terminal domain of nsP3 and this interaction was not dependent on phosphorylation. Therefore, PARP-1 is recruited to SINV replication complexes by interaction with the C-terminal domain of nsP3 during the phase of rapid virus replication.
A primary function for nsP3 is postulated to be its ability to recruit cellular proteins – presumed to be important for RNA synthesis – to SINV replication complexes. In support of this idea, mass spectroscopy analyses of immunoprecipitated proteins from SINV-infected Rat-1 and BHK cells have identified time-dependent nsP3-associated recruitment of a variety of cellular proteins (Cristea et al., 2006; Frolova et al., 2006). These host proteins include Ras GTPase-activating protein SH3 domain-binding proteins (G3BP1 and G3BP2), 14-3-3 and heterogeneous nuclear ribonucleoproteins. We used a candidate protein approach to search for nsP3 binding partners in neuronal cells and identified PARP-1 as an additional nsP3-interacting cellular protein recruited to SINV replication complexes.
The domains of nsP3 that interact with specific host-cell proteins have not been identified previously. Both the N-terminal macro domain and the C-terminal phosphorylated domain are candidates for mediating interactions with cellular proteins. We have shown that it is the C-terminal domain that interacts with PARP-1. It has been suggested previously that this region optimizes replication in different host cells by interacting with host-specific factors (LaStarza et al., 1994a). Although phosphorylation is increasingly recognized to be important for regulation of plus-strand RNA virus replication (Jakubiec & Jupin, 2007) and elimination of all nsP3 phosphorylation sites affects SFV neurovirulence (Vihinen et al., 2001), phosphorylation was not necessary for nsP3 interaction with PARP-1.
PARP-1 is transiently recruited to virus replication complexes during active virus replication in neuronal cells, but its function in these complexes is not known. PARP-1 is usually found in the nucleus and plays an important role in many physiological and pathophysiological cellular processes, including DNA repair, regulation of transcription and induction of cell death (Amé et al., 2004; Andrabi et al., 2008; Kim et al., 2005; Kraus & Lis, 2003). It functions both through modification of target proteins with PAR (PARylation) (Hassa & Hottiger, 2002; Kraus & Lis, 2003) and by direct protein interaction, independent of its enzymic activity (Cervellera & Sala, 2000; Haince et al., 2006; Hassa et al., 2001, 2005; Pavri et al., 2005). For instance, cellular RNA transcription is regulated both by modifying transcription factors with PAR (Schreiber et al., 2006) and through PAR-independent mechanisms involving protein–protein interactions (Cervellera & Sala, 2000; Haince et al., 2006; Hassa et al., 2001, 2005; Pavri et al., 2005; Schreiber et al., 2006). Inhibition of PAR synthesis did not affect SINV replication or the recruitment of PARP-1, indicating that the function of PARP in SINV replication complexes is probably independent of its enzymic activity and is most likely to be dependent on protein interaction. Both the DNA-binding and automodification domains can mediate interaction with other cellular proteins (Griesenbeck et al., 1999; Li et al., 2006; Masutani & Miwa, 2002; Meder et al., 2005; Monaco et al., 2005; Simbulan-Rosenthal et al., 2003).
As other cellular proteins are recruited to SINV replication complexes (Cristea et al., 2006; Despres et al., 1995; Frolova et al., 2006; Gorchakov et al., 2008), PARP-1 might interact with these proteins and activate or inhibit them to promote SINV replication. It is also possible that PARP-1 is involved in stabilizing SINV replication complexes. SINV is able to replicate in fibroblasts from mice lacking PARP-1 (Nargi-Aizenman et al., 2002), so PARP-1 is not required for replication.
PARP activation was previously observed after SINV infection of fibroblasts and N18 neuroblastoma cells (Nargi-Aizenman et al., 2002; Ubol et al., 1996). In the current study, PAR synthesis was also observed after SINV infection of NSC34 neuronal cells, although this occurred after peak interaction with nsP3. Classically, PARP-1 activation is induced by DNA damage as a result of oxidative stress, excitotoxicity or inflammation (Bryant & Helleday, 2004), but SINV-induced activation of PARP occurs prior to the DNA damage associated with apoptosis (Nargi-Aizenman et al., 2002). A number of alternative pathways for PARP-1 activation have been described. PARP-1 can be activated by binding to stem–loop DNA structures (Kun et al., 2002), by phosphorylation (Ju et al., 2004; Walker et al., 2006), by interaction with other proteins (Cohen-Armon et al., 2007; Griesenbeck et al., 1999; Kun et al., 2004) and by membrane depolarization followed by an increase in intracellular Ca2+ (Homburg et al., 2000; Meli et al., 2005; Visochek et al., 2005). Membrane depolarization occurs in alphavirus-infected cells (Nargi-Aizenman & Griffin, 2001) and could potentially induce PARP activation.
In conclusion, this study has shown that PARP-1 is recruited to virus replication complexes by nsP3 and that this interaction is independent of PARP's enzymic activity and is mediated primarily by the C-terminal domain of nsP3, independent of its phosphorylation status.
Acknowledgments
This work was supported by research grant R01 NS18596 from the National Institutes of Health. We would like to thank Dr Richard Kuhn for helpful discussions and Debra Hauer and Marcia Lyons for their technical support.
References
- Amé, J. C., Spenlehauer, C. & de Murcia, G. (2004). The PARP superfamily. Bioessays 26, 882–893. [DOI] [PubMed] [Google Scholar]
- Andrabi, S. A., Dawson, T. M. & Dawson, V. L. (2008). Mitochondrial and nuclear cross talk in cell death: parthanatos. Ann N Y Acad Sci 1147, 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, H. E. & Helleday, T. (2004). Poly(ADP-ribose) polymerase inhibitors as potential chemotherapeutic agents. Biochem Soc Trans 32, 959–961. [DOI] [PubMed] [Google Scholar]
- Calisher, C. H. (1994). Medically important arboviruses of the United States and Canada. Clin Microbiol Rev 7, 89–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashman, N. R., Durham, H. D., Blusztajn, J. K., Oda, K., Tabira, T., Shaw, I. T., Dahrouge, S. & Antel, J. P. (1992). Neuroblastoma × spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev Dyn 194, 209–221. [DOI] [PubMed] [Google Scholar]
- Cervellera, M. N. & Sala, A. (2000). Poly(ADP-ribose) polymerase is a B-MYB coactivator. J Biol Chem 275, 10692–10696. [DOI] [PubMed] [Google Scholar]
- Cohen-Armon, M., Visochek, L., Rozensal, D., Kalal, A., Geistrikh, I., Klein, R., Bendetz-Nezer, S., Yao, Z. & Seger, R. (2007). DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol Cell 25, 297–308. [DOI] [PubMed] [Google Scholar]
- Comstock, L. R. & Denu, J. M. (2007). Synthesis and biochemical evaluation of O-acetyl-ADP-ribose and N-acetyl analogs. Org Biomol Chem 5, 3087–3091. [DOI] [PubMed] [Google Scholar]
- Cristea, I. M., Carroll, J. W., Rout, M. P., Rice, C. M., Chait, B. T. & MacDonald, M. R. (2006). Tracking and elucidating alphavirus–host protein interactions. J Biol Chem 281, 30269–30278. [DOI] [PubMed] [Google Scholar]
- De, I., Sawicki, S. G. & Sawicki, D. L. (1996). Sindbis virus RNA-negative mutants that fail to convert from minus-strand to plus-strand synthesis: role of the nsP2 protein. J Virol 70, 2706–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De, I., Fata-Hartley, C., Sawicki, S. G. & Sawicki, D. L. (2003). Functional analysis of nsP3 phosphoprotein mutants of Sindbis virus. J Virol 77, 13106–13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot, R. J., Hardy, W. R., Shirako, Y. & Strauss, J. H. (1990). Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J 9, 2631–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres, P., Griffin, J. W. & Griffin, D. E. (1995). Effects of anti-E2 monoclonal antibody on Sindbis virus replication in AT3 cells expressing bcl-2. J Virol 69, 7006–7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, M. X. & Schlesinger, M. J. (1989). Evidence that Sindbis virus nsP2 is an autoprotease which processes the virus nonstructural polyprotein. Virology 171, 280–284. [DOI] [PubMed] [Google Scholar]
- Durham, H. D., Dahrouge, S. & Cashman, N. R. (1993). Evaluation of the spinal cord neuron × neuroblastoma hybrid cell line NSC-34 as a model for neurotoxicity testing. Neurotoxicology 14, 387–395. [PubMed] [Google Scholar]
- Egloff, M. P., Malet, H., Putics, A., Heinonen, M, Dutartre, H., Frangeul, A., Gruez, A., Campanacci, V., Cambillau, C. & other authors (2006). Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J Virol 80, 8493–8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova, E., Gorchakov, R., Garmashova, N., Atasheva, S., Vergara, L. A. & Frolov, I. (2006). Formation of nsP3-specific protein complexes during Sindbis virus replication. J Virol 80, 4122–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez de Cedrón, M., Ehsani, N., Mikkola, M. L., García, J. A. & Kääriäinen, L. (1999). RNA helicase activity of Semliki Forest virus replicase protein NSP2. FEBS Lett 448, 19–22. [DOI] [PubMed] [Google Scholar]
- Gorbalenya, A. E., Koonin, E. V. & Lai, M. M. (1991). Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett 288, 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov, R., Garmashova, N., Frolova, E. & Frolov, I. (2008). Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J Virol 82, 10088–10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesenbeck, J., Ziegler, M., Tomilin, N., Schweiger, M. & Oei, S. L. (1999). Stimulation of the catalytic activity of poly(ADP-ribosyl) transferase by transcription factor Yin Yang 1. FEBS Lett 443, 20–24. [DOI] [PubMed] [Google Scholar]
- Griffin, D. E. (2007). Alphaviruses. In Fields Virology, 5th edn, pp. 1023–1067. Edited by D. L. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman & S. E. Straus. Philadelphia, PA: Lippincott Williams & Wilkins.
- Haince, J. F., Rouleau, M. & Poirier, G. G. (2006). Transcription. Gene expression needs a break to unwind before carrying on. Science 312, 1752–1753. [DOI] [PubMed] [Google Scholar]
- Hardy, W. R. & Strauss, J. H. (1989). Processing the nonstructural polyproteins of Sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J Virol 63, 4653–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa, P. O. & Hottiger, M. O. (2002). The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-κB in inflammatory disorders. Cell Mol Life Sci 59, 1534–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa, P. O., Covic, M., Hasan, S., Imhof, R. & Hottiger, M. O. (2001). The enzymatic and DNA binding activity of PARP-1 are not required for NF-κB coactivator function. J Biol Chem 276, 45588–45597. [DOI] [PubMed] [Google Scholar]
- Hassa, P. O., Haenni, S. S., Buerki, C., Meier, N. I., Lane, W. S., Owen, H., Gersbach, M., Imhof, R. & Hottiger, M. O. (2005). Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-κB-dependent transcription. J Biol Chem 280, 40450–40464. [DOI] [PubMed] [Google Scholar]
- Homburg, S., Visochek, L., Moran, N., Dantzer, F., Priel, E., Asculai, E., Schwartz, D., Rotter, V., Dekel, N. & Cohen-Armon, M. (2000). A fast signal-induced activation of poly(ADP-ribose) polymerase: a novel downstream target of phospholipase C. J Cell Biol 150, 293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubiec, A. & Jupin, I. (2007). Regulation of positive-strand RNA virus replication: the emerging role of phosphorylation. Virus Res 129, 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju, B. G., Solum, D., Song, E. J., Lee, K. J., Rose, D. W., Glass, C. K. & Rosenfeld, M. G. (2004). Activating the PARP-1 sensor component of the groucho/TLE1 corepressor complex mediates a CaMKinase IIδ-dependent neurogenic gene activation pathway. Cell 119, 815–829. [DOI] [PubMed] [Google Scholar]
- Kamer, G. & Argos, P. (1984). Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res 12, 7269–7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras, G. I., Kustatscher, G., Buhecha, H. R., Allen, M. D., Pugieux, C., Sait, F., Bycroft, M. & Ladurner, A. G. (2005). The macro domain is an ADP-ribose binding module. EMBO J 24, 1911–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. Y., Zhang, T. & Kraus, W. L. (2005). Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev 19, 1951–1967. [DOI] [PubMed] [Google Scholar]
- Koonin, E. V., Gorbalenya, A. E., Purdy, M. A., Rozanov, M. N., Reyes, G. R. & Bradley, D. W. (1992). Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc Natl Acad Sci U S A 89, 8259–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus, W. L. & Lis, J. T. (2003). PARP goes transcription. Cell 113, 677–683. [DOI] [PubMed] [Google Scholar]
- Kun, E., Kirsten, E. & Ordahl, C. P. (2002). Coenzymatic activity of randomly broken or intact double-stranded DNAs in auto and histone H1 trans-poly(ADP-ribosylation), catalyzed by poly(ADP-ribose) polymerase (PARP I). J Biol Chem 277, 39066–39069. [DOI] [PubMed] [Google Scholar]
- Kun, E., Kirsten, E., Mendeleyev, J. & Ordahl, C. P. (2004). Regulation of the enzymatic catalysis of poly(ADP-ribose) polymerase by dsDNA, polyamines, Mg2+, Ca2+, histones H1 and H3, and ATP. Biochemistry 43, 210–216. [DOI] [PubMed] [Google Scholar]
- Kustatscher, G., Hothorn, M., Pugieux, C., Scheffzek, K. & Ladurner, A. G. (2005). Splicing regulates NAD metabolite binding to histone macroH2A. Nat Struct Mol Biol 12, 624–625. [DOI] [PubMed] [Google Scholar]
- Laine, M., Luukkainen, R. & Toivanen, A. (2004). Sindbis viruses and other alphaviruses as cause of human arthritic disease. J Intern Med 256, 457–471. [DOI] [PubMed] [Google Scholar]
- LaStarza, M. W., Grakoui, A. & Rice, C. M. (1994a). Deletion and duplication mutations in the C-terminal nonconserved region of Sindbis virus nsP3: effects on phosphorylation and on virus replication in vertebrate and invertebrate cells. Virology 202, 224–232. [DOI] [PubMed] [Google Scholar]
- LaStarza, M. W., Lemm, J. A. & Rice, C. M. (1994b). Genetic analysis of the nsP3 region of Sindbis virus: evidence for roles in minus-strand and subgenomic RNA synthesis. J Virol 68, 5781–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm, J. A., Rumenapf, T., Strauss, E. G., Strauss, J. H. & Rice, C. M. (1994). Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J 13, 2925–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm, J. A., Bergqvist, A., Read, C. M. & Rice, C. M. (1998). Template-dependent initiation of Sindbis virus RNA replication in vitro. J Virol 72, 6546–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G. P., La Starza, M. W., Hardy, W. R., Strauss, J. H. & Rice, C. M. (1990). Phosphorylation of Sindbis virus nsP3 in vivo and in vitro. Virology 179, 416–427. [DOI] [PubMed] [Google Scholar]
- Li, Y., Oh, H. J. & Lau, Y. F. (2006). The poly(ADP-ribose) polymerase 1 interacts with Sry and modulates its biological functions. Mol Cell Endocrinol 257–258, 35–46. [DOI] [PubMed] [Google Scholar]
- Liang, Y., Yao, J. & Gillam, S. (2000). Rubella virus nonstructural protein protease domains involved in trans- and cis-cleavage activities. J Virol 74, 5412–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig, S., Jackson, A. C., Hahn, C. S., Griffin, D. E., Strauss, E. G. & Strauss, J. H. (1988). The molecular basis of Sindbis virus neurovirulence in mice. J Virol 62, 2329–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani, M. & Miwa, M. (2002). Poly(ADP-ribose) polymerase and cancer: in relation to the lectures presented by Dr Gilbert de Murcia. Jpn J Clin Oncol 32, 483–487. [DOI] [PubMed] [Google Scholar]
- Meder, V. S., Boeglin, M., de Murcia, G. & Schreiber, V. (2005). PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J Cell Sci 118, 211–222. [DOI] [PubMed] [Google Scholar]
- Meli, E., Baronti, R., Pangallo, M., Picca, R., Moroni, F. & Pellegrini-Giampietro, D. E. (2005). Group I metabotropic glutamate receptors stimulate the activity of poly(ADP-ribose) polymerase in mammalian mGlu1-transfected cells and in cortical cell cultures. Neuropharmacology 49 (Suppl. 1), 80–88. [DOI] [PubMed] [Google Scholar]
- Mi, S., Durbin, R., Huang, H. V., Rice, C. M. & Stollar, V. (1989). Association of the Sindbis virus RNA methytransferase activity with the nonstructural protein nsP1. Virology 170, 385–391. [DOI] [PubMed] [Google Scholar]
- Monaco, L., Kolthur-Seetharam, U., Loury, R., Murcia, J. M., de Murcia, G. & Sassone-Corsi, P. (2005). Inhibition of Aurora-B kinase activity by poly(ADP-ribosyl)ation in response to DNA damage. Proc Natl Acad Sci U S A 102, 14244–14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargi-Aizenman, J. L. & Griffin, D. E. (2001). Sindbis virus-induced neuronal death is both necrotic and apoptotic and is ameliorated by N-methyl-d-aspartate receptor antagonists. J Virol 75, 7114–7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargi-Aizenman, J. L., Simbulan-Rosenthal, C. M., Kelly, T. A., Smulson, M. E. & Griffin, D. E. (2002). Rapid activation of poly(ADP-ribose) polymerase contributes to Sindbis virus and staurosporine-induced apoptotic cell death. Virology 293, 164–171. [DOI] [PubMed] [Google Scholar]
- Neuvonen, M. & Ahola, T. (2009). Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J Mol Biol 385, 212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow, D. A., Hernández-Muñoz, I., Fazzio, T. G., Shah, G. M., Kraus, W. L. & Panning, B. (2007). Poly(ADP-ribose) polymerase 1 is inhibited by a histone H2A variant, MacroH2A, and contributes to silencing of the inactive X chromosome. J Biol Chem 282, 12851–12859. [DOI] [PubMed] [Google Scholar]
- Park, E. & Griffin, D. E. (2009). The nsP3 macro domain is important for Sindbis virus replication in neurons and neurovirulence in mice. Virology 388, 305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri, R., Lewis, B., Kim, T. K., Dilworth, F. J., Erdjument-Bromage, H., Tempst, P., de Murcia, G., Evans, R., Chambon, P. & Reinberg, D. (2005). PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell 18, 83–96. [DOI] [PubMed] [Google Scholar]
- Pehrson, J. R. & Fried, V. A. (1992). MacroH2A, a core histone containing a large nonhistone region. Science 257, 1398–1400. [DOI] [PubMed] [Google Scholar]
- Pehrson, J. R. & Fuji, R. N. (1998). Evolutionary conservation of histone macroH2A subtypes and domains. Nucleic Acids Res 26, 2837–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peranen, J. (1991). Localization and phosphorylation of Semliki Forest virus nonstructural protein nsP3 expressed in COS cells from a cloned cDNA. J Gen Virol 72, 195–199. [DOI] [PubMed] [Google Scholar]
- Peranen, J. & Kaariainen, L. (1991). Biogenesis of type I cytopathic vacuoles in Semliki Forest virus-infected BHK cells. J Virol 65, 1623–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peranen, J., Takkinen, K., Kalkkinen, N. & Kaariainen, L. (1988). Semliki Forest virus-specific non-structural protein nsP3 is a phosphoprotein. J Gen Virol 69, 2165–2178. [DOI] [PubMed] [Google Scholar]
- Scheidel, L. M., Durbin, R. K. & Stollar, V. (1987). Sindbis virus mutants resistant to mycophenolic acid and ribavirin. Virology 158, 1–7. [DOI] [PubMed] [Google Scholar]
- Schreiber, V., Dantzer, F., Ame, J. C. & de Murcia, G. (2006). Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 7, 517–528. [DOI] [PubMed] [Google Scholar]
- Shirako, Y. & Strauss, J. H. (1994). Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol 68, 1874–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbulan-Rosenthal, C. M., Rosenthal, D. S., Luo, R., Samara, R., Espinoza, L. A., Hassa, P. O., Hottiger, M. O. & Smulson, M. E. (2003). PARP-1 binds E2F-1 independently of its DNA binding and catalytic domains, and acts as a novel coactivator of E2F-1-mediated transcription during re-entry of quiescent cells into S phase. Oncogene 22, 8460–8471. [DOI] [PubMed] [Google Scholar]
- Southan, G. J. & Szabo, C. (2003). Poly(ADP-ribose) polymerase inhibitors. Curr Med Chem 10, 321–340. [DOI] [PubMed] [Google Scholar]
- Tomar, S., Hardy, R. W., Smith, J. L. & Kuhn, R. J. (2006). Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J Virol 80, 9962–9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubol, S., Park, S., Budihardjo, I., Desnoyers, S., Montrose, M. H., Poirier, G. G., Kaufmann, S. H. & Griffin, D. E. (1996). Temporal changes in chromatin, intracellular calcium, and poly(ADP-ribose) polymerase during Sindbis virus-induced apoptosis of neuroblastoma cells. J Virol 70, 2215–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva, L., Merits, A., Auvinen, P. & Kaariainen, L. (2000). Identification of a novel function of the alphavirus capping apparatus. RNA 5′-triphosphatase activity of nsP2. J Biol Chem 275, 17281–17287. [DOI] [PubMed] [Google Scholar]
- Vihinen, H. & Saarinen, J. (2000). Phosphorylation site analysis of Semliki Forest virus nonstructural protein 3. J Biol Chem 275, 27775–27783. [DOI] [PubMed] [Google Scholar]
- Vihinen, H., Ahola, T., Tuittila, M., Merits, A. & Kaariainen, L. (2001). Elimination of phosphorylation sites of Semliki Forest virus replicase protein nsP3. J Biol Chem 276, 5745–5752. [DOI] [PubMed] [Google Scholar]
- Visochek, L., Steingart, R. A., Vulih-Shultzman, I., Klein, R., Priel, E., Gozes, I. & Cohen-Armon, M. (2005). PolyADP-ribosylation is involved in neurotrophic activity. J Neurosci 25, 7420–7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J. W., Jijon, H. B. & Madsen, K. L. (2006). AMP-activated protein kinase is a positive regulator of poly(ADP-ribose) polymerase. Biochem Biophys Res Commun 342, 336–341. [DOI] [PubMed] [Google Scholar]
- Wang, Y. F., Sawicki, S. G. & Sawicki, D. L. (1994). Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J Virol 68, 6466–6475. [DOI] [PMC free article] [PubMed] [Google Scholar]