Abstract
Previous experiments established that when the unicellular green alga Chlorella NC64A is inoculated with two viruses, usually only one virus replicates in a single cell. That is, the viruses mutually exclude one another. In the current study, we explore the possibility that virus-induced host membrane depolarization, at least partially caused by a virus-encoded K+ channel (Kcv), is involved in this mutual exclusion. Two chlorella viruses, PBCV-1 and NY-2A, were chosen for the study because (i) they can be distinguished by real-time PCR and (ii) they exhibit differential sensitivity to Cs+, a well-known K+ channel blocker. PBCV-1-induced host membrane depolarization, Kcv channel activity and plaque formation are only slightly affected by Cs+, whereas all three NY-2A-induced events are strongly inhibited by Cs+. The addition of one virus 5–15 min before the other results primarily in replication of the first virus. However, if virus NY-2A-induced membrane depolarization of the host is blocked by Cs+, PBCV-1 is not excluded. We conclude that virus-induced membrane depolarization is at least partially responsible for the exclusion phenomenon.
INTRODUCTION
Mutual exclusion occurs when a host cell is simultaneously inoculated with two competing viruses but only one virus replicates. This phenomenon was originally described in bacteriophage by Delbrück (1945). Mutual exclusion implies that the virus particle that infects first alters the host cell in such a way that a second infection is unlikely. Subsequent studies found that mutual exclusion occurs not only between different viruses but also with nearly identical viruses, presumably as a way to avoid ‘super-infection’ (Dulbecco, 1952). While the virus that wins prevents infection by additional viruses, the excluded viruses often interfere with replication of the former, referred to as a ‘depressor effect’ (Delbrück, 1945). There are different mechanisms underlying mutual exclusion and depression among bacteriophages, not all of which are understood. In some instances, exclusion causes the out-competed virus to be rapidly degraded (Dulbecco, 1952). For example, with phage T3, degradation is initiated after adsorption of the primary infecting phage but before the viral genome is expressed (Hirsch-Kauffmann et al., 1976). There is indirect evidence with phage λ that host membrane depolarization is triggered by the successful infecting particle, which is mediated by the RexB channel, in such a way as to initiate exclusion (Li & Bockrath, 1993; Snyder, 1995; Snyder & McWilliams, 1989).
Mutual exclusion occurs among not only bacteriophages but also viruses infecting certain unicellular, eukaryotic chlorella-like green algae (called chlorella viruses). That is, plaques arising from single cells simultaneously inoculated with two different chlorella viruses usually only contain one of the two viruses (Chase et al., 1989). Among the various chlorella viruses used in these competition experiments, some effectively out-compete others in the infection process, i.e. they are considered to be more fit. The mechanism underlying chlorella virus mutual exclusion is unknown. Chlorella viruses often encode DNA restriction endonucleases (e.g. Xia et al., 1986, 1987, 1988; Chan et al., 2004) and it was originally suggested that one function of the DNA restriction endonucleases might be to exclude infection by other viruses. However, experimental results did not support this suggestion (Chase et al., 1989).
In the current study, we have examined another explanation for chlorella virus mutual exclusion. The icosahedral-shaped chlorella viruses initiate infection by attaching rapidly, specifically and irreversibly to their host cell wall (Meints et al., 1984, 1988), probably at a unique virus vertex (Onimatsu et al., 2006; Cherrier et al., 2009). Attachment is immediately followed by cell wall degradation at the point of contact by a virus-packaged enzyme(s). Following wall degradation, the viral internal membrane presumably fuses with the host membrane, facilitating entry of the viral DNA and virion-associated proteins into the cell, leaving an empty virus capsid attached to the cell wall. This process initiates rapid depolarization of the host membrane, presumably triggered by a virus-encoded K+ channel (Kcv) located in the virus internal membrane (Frohns et al., 2006; Plugge et al., 2000) and the rapid release of K+ from the cell (Neupärtl et al., 2008). The rapid loss of K+ and associated water fluxes from the host reduce its turgor pressure, which may aid ejection of viral DNA into the host (Neupärtl et al., 2008; Frohns et al., 2006). In the current manuscript, we explore the possibility that the virus-induced membrane depolarization is also involved in excluding a second virus.
The chlorella viruses PBCV-1 and NY-2A were used to examine the role of membrane depolarization and Kcv in mutual exclusion. These two viruses were chosen because they are genetically different enough that quantitative PCR (qPCR) can distinguish them. Another benefit of this pair of viruses is their differential sensitivity to Cs+. Virus PBCV-1-induced host membrane depolarization, Kcv channel activity and plaque formation are only slightly affected by Cs+. In contrast, virus NY-2A-induced host membrane depolarization, Kcv channel activity and plaque formation are strongly inhibited by Cs+ (Frohns et al., 2006; Gazzarrini et al., 2004; Kang et al., 2004a).
METHODS
Growth of viruses and cells.
Growth of Chlorella NC64A, infection assays and the production of PBCV-1 and NY-2A viruses were carried out as described previously (Van Etten et al., 1981, 1983a; Schuster et al., 1986).
Infection.
To establish that the results from the two viruses are comparable, the percentage of infected cells (m.o.i. of 10) was determined for both viruses. Cells were infected and the number of surviving cells after one replication cycle (8 h for PBCV-1 and 18 h for NY-2A) was measured using the cell counter function of a NanoDrop-1000 Spectrophotometer (Thermo Scientific). In both infections, more than 80 % of the cells lysed; these results were confirmed by counting the cells under a microscope.
To determine whether infection of a single cell by both viruses occurs, Chlorella NC64A cells were infected with an m.o.i. of 10 for each virus. After 1 h, Chlorella NC64A cell wall fragments (Meints et al., 1988) were added to remove unattached virus particles and the cells were immediately plaqued (Van Etten et al., 1983b). Fifty plaques, which should represent single infections, were picked randomly and assayed by qPCR (see below) to determine if one or both viruses were present.
Virus attachment.
The assay for measuring virus attachment to host cell wall fragments followed by a plaque assay was carried out as described previously (Meints et al., 1988; Van Etten et al., 1983b).
qPCR.
The concentration of virus-specific DNA was estimated by qPCR (Jansohn, 2007) using a StepOne system (Applied Biosystems) according to the manufacturer's manual. Reactions were conducted in MicroAmp fast optical 48-well reaction plates sealed with MicroAmp 48-well optical adhesive tape (Applied Biosystems).
Viral DNA was extracted from the supernatant by phenol/chloroform (Sambrook et al., 1989) after removal of cell debris by low speed centrifugation (400 g, 5 min) 24 h after infection.
The reaction mixtures contained 5 μl qPCR ROX-GO Green Mastermix (MP Biomedicals), 0.5 μl forward primer (1 μM), 0.5 μl reverse primer (1 μM), 0.4 μl ROX (25 μM), 10 μl template (1 : 10) and 8.6 μl water. Specific virus genes were used to design primers. We chose gene a162l from PBCV-1 and gene b083l from NY-2A. The amplified fragments were each 90 bp. The primers for PBCV-1 were a162l-forward, 5′-CACCTACGTGTACGGAACAACCT-3′, and a162l-reverse, 5′-ACAGGAGTGTCATGGGAATGAAA-3′, and for NY-2A were b083l-forward, 5′-GACGATGGGCGGAAACG-3′, and b083l-reverse, 5′-GAACGACGCCGAAAAGGTT-3′ (Biomers.net).
The expected specificity of the primers was verified by conventional PCR. Control experiments revealed a linear correlation between template concentration and DNA fluorescence.
Calibration.
The viral DNA fragments obtained from the PCR were amplified and extracted by phenol/chloroform. The concentration of DNA was measured on a NanoDrop-1000 Spectrophotometer (Thermo Scientific). The number of amplified DNA molecules was calculated by using the measured DNA concentration, the molecular mass of the fragments and Avogadro's number.
The critical threshold value (Ct), which represents the number of cycles required for a fluorescence signal to pass background fluorescence, was obtained from an amplification plot. Each data point was the mean value±sem of 3 or more independent experiments in which data were measured in triplicate. At the end of a qPCR measurement, a melting curve was performed to verify the specificity of the reaction. These curves all revealed single peaks, meaning that the PCR products were specific. The average Ct values were plotted as a function of the DNA fragment concentration. The plot exhibited a quasi-linear correlation.
Electron microscopy.
Cells were concentrated by centrifugation 5 h post-infection (p.i.) and immediately fixed with a cacodylate-buffered (pH 6.8) 2 % glutaraldehyde, 2 % formaldehyde (freshly prepared from paraformaldehyde) solution. After washing in buffer, samples were post-fixed in OsO4 (2 % in the same buffer), dehydrated in a graded acetone series, and embedded in Spurr's resin. Ultrathin sections were obtained with diamond knives, post-stained with uranyl acetate and lead citrate and examined with a Zeiss EM 109 transmission electron microscope. For each treatment, more than 50 individual cells were examined for progeny viruses. For each cell, only one cross section was examined.
Measurements of host membrane potential.
Changes in host membrane potential were monitored as reported previously (Frohns et al., 2006) with the voltage-sensitive fluorescent dye bis-(1,3-diethylthiobarbituric acid) trimethine oxonol (named bisoxonol) (Molecular Probes). Chlorella NC64A cells (7×106 cells ml−1) from an actively growing culture were incubated in modified Bold's basal medium (Van Etten et al., 1983), adding dye (1 μM) approximately 15 min prior to the measurements; cells were inoculated with viruses at an m.o.i. of 10. Fluorescence was monitored with a spectrofluorophotometer (FP-6200; Jasko) with excitation at 540±5 nm and emission at 560±10 nm. Additional details are given in the paper by Frohns et al. (2006).
RESULTS AND DISCUSSION
Single inoculation
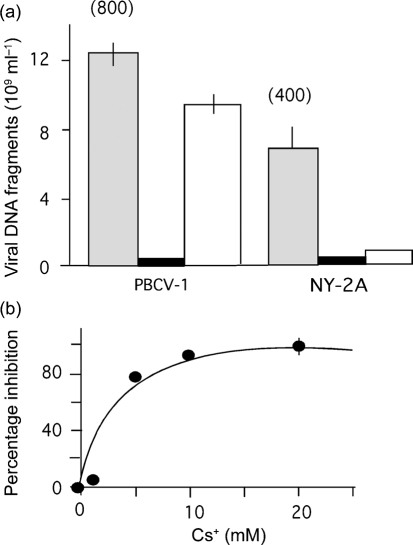
To quantify viral-specific DNA replication in the host cells, 1.5×107 Chlorella NC64A cells were inoculated with either virus PBCV-1 or virus NY-2A at an m.o.i. of 10. Virus-specific DNA was quantified by qPCR at 24 h p.i. From the linear calibration curve, the qPCR signal was converted into DNA fragment equivalents. The data in Fig. 1(a) show that PBCV-1 infection of Chlorella NC64A cells resulted in about 800 DNA fragments. Assuming that 25–50 % of the particles are infectious (Van Etten et al., 1983a), this number translates into 200–400 p.f.u. per cell for PBCV-1. This number agrees with previous estimates of 200–350 p.f.u. per cell (Van Etten et al., 1983a) and indicates that the qPCR method is suitable for quantifying viral DNA in Chlorella NC64A cells.
Fig. 1.
Concentration of PBCV-1 and NY-2A DNA fragments in infected Chlorella NC64A cells. (a) Concentration of DNA fragments in cells inoculated with either virus PBCV-1 or virus NY-2A under control conditions (shaded bars), in the presence of 20 mM BaCl2 (solid bars) or 20 mM CsCl (open bars). The numbers above the bars give the estimated number of DNA fragments per infected cell. (b) Inhibition of virus-specific DNA fragment synthesis in Chlorella cells inoculated with virus NY-2A as a function of Cs+ concentration. Data are means±sem of ≥3 independent experiments. In some cases, the sem is smaller than the symbols.
The same analysis performed with cells inoculated with NY-2A indicated that this virus generates a signal approximately twofold smaller (Fig. 1a). Again, this is in agreement with previous results showing that NY-2A has a burst size that is two- to threefold less than PBCV-1 (Van Etten et al., 1988).
DNA replication is sensitive to inhibitors
Previous investigations indicate that host membrane depolarization, infection (DNA ejection) and replication of both viruses are sensitive to Ba2+; in contrast, Cs+ only prevents these processes in NY-2A (Frohns et al., 2006). The PCR data with the two viruses and inhibitors verified these results. Addition of 20 mM Ba2+ to the incubation medium prevented DNA synthesis of both viruses (Fig. 1a).
Cs+ had a virus-specific effect (Fig. 1a). PBCV-1 DNA replication was reduced by about 20 % in 20 mM Cs+. The effect was much stronger in virus NY-2A; DNA replication was inhibited by approximately 90 %. The Cs+ effect was concentration-dependent (Fig. 1b). Half maximal inhibition was achieved with about 2 mM Cs+, which is the same Cs+ concentration required to achieve half maximal inhibition of NY-2A-triggered host membrane depolarization (Frohns et al., 2006). These similarities suggest that exclusion and membrane depolarization might be causally linked. Separate experiments established that neither Cs+ nor Ba2+ had any effect on PBCV-1 and NY-2A attachment to the host.
Virus PBCV-1 dominates over NY-2A
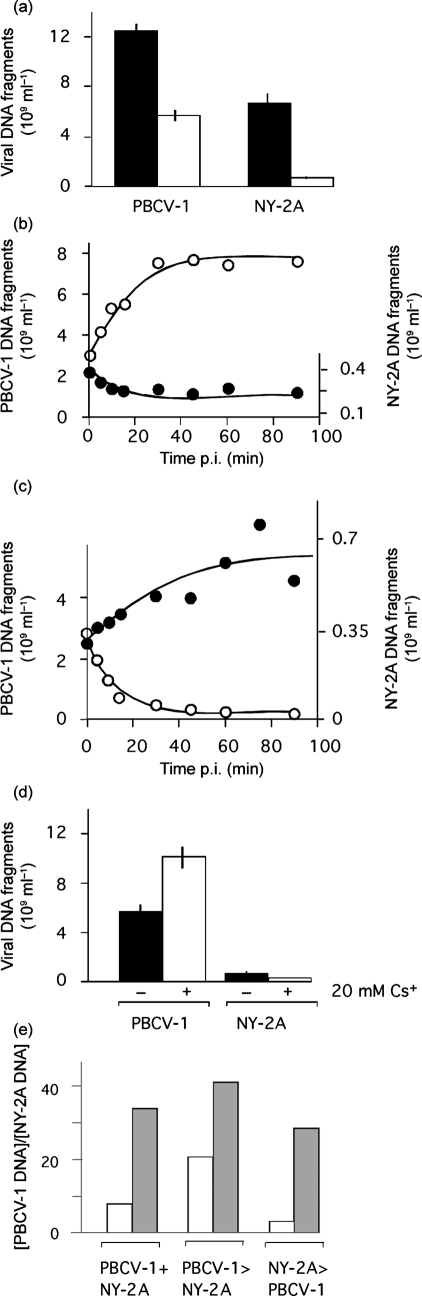
To examine competition between the two viruses during infection, we inoculated Chlorella NC64A simultaneously with both viruses and monitored infection by virus DNA replication. This experiment led to two conclusions. (i) PBCV-1 out-competes NY-2A. In single inoculation experiments, PBCV-1 generated about twofold more DNA fragments than NY-2A. In mixed inoculations, this ratio increased to about sevenfold (Fig. 2a). This result clearly indicates that PBCV-1 out-competes NY-2A in the infection process. This supports previous data that indicate some chlorella viruses can out-compete others (Chase et al., 1989, note, NY-2A was not used in this study). (ii) Mixed inoculations resulted in depression of both viruses compared with a single inoculation; PBCV-1 DNA fragments were reduced by ∼55 % and NY-2A DNA fragments were reduced by ∼90 % (compare open bars with corresponding solid bars in Fig. 2a). This result suggests that the second virus depresses the replicating virus.
Fig. 2.
PBCV-1 out-competes NY-2A in double inoculation experiments. (a) Mean virus-specific DNA fragment concentration in Chlorella NC64A cells inoculated with either a single virus (solid bars) or both viruses simultaneously (open bars). (b) Amount of PBCV-1- (○) over NY-2A- (•) specific DNA fragments as a function of the time difference (Δt) between primary inoculation with virus PBCV-1 and subsequent inoculation with virus NY-2A. (c) Amount of NY-2A- (•) over PBCV-1- (○) specific DNA fragments as a function of Δt between primary inoculation with virus NY-2A and subsequent inoculation with virus PBCV-1. (d) Mean virus-specific DNA fragment concentration in Chlorella NC64A cells inoculated simultaneously with PBCV-1 and NY-2A in the absence (solid bars) and presence (open bars) of 20 mM Cs+. (e) Ratio of the concentration of PBCV-1- over NY-2A-specific DNA fragments obtained in double inoculation experiments in which NC64A cells were inoculated simultaneously or sequentially with PBCV-1 and virus NY-2A in the absence (open bars) and presence (solid bars) of 20 mM Cs+. PBCV-1+NY-2A, cells were inoculated simultaneously; PBCV-1>NY-2A, cells were inoculated first with PBCV-1 and with NY-2A 20 min later; PBCV-1<NY-2A, cells were inoculated first with NY-2A and with PBCV-1 20 min later. Data in (a), (d) and (e) are means±sem of ≥3 independent experiments. In some cases the sem is smaller than the symbols. The data in (b) and (c) are ratios from ≥3 independent experiments.
The first few minutes determine the outcome of competition between viruses
Previous results indicate that the initial infection phase of virus PBCV-1 is completed within the first few minutes p.i. (e.g. Neupärtl et al., 2008; Frohns et al., 2006). Therefore, we examined the possibility that this time-frame is critical for competition between the two viruses. To test this possibility, we inoculated Chlorella NC64A cells with PBCV-1 and then added NY-2A at intervals between 0 and 90 min. The fitness of the two viruses to infect the algae was monitored by replication of virus DNA fragments (Fig. 2b). As expected, PBCV-1 dominated when both viruses were added simultaneously. This dominance increased with the delay with which NY-2A followed PBCV-1. Likewise, in the reverse experiment, adding PBCV-1 after NY-2A resulted in NY-2A dominating (Fig. 2c). In both situations, exclusion began within 5 min of addition of the first virus.
Cs+ in the medium favours virus PBCV-1
The differential sensitivity of PBCV-1 and NY-2A to Cs+ (Frohns et al., 2006) raises the question of whether an NY-2A-specific Cs+-sensitive mechanism is involved in the competition between the viruses. Chlorella NC64A cells were simultaneously inoculated with both viruses in the presence of 20 mM Cs+; this concentration is sufficient to obtain maximal Cs+ inhibition (Fig. 1b; Frohns et al., 2006). The results of these experiments led to two conclusions (Fig. 2d). (i) The synthesis of NY-2A-specific DNA was reduced further, indicating that almost no NY-2A infected the host under these conditions; the majority of the cells were infected by PBCV-1. (ii) Cs+ had no inhibitory effect on PBCV-1 replication when co-inoculated with NY-2A; in fact, PBCV-1 DNA increased by approximately 77 %. This result suggests that the depression of PBCV-1 replication, which was observed in mixed inoculations, is released; i.e. the negative effect of NY-2A on PBCV-1 replication is abolished.
To further test whether a Cs+-sensitive mechanism is responsible for this competition, we studied the effect of sequential inoculation of both viruses in the presence and absence of Cs+. In this set of experiments, one virus was added to Chlorella NC64A cells 20 min after the other; this double inoculation was carried out in the presence or absence of 20 mM Cs+. To compare the fitness of one virus over the other, the data are expressed as a ratio of PBCV-1 DNA : NY-2A DNA (Fig. 2e). When the host was inoculated with PBCV-1 first, the ratio was high, meaning that PBCV-1 out-competes NY-2A; this ratio was independent of Cs+. The results differed when cells were inoculated with NY-2A first. In this case, the ratio was low, meaning that NY-2A out-competes PBCV-1 in the absence of Cs+. In the presence of Cs+, NY-2A no longer dominates replication. PBCV-1 achieves maximal replication even when added 20 min later than NY-2A.
Virus particles in Chlorella cells
The aforementioned data are consistent with the view that Chlorella NC64A cells are only infected by one virus and that only this virus replicates in the host cells. Two additional experiments support this hypothesis. First, cells were inoculated with both viruses, each at an m.o.i. of 10. After 1 h, Chlorella NC64A cell walls (Meints et al., 1988) were added to remove unattached virus particles and the cells were then plaqued as infective centres (Van Etten et al., 1983b). Fifty plaques of various sizes were randomly picked and assayed by standard PCR for virus DNA. All 50 plaques contained only one of the two viruses.
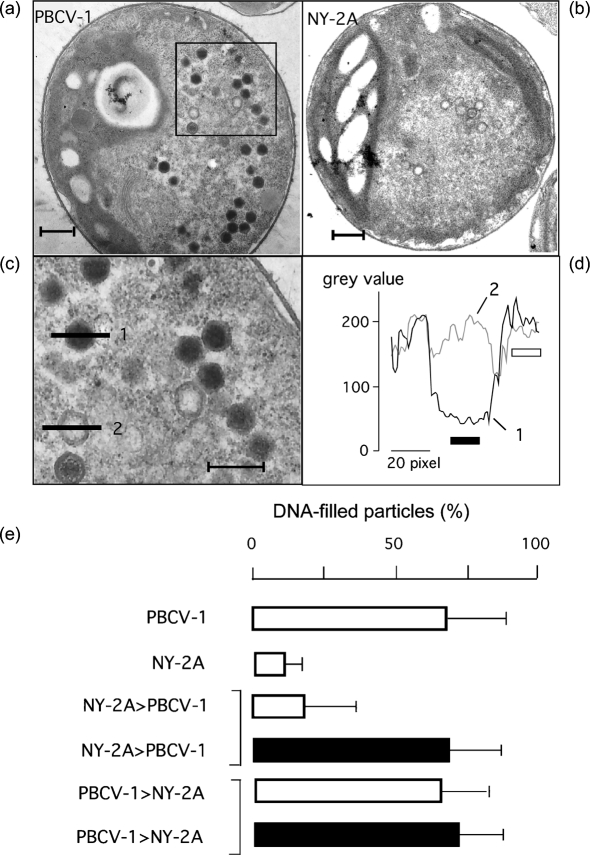
Second, we employed quantitative electron microscopy to determine which virus replicates in host cells following mixed inoculation. Identification of viral progeny in the host cells is possible because NY-2A morphogenesis is slower than PBCV-1 (Van Etten et al., 1988). Fig. 3 shows typical electron micrographs of Chlorella NC64A cells 5 h after infection with either PBCV-1 or NY-2A (Fig. 3a, b, respectively). The majority of the nascent PBCV-1 particles were electron-dense, i.e. they were filled with DNA. At the same time, NY-2A particles lacked DNA, i.e. they were largely empty. An analysis of 50 micrographs from cells infected with virus PBCV-1 revealed that, on average, 69±20 % of viral PBCV-1 particles were filled with DNA at 5 h p.i. At the same time, only 11±7 % of the nascent NY-2A particles were filled with DNA. This clear difference in the kinetics of virus morphogenesis allowed the identification of the progeny when Chlorella cells were inoculated with both viruses.
Fig. 3.
Chlorella NC64A cells infected with PBCV-1 (a, c) and NY-2A (b) can be distinguished by electron microscopy. (a–c) At 5 h p.i., the progeny of PBCV-1 are mostly electron-dense because of completed DNA packing (a, c). At the same time point, the progeny of NY-2A are still devoid of DNA (b). Magnification of the boxed area in (a) is shown in (c). The degree of DNA packing can be estimated from the analysis of grey values of the electron microscopic images. An intensity profile of a DNA-filled particle (1) in (c) reveals a much lower grey value in the centre of the particle than in the overall background of the cell (d); however, the same value in the centre of an empty particle (2) is similar to the background (d). For an analysis of DNA packing, we obtained the intensity profiles for each treatment (as in d) from more than 30 randomly selected viral particles in more than 50 cells. We considered a particle electron-dense when the mean value measured in the centre of the particle (indicated by solid bar) was more than two times lower than the mean of the background (indicated by open bar). Bars, 500 nm in (a) and (b), 300 nm in (c). (e) Percentage of DNA-filled particles estimated as in (a–d) from a total of 2000 particles from 49 cell sections per treatment. This analysis shows that the percentage of DNA-packed particles was high in cells infected only with virus PBCV-1 and low in cells inoculated with only NY-2A. In dual inoculation experiments, Chlorella cells were first inoculated with one virus and then with the other virus 20 min later in the presence (solid bars) or absence (open bars) of 20 mM Cs+. The percentage was low, i.e. dominated by NY-2A, when virus NY-2A was added in the absence of Cs+ 20 min before virus PBCV-1. In the two other experiments when cells were inoculated first with either PBCV-1 or NY-2A in the presence of Cs+, the percentage was high, indicating that PBCV-1 dominates.
The percentage of DNA packaged at 5 h p.i. was low (20±17 %) when NY-2A was added 20 min before PBCV-1 (Fig. 3e). This implies that replication was dominated by NY-2A, i.e. the virus added first. Likewise, when PBCV-1 was added first, it dominated (68±16 %) (Fig. 3e). These results confirm the data reported in Fig. 2(e).
The experiment was repeated in the presence and absence of Cs+. The majority of the particles were filled with DNA when PBCV-1 was added prior to NY-2A in the presence of Cs+ (Fig. 3e). The same percentage of DNA-packed particles was observed when NY-2A was added 20 min before PBCV-1 in the presence of Cs+ (Fig. 3e). Thus, NY-2A alone did not prevent subsequent infection by PBCV-1.
Collectively, these experiments establish that the two viruses mutually exclude one another. The mechanism of exclusion is associated with some early steps during NY-2A infection that are sensitive to Cs+. The presence of a competing virus alone is not sufficient to produce exclusion.
Ideally, we would like to disrupt the Kcv gene (kcv) in one or both of the viruses and determine whether cells infected with these viruses lose the ability to exclude one another. However, we do not currently have the molecular methods needed to manipulate the viral genomes.
Virus-induced host membrane depolarization is the primary mechanism for exclusion
Previous experiments established that both NY-2A and PBCV-1 cause a similar depolarization of the host cell membrane within the first few minutes of infection (Mehmel et al., 2003; Frohns et al., 2006). While PBCV-1-induced depolarization was only slightly inhibited by Cs+, the depolarization caused by NY-2A was severely reduced by Cs+ (Frohns et al., 2006). To examine the role of depolarization in exclusion, we monitored the change in host membrane voltage in response to inoculation with the two viruses.
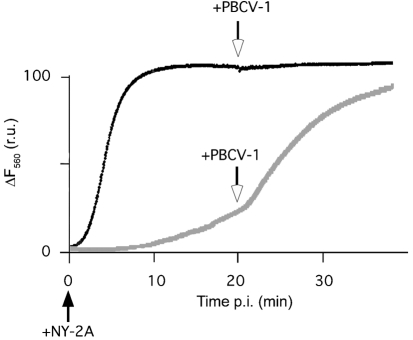
NY-2A inoculation of Chlorella NC64A resulted in a rapid rise in bisoxonol fluorescence, indicating that the membrane voltage of the cells depolarized. Subsequent addition of PBCV-1 at 20 min had no additional impact on fluorescence (Fig. 4). The scenario was different in the presence of Cs+. NY-2A only had a small effect on the host membrane voltage (Frohns et al., 2006); however, PBCV-1 addition elicited a strong membrane depolarization even when added 20 min after NY-2A (Fig. 4). The final steady state of the fluorescence was similar to that induced by NY-2A in the absence of Cs+. Similar results were obtained in nine independent experiments.
Fig. 4.
Representative recordings of membrane voltage in Chlorella NC64A cells during sequential inoculations with PBCV-1 and NY-2A in the presence (grey line) or absence (black line) of Cs+. When a suspension of 7×106 Chlorella NC64A cells was inoculated with virus NY-2A, the fluorescence increased due to membrane depolarization of the host (black line). Addition of virus PBCV-1 (arrow) 20 min after the primary infection had no further effect on fluorescence. When cells were inoculated under the same conditions with Cs+ in the medium, NY-2A inoculation only produced a small increase in fluorescence (grey line). However, when the Cs+-insensitive virus PBCV-1 was added 20 min later, fluorescence increased rapidly. r.u., Relative units.
Collectively, these experiments indicate that NY-2A was unable to exclude PBCV-1 in the presence of Cs+ because Cs+ prevents NY-2A from depolarizing the host membrane. Therefore, we believe that host membrane depolarization plays an essential role early in chlorella virus infection and that it is required for mutual exclusion.
Conclusions
The present data are consistent with the following model: all chlorella viruses induce depolarization of the host cell membrane during infection (Frohns et al., 2006). This depolarization results in an immediate loss of K+ from the host, a process which energetically favours ejection of viral DNA into the host (Neupärtl et al., 2008). The depolarization event also prevents infection of the host by another virus.
Virus-induced depolarization of the host membrane is probably generated by the viral-encoded K+ channel, Kcv (Frohns et al., 2006; Plugge et al., 2000). In this sense, Kcvs have a functional analogy with the RexB protein from phage λ (Snyder & McWilliams, 1989); RexB protein induces a rapid depolarization of the host cell membrane that is an effective mechanism for achieving λ exclusion. Since the chlorella virus Kcvs are apparently essential for virus-triggered membrane depolarization, their genes are probably under high selective pressure (Kang et al., 2004b). The fitness of the virus is probably enhanced by any mutation which modifies the channel in such a way that it increases membrane depolarization and/or decreases sensitivity to potential channel blockers that might exist in the environment.
Acknowledgments
This investigation was supported in part by Public Health Service grant GM32441 (J. L. V. E.), NIH grant P20RR15635 from the COBRE program of the National Center for Research Resources (J. L. V. E.), the Deutsche Forschungsgemeinschaft (G. T.) and the European Drug Initiative on Channels and Transporters (EDICT) project EU FP7 (201924) to A. M. We are grateful to Dr D. Kramer (Darmstadt) for help with the electron microscopy experiments.
References
- Chan, S. H., Zhu, Z., Van Etten, J. L. & Xu, S. Y. (2004). Cloning of CviPII nicking and modification system from chlorella virus NYs-1 and application of Nt.CviPII in random DNA amplification. Nucleic Acids Res 32, 6187–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase, T. E., Nelson, J. A., Burbank, D. E. & Van Etten, J. L. (1989). Mutual exclusion occurs in a chlorella-like green alga inoculated with two viruses. J Gen Virol 70, 1829–1836. [DOI] [PubMed] [Google Scholar]
- Cherrier, M. V., Kostyuchenko, V. A., Xiao, C., Bowman, V. D., Battisti, A. J., Yan, X., Chipman, P. R., Baker, T. S., Van Etten, J. L. & Rossmann, M. G. (2009). An icosahedral algal virus has a complex unique vertex decorated by a spike. Proc Natl Acad Sci U S A in press [DOI] [PMC free article] [PubMed]
- Delbrück, M. (1945). Interference between bacterial viruses. III. The mutual exclusion effect and the depressor effect. J Bacteriol 50, 151–170. [DOI] [PubMed] [Google Scholar]
- Dulbecco, R. (1952). Mutual exclusion between related phages. J Bacteriol 63, 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohns, F., Käsmann, A., Kramer, D., Schäfer, B., Mehmel, M., Kang, M., Van Etten, J. L., Gazzarrini, S., Moroni, A. & Thiel, G. (2006). Potassium ion channels of chlorella viruses cause rapid depolarization of host cell during infection. J Virol 80, 2437–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini, S., Kang, M., Van Etten, J. L., Tayefeh, S., Kast, S. M., DiFrancesco, D., Thiel, G. & Moroni, A. (2004). Long-distance interactions within the potassium channel pore are revealed by molecular diversity of viral proteins. J Biol Chem 279, 28443–28449. [DOI] [PubMed] [Google Scholar]
- Hirsch-Kauffmann, M., Pfenning-Yeh, M., Ponta, H., Herrlich, P. & Schweiger, M. (1976). A virus-specified mechanism for the prevention of multiple infection – T7- and T3-mutual and superinfection exclusion. Mol Gen Genet 149, 243–249. [DOI] [PubMed] [Google Scholar]
- Jansohn, M. (2007). Gentechnische Methoden, 4. Auflage. München: Elsevier.
- Kang, M., Moroni, A., Gazzarrini, S., DiFrancesco, D., Thiel, G., Severino, M. & Van Etten, J. L. (2004a). Small potassium ion channel protein encoded by chlorella viruses. Proc Natl Acad Sci U S A 101, 5318–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, M., Graves, M., Mehmel, M., Moroni, A., Gazzarrini, S., Thiel, G., Gurnon, J. & Van Etten, J. L. (2004b). Genetic diversity in chlorella viruses flanking kcv, a gene that encodes a potassium ion channel protein. Virology 326, 150–159. [DOI] [PubMed] [Google Scholar]
- Li, B. H. & Bockrath, R. (1993). Photolyase-dimer-DNA complexes and exclusion stimulation in Escherichia coli: depolarization of the plasma membrane. Mol Gen Genet 240, 450–454. [DOI] [PubMed] [Google Scholar]
- Mehmel, M., Rothermel, M., Meckel, T., Van Etten, J. L., Moroni, A. & Thiel, G. (2003). Possible function for virus encoded K+ channel Kcv in the replication of chlorella virus PBCV-1. FEBS Lett 552, 7–11. [DOI] [PubMed] [Google Scholar]
- Meints, R. H., Lee, K., Burbank, D. E. & Van Etten, J. L. (1984). Infection of a chlorella-like alga with the virus, PBCV-1: ultrastructure studies. Virology 138, 341–346. [DOI] [PubMed] [Google Scholar]
- Meints, R. H., Burbank, D. E., Van Etten, J. L. & Lamport, D. T. A. (1988). Properties of the chlorella receptor for the virus PBCV-1. Virology 164, 15–21. [DOI] [PubMed] [Google Scholar]
- Neupärtl, M., Meyer, C., Woll, I., Frohns, F., Kang, M., Van Etten, J. L., Kramer, D., Hertel, B., Moroni, A. & Thiel, G. (2008). Chlorella viruses evoke a rapid release of K+ from host cells during early phase of infection. Virology 372, 340–348. [DOI] [PubMed] [Google Scholar]
- Onimatsu, H., Suganuma, K., Uenoyama, S. & Yamada, T. (2006). C-terminal repetitive motifs in Vp130 present at the unique vertex of the Chlorovirus capsid are essential for binding to the host Chlorella cell wall. Virology 353, 433–442. [DOI] [PubMed] [Google Scholar]
- Plugge, B., Gazzarini, S., Nelson, M., Cerana, R., Van Etten, J. L., Derst, C., DiFrancesco, D., Moroni, A. & Thiel, G. (2000). A potassium ion channel protein encoded by chlorella virus PBCV-1. Science 287, 1641–1644. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989). Molecular Cloning – a Laboratory Manual, 2nd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
- Schuster, A. M., Burbank, D. E., Meister, B., Skrdla, M. P., Meints, R. H., Hattman, S., Swinton, D. & Van Etten, J. L. (1986). Characterization of viruses infecting a eukaryotic chlorella-like green alga. Virology 150, 170–177. [DOI] [PubMed] [Google Scholar]
- Snyder, L. (1995). Phage-exclusion enzymes: a bonanza of biochemical and cell biology reagents? Mol Microbiol 15, 415–420. [DOI] [PubMed] [Google Scholar]
- Snyder, L. & McWilliams, K. (1989). The rex genes of bacteriophage lambda can inhibit cell function without phage superinfection. Gene 81, 17–24. [DOI] [PubMed] [Google Scholar]
- Van Etten, J. L., Meints, R. H., Burbank, D. E., Kuczmarski, D., Cuppels, D. A. & Lane, L. C. (1981). Isolation and characterization of a virus from the intracellular green alga symbiotic with Hydra viridis. Virology 113, 704–711. [DOI] [PubMed] [Google Scholar]
- Van Etten, J. L., Burbank, D. E., Xia, Y. & Meints, R. H. (1983a). Growth cycle of a virus, PBCV-1, that infects chlorella-like algae. Virology 126, 117–125. [DOI] [PubMed] [Google Scholar]
- Van Etten, J. L., Burbank, D. E., Kuczmarski, D. & Meints, R. H. (1983b). Virus infection of culturable chlorella-like algae and development of a plaque assay. Science 219, 994–996. [DOI] [PubMed] [Google Scholar]
- Van Etten, J. L., Schuster, A. M. & Meints, R. H. (1988). Viruses of eukaryotic chlorella-like algae. In Viruses of Fungi and Simple Eukaryotes, pp. 411–428. Edited by Y. Koltin & M. J. Leibowitz. Boca Raton, FL: CRC Press.
- Xia, Y., Burbank, D. E. & Van Etten, J. L. (1986). Restriction endonuclease activity induced by NC-1A virus infection of a chlorella-like green alga. Nucleic Acids Res 14, 6017–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Y., Burbank, D. E., Uher, L., Rabussay, D. & Van Etten, J. L. (1987). IL-3A virus infection of a chlorella-like green alga induces a DNA restriction endonuclease with a novel sequence specificity. Nucleic Acids Res 15, 6075–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Y., Morgan, R., Schildkraut, I. & Van Etten, J. L. (1988). A site-specific single strand endonuclease activity induced by NYs-1 virus infection of a chlorella-like green alga. Nucleic Acids Res 16, 9477–9487. [DOI] [PMC free article] [PubMed] [Google Scholar]