Abstract
MicroRNAs (miRNAs) are noncoding RNAs that regulate numerous target genes through a posttranscriptional mechanism and thus control major developmental pathways. The phylogenetically conserved let-7 miRNA regulates cell proliferation and differentiation, thus functioning as a key regulator of developmental timing in C. elegans and a tumor suppressor gene in humans. Using a reverse genetic screen, we have identified genetic interaction partners of C. elegans let-7, including known and novel potential target genes. Initial identification of several translation initiation factors as suppressors of a let-7 mutation led us to systematically examine genetic interaction between let-7 and the translational machinery, which we found to be widespread. In the presence of wild-type let-7, depletion of the translation initiation factor eIF3 resulted in precocious cell differentiation, suggesting that developmental timing is translationally regulated, possibly by let-7. As overexpression of eIF3 in humans promotes translation of mRNAs that are also targets of let-7-mediated repression, we suggest that eIF3 may directly or indirectly oppose let-7 activity. This might provide an explanation for the opposite functions of let-7 and eIF3 in regulating tumorigenesis.
Keywords: miRNA, let-7, translation factor, heterochronic, C. elegans, RNAi, eIF3, eIF6
Introduction
MicroRNAs (miRNAs) are small, untranslated RNAs involved in numerous developmental pathways (reviewed in ref. 1). They function through an antisense mechanism where binding of an miRNA to complementary sequences in its target mRNAs (‘cognate mRNAs’) causes cognate mRNA repression, but the mechanisms of target mRNA repression are less clear. Many different, and some-times contradictory, miRNA modes of action have been proposed (reviewed in refs. 2 and 3). These include inhibition of target mRNA translation either at the initiation or elongation step, target mRNA degradation in a non-endonucleolytic fashion, which may or may not result from deadenylation, and co-translational protein degradation. MicroRNAs may thus act through multiple mechanisms. These mechanisms may either function redundantly or as alternate pathways that affect only individual subsets of miRNAs and/or cognate mRNAs.2,3
The C. elegans let-7 miRNA was originally identified as a component of the heterochronic pathway,4 which controls the temporal fate of cells during postembryonic development (reviewed in ref. 5). Postembryonic development proceeds through the four larval stages, L1 through L4, followed by the sexually mature, adult stage. During normal development, cells adopt fates that are characteristic of the developmental stage of the animal, e.g., certain cells divide while others may exit the cell cycle and differentiate. Mutations in heterochronic genes may cause cells to prematurely adopt fates that are normally observed at a later developmental stage, i.e., cause precocious phenotypes. Alternatively, the mutant cells may display retarded phenotypes, i.e., characteristics typical of cells in earlier developmental stages. Partial loss of let-7 activity causes retarded phenotypes, i.e., repetition of fourth larval stage (L4) cell fates, while more complete loss of activity causes animals to die by bursting through the vulva at the larval-to-adult transition.4 These phenotypes are due to overexpression of let-7 target genes and can be partially suppressed by knock-down of individual let-7 target genes.6-10
let-7 is conserved in higher eukaryotes, with a striking 100% sequence identity in the case of the mature let-7 of C. elegans and humans.11,12 This, and the observation that let-7 expression is temporally regulated in invertebrates as well as vertebrates,11 suggests that let-7 function may also be conserved. This view is supported by our recent finding that C. elegans let-7 regulates let-60/ras expression, while human let-7 regulates the let-60 orthologue RAS.7,13 Human let-7 also regulates the chromatin-binding factor HMGA2, and failure of let-7-mediated HMGA2 repression promotes oncogenic transformation.14-18 Consistent with overproliferation of cells with reduced let-7 expression, let-7 also represses the expression of the cell cycle regulator CDC25A (refs. 19 and 20). Reduced let-7 expression in lung cancer13,21 may contribute to tumorigenic transformation through upregulation of these oncogenes,22,23 and reduced let-7 expression levels are prognostic for poor patient survival.21,24 let-7 has also been shown to function as a tumor suppressor in breast cancer, where it controls proliferation and differentiation of tumor initiating cells.25 The converging results from these different experimental systems have supported a model of let-7 functioning as an important regulator of stem cell fates in both normal and tumor cells.26 To achieve this function, let-7 expression is highly regulated not only at the transcriptional level, but, as recent data suggest, also posttranscriptionally (reviewed in ref. 26).
To identify interaction partners of the let-7 miRNA, which might include novel let-7 targets, regulators of let-7 expression, mediators of let-7 activity, or heterochronic genes, we devised a high-throughput, functional genomics screen based on RNA interference (RNAi). Through this screen, we identified 41 known and novel interaction partners of let-7. As several genes directly or indirectly involved in translation were found among the novel let-7 suppressors, we systematically examined genetic interactions between let-7 and the core translational machinery and found them to be widespread. Consistent with translational control of the heterochronic pathway, we found that depletion of several of these genes, in particular subunits of the tumor promoting translation initation factor eIF3, caused abnormal timing of cell differentiation in the presence of wild-type let-7.
Results
A reverse genetics screen reveals translation factors as suppressors of let-7
The temperature sensitive let-7(n2853) allele contains a point mutation in the mature let-7 sequence that impairs target binding.4,27 In addition, reduced accumulation of the mutant let-7 RNA28 further impairs target repression and as a result, mutant animals die by bursting through the vulva at the larval-to-adult transition when grown at or above 20°C (reviewed in ref. 4). RNAi-mediated knockdown of individual let-7 targets can partially suppress this lethality.6-10 To identify novel interaction partners of the let-7 miRNA, we carried out a feeding RNAi screen to uncover additional suppressors of the let-7(n2853) bursting phenotype. We performed this screen by RNAi on synchronized L1 stage larvae to avoid missing factors whose efficient depletion would cause sterility and/or embryonic lethality. Using a previously described feeding library of bacteria producing double-strand RNA,29 we individually tested the suppressing effect of RNAi on almost 90% of the genes on chromosome I, i.e., ca. 2,400 genes (Fig. 1). We found that 41 genes could efficiently suppress the conditional lethality of the let-7 mutation when knocked down through RNAi by feeding (Table 1). Some but not all of the suppressor genes contained let-7 complementary sites, as defined previously,7 in their 3′ untranslated regions (UTR) suggesting that these genes may be targets of the let-7 RNA (Table 1).
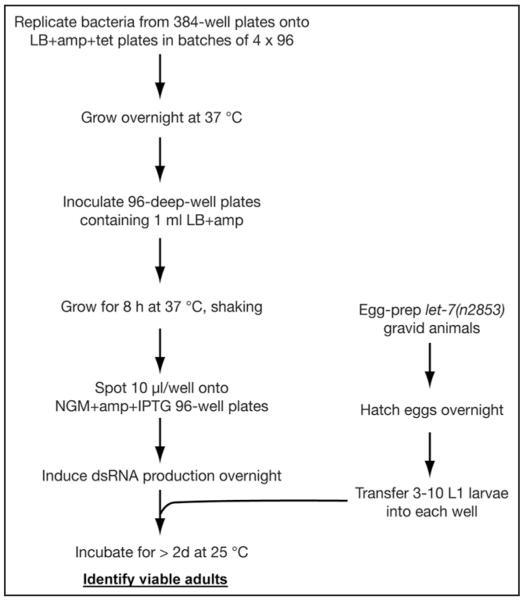
Figure 1.
A high-throughput reverse genetics screen to identify suppressors of let-7(n2853) lethality. See main text and Materials and Methods for details.
Table 1.
Suppressors of let-7(n2853) lethality identified in a screen
| ORFa | Locusb | Function/Homologiesb,c | LCSd | Suppressione |
|---|---|---|---|---|
| DNA synthesis | ||||
| W02D9.1 | pri-2 | DNA primase | − | +++ |
| Y54E10A.15 | cdt-1 | Hs CDT1, Dm dup | − | +++ |
| RNA metabolism | ||||
| B0511.6 | DEAD box helicase, Dm pit | − | ++ | |
| C17E4.5 | pabp-2 | Poly(A)-binding protein, Hs PABN1 | − | ++ |
| C36B1.3 | rbp-3 | RNA polymerase II subunit | − | ++ |
| C53H9.2 | Nucleolar GTPase, Sc Lsg1p | 1 | + | |
| F14B4.3 | RNA Polymerase I, second largest subunit | 1 | ++ | |
| T19A6.2 | ngp-1 | Nucleolar GTPase, Sc Nog2p | − | +++ |
| W01B11.3 | nol-5 | snoRNP associated, Sc Nop5p | 1 | ++ |
| W04A4.5 | Hs Integrator isoform 1 (U1, U2 RNA processing) | − | +++ | |
| Y48G1A.4 | snoRNP associated, Sc Nop14p | nd | ++ | |
| Y54E10BR.6 | rbp-7 | RNA polymerase II subunit | − | ++ |
| Y71F9B.4 | snr-7 | Hs SNRPG (spliceosome subunit) | − | ++ |
| Y106G6H.2 | pab-1 | Poly(A)-binding protein, Sc Pab1p | − | ++ |
| Protein metabolism | ||||
| B0511.10 | eif-3.E | Translational initiation factor eIF3 subunit | − | ++ |
| C03D6.8 | rpl-24.2 | Ribosomal protein L24-family, Sc Rlp24p | − | ++ |
| C12C8.3 | lin-41 | NHL domains | 2 | +++ |
| C41D11.2 | eif-3.H | Translational initiation factor eIF3 subunit | − | ++ |
| C47B2.5 | eif-6 | Translational initiation factor eIF6 | − | ++ |
| Y47G6A.10 | spg-7 | AAA-ATPase, protease | − | ++ |
| F11A3.2f | Translational initiation factor, eIF2Bδ | − | +++ | |
| Y65B4BR.5 | Nascent polypeptide associated complex α-chain | − | ++ | |
| Energy/metabolism | ||||
| T09B4.9 | Mitochondrial inner membrane translocase, Hs TIM44 | − | ++ | |
| W09C5.8 | Subunit IV of cytochrome c oxidase | − | + | |
| Chromosome dynamics | ||||
| C45G3.1 | aspm-1 | Dm Asp | − | + |
| T03F1.9 | hcp-4 | CENP-C homologue | − | ++ |
| Cell structure | ||||
| F56A3.3 | npp-6 | Nuclear pore protein | − | +++ |
| T19B4.2 | npp-7 | Nuclear pore protein | 2 | +++ |
| T21E12.4 | dhc-1 | Dynein heavy chain | − | ++ |
| Y48G1A.5 | imb-5 | Sc Cse1p, Hs CAS/CSE1 | 1 | ++ |
| Y71F9AM.5 | nxt-1 | NTF2-family | − | + |
| Y105E8A.9 | apg-1 | γ-adaptin AP-1 | 1 | ++ |
| H15N14.2 | nsf-1 | Vesicle fusion | − | + |
| Specific transcription | ||||
| C01G8.9 | lss-4 | Dm osa/eld | 3 | +++ |
| F57B10.1 | CREB/ATF family transcription factor | − | ++ | |
| Signalling | ||||
| K12C11.2 | smo-1 | Hs SUMO-1 | − | + |
| ZC581.1 | nekl-2 | NEK kinase family | − | ++ |
| Unknown | ||||
| F20G4.1 | smgl-1 | Hs Neuroblastoma amplified gene protein | − | ++ |
| F56A3.4 | spd-5 | Coiled coils | − | ++ |
| T23D8.3 | Hs LTV1; in operon with eif-3C | − | ++ | |
| Y63D3A.5 | tfg-1 | Hs TFG1 (TrkA-fused gene) | − | ++ |
Some ORFs were targeted by more than one dsRNA construct and/or construct names might differ from those of the target ORF indicated here.
Gene loci names according to Wormbase, Release 188. Gene names were not considered when assigning functional classes in cases where no published information or sequence homologies were available to support the gene designation.
Sc: Saccharomyces cerevisiae, Dm: Drosophila melanogaster, Hs: Homo sapiens. In most cases functions are predicted from homologies.
LCS: let-7 complementary site as identified in7. nd: 3′ UTR was not included in dataset used for LCS prediction.
+, >30%, ++, >50%, +++, >80% survival.
Sequencing revealed plasmid contained other than predicted insert, targeting the indicated ORF (chromosome V).
Our screening procedure was validated by two observations. First, we blindly identified lin-41, the only known heterochronic gene in the chromosome I library, as a potent suppressor of let-7(n2853) when depleted. lin-41 is a known downstream target of the let-7 miRNA whose depletion had previously been shown to suppress let-7(n2853) (refs. 4 and 10). No RNAi construct targeting lin-28, another heterochronic gene and known suppressor of let-7 encoded on chromosome I, was included in the RNAi library.29 A second gene, lss-4 was identified independently by us through a computational approach and also subsequently validated as a let-7 target.7 Second, seven genes in the library are targeted by two independent RNAi constructs,29 and we identified both clones for four of these genes, rpl-24.2, Y65B4BR.5, imb-5/xpo-2 and spg-7. In the remaining three cases differences in the RNAi phenotypes elicited by each pair of constructs were already previously noted.29
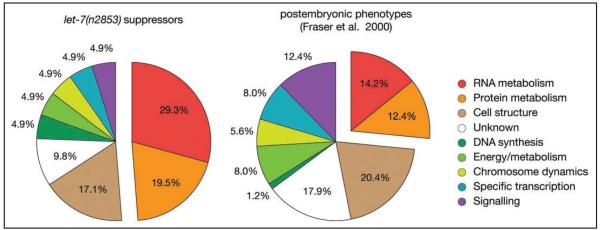
The largest class of suppressors identified in our screen is comprised of genes with a predicted function in the metabolism of RNA or protein, which account for nearly half (20/41) of the suppressors (Fig. 2). Genes from this category showed a 50% increase over the frequency found by Fraser et al.,29 who queried the library for genes eliciting any phenotype when depleted in wild-type animals (Fig. 2). This observation may suggest that genes of this functional class are particularly important as targets and/or mediators of let-7 function or are heterochronic genes. Given the tight posttranscriptional regulation of let-7 expression,26 it will also be of considerable interest to test in future work whether any of these novel let-7 interactors control let-7 maturation.
Figure 2.
let-7 suppressors are enriched for factors involved in RNA and protein metabolism. Genes involved in RNA and protein metabolism are enriched among the suppressors of the let-7(n2853) mutation. Indicated are the distributions across functional classes of genes causing suppression of the let-7(n2853)-associated lethality (left chart) or visible phenotypes in otherwise wild-type animals (right chart; assembled from data in ref. 29). Assignments to functional classes are from Wormbase Release WS188. Where gene assignments had changed from those used by Fraser et al., their data were adjusted accordingly. Note that both studies used an identical RNAi library.
Our list of suppressors contained several translation initiation factors: two putative subunits of the eukaryotic translation initiation factor (eIF) 3, eif-3.H and eif-3.E (C41D11.2 and B0511.10, respectively) and the delta subunit of eIF2B (F11A3.2). We also identified eif-6 (C47B2.5) as a let-7 suppressor, an unexpected result given previous data showing that depletion of eIF6 abrogated miRNA target repression.30 However, recent data from Drosophila S2 cells suggest that eIF6 may not be widely required for miRNA activity.31,32
As our initial screen did not cover the whole genome, we tested additional translation factors for a genetic interaction with let-7. Many translation factors have identifiable homologues in C. elegans33 and we performed a systematic RNAi screen of these factors for suppression of the let-7(n2853) phenotype (Table 2). In most cases, knock-down of translation factors induced a slow growth or developmental arrest phenotype. We could frequently avoid developmental arrest by mixing bacteria that carried the dsRNA producing plasmid with those carrying a plasmid without insert. We found that additional translation factors could suppress the let-7 mutation, in fact, most of the translation factors tested, including initiation, elongation, as well as termination factors, showed partial suppression. Many, but not all suppressors caused slow growth (Table 2), ruling out for at least a subset of translation factors that delayed development is the cause for let-7 suppression. Moreover, an approximately wild-type rate of development also suggests that at least this subset of suppressors affects general translation only weakly. As a corollary, let-7 function and/or the heterochronic pathway appear to be highly sensitive to altered translation levels.
Table 2.
Genetic interactions between let-7 and the translation machinery
| Translation factor | locus | ORF | let-7(n2853) suppression |
|---|---|---|---|
| Initiation factors | |||
| eIF1 | T27F7.3b | − | |
| eIF1Aa | H06H21.3 | ++ | |
| eIF2βb | iftb-1 | K04G2.1 | ++ |
| eIF2γd | Y39G10AR.8 | × | |
| eIF2A | E04D5.1 (a, b) | − | |
| eIF2Bα | ZK1098.4 | − | |
| eIF2Bβ | Y47H9C.7 | ++ | |
| eIF2Bγ | ppp-1 | C15F1.4 | ++ |
| eIF2B∂ | F11A3.2 | +++ | |
| eIF2Bεd | D2085.3 | +++ | |
| eIF3ac,d | egl-45 | C27D11.1 | × |
| eIF3bc,d | eif-3.B | Y54E2A.11a | +++ |
| eIF3cc,d | eif-3.C | T23D8.4 | +++ |
| eIF3dd | eif-3.D | R08D7.3 | ++ |
| eIF3ed | eif-3.E | B0511.10 | +++ |
| eIF3fd | eif-3.F | D2013.7 | +++ |
| eIF3gd | eif-3.G | F22B5.2 | × |
| eIF3h | eif-3.H | C41D11.2 | ++ |
| eIF3ib | eif-3.I | Y74C10AR.1 | × |
| eIF3k | eif-3.K | T16G1.11 | − |
| eIF3ma | cif-1 | K08F11.3 | +++ |
| eIF4Ab,d | inf-1 | F57B9.6 | × |
| eIF4Aa,d | F57B9.3 | +++ | |
| eIF4E-1+5 | ife-1 | F53A2.6 | − |
| ife-5 | Y57A10A.30 (a, b) | ||
| eIF4E-2 | ife-2 | R04A9.4 | − |
| eIF4E-3 | ife-3 | B0348.6 (a, b, c) | − |
| eIF4E-4 | ife-4 | C05D9.5 | − |
| eIF4Gb,d | ifg-1 | M110.4 (a, b) | × |
| eIF4H | drr-2 | T12D8.2 | − |
| eIF5b | C37C3.2 (a, b, c) | ++ | |
| eIF5Ad | iff-2 | F54C9.1 | +++ |
| eIF5B | iffb-1 | Y54F10BM.2 | +++ |
| eIF6 | eif-6 | C47B2.5 | ++ |
| Elongation factors | |||
| eEF1Ab,d,e | eft-3 | F31E3.5 | ++ |
| eft-4 | R03G5.1 (a, b, c, d) | ||
| eEF1Be | Y41E3.10 | ++ | |
| F54H12.6 | |||
| eEF2b,d | eft-2 | F25H5.4 | +++ |
| eEF2c,d | eft-1 | ZK328.2 | ++ |
| Release factors | |||
| eRF1c,d | T05H4.6 (a, b) | × | |
| eRF3b,d | H19N07.1 | ++ |
In some cases, RNAi titration was performed to overcome developmental block by mixing bacteria that carried the dsRNA producing plasmid with those carrying a plasmid without insert.
1:1 dilution
1:5 dilution
1:10 dilution
slow growth
multiple RNAi targets. Suppression: −,<20%, +, >20%, ++, >40%, +++, >80%, x, developmental block, n ≥ 60 worms for each. Control animals fed with bacteria carrying empty L4440 vector showed never more than 10% survival, whereas daf-12(RNAi), our positive control, showed never less than 90% survival.
Depletion of translation factors affects the heterochronic pathway
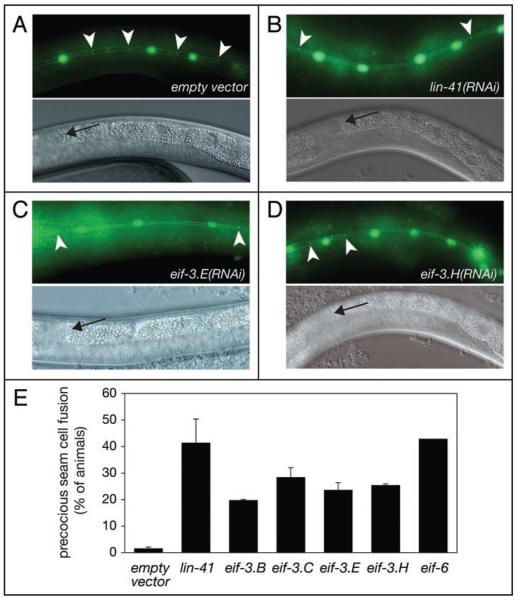
To ascertain that the suppression of the let-7 vulval bursting phenotype through impaired translation was specific, i.e., mediated through a modulation of the heterochronic pathway rather than an indirect consequence of, for instance, aberrant vulval development, we investigated whether depletion of individual translation factors caused heterochronic phenotypes in the seam cells. Seam cells are a subset of hypodermal cells that display a stem-cell-like division pattern during larval stages. At the larval to adult transition, seam cells exit the cell cycle and fuse to form a syncytium, which subsequently secretes collagenous structures termed alae. let-7 is expressed in the seam cells34 and in its absence, seam cells fail to terminally differentiate at this time and instead divide again.4 By contrast, overexpression of let-7 causes the opposite, precocious phenotype and seam cells fuse after the L3-to-L4 molt.4 We examined the effect of RNAi against eif-3.B, eif-3.C, eif-3.E, eif-3.H and eif-6 in animals carrying a wild-type allele of let-7. We selected these factors because they were amongst the first suppressors we had identified. While seam cell fusion is not observed in mock-treated animals at early L4 stage, significant numbers of eif-3(RNAi) and eif-6(RNAi) animals displayed precocious seam cell fusion, as did the lin-41(RNAi) positive control animals (Fig. 3). In the case of eif-3.B(RNAi) and eif-3.C(RNAi) we again had to dilute the RNAi-inducing bacteria with inert bacteria to avoid developmental arrest of the affected animals. These findings directly demonstrate that knock-down of this subset of the suppressing translation factors causes heterochronic defects.
Figure 3.
Reduced levels of eIF3-subunits cause precocious seam cell fusion. Synchronized N2; wIs79 L1-stage larvae were grown on bacteria producing the indicated dsRNA and examined for presence of precocious seam cell fusion upon reaching early-L4 stage. (A–D) Photomicrographs of animals grown on the indicated bacteria. Arrowheads point to AJM-1/GFP signal between seam cells observed in the absence of cell fusion. Arrows in lower panels indicate the distal tips of the gonads, visualized through Nomarski optics, which demonstrate the appropriate early L4 developmental stage. Anterior of each animal is left, ventral down. Note that GFP and Nomarski micrographs show different parts of the same animal. (E) Percentages of animals with precocious seam cell fusion were averaged from at least two independent experiments. To avoid developmental arrest in the case of eif-3.B and eif-3.C subunits, and gonadal migration defects in the case of the eif-3.E, animals were fed bacteria expressing the appropriate dsRNA, diluted appropriately (1:2 to 1:6) with bacteria producing mock dsRNA. n ≥ 82 for each. Error bars correspond to SEM.
To obtain further evidence for a role of translation factors in the heterochronic pathway we analyzed the genetic interaction between eif-3.E or eif-3.H and lin-41. lin-41 codes for a protein that prevents premature execution of adult fates by repressing production of the transcription factor LIN-29 until the L4 stage.10 LIN-41 protein levels themselves are regulated through the interaction of let-7 miRNA with the let-7 complementary sites in the 3′ UTR of lin-41 mRNA.4,10,27 lin-41 loss-of-function mutations lead to partially penetrant precocious phenotypes in the seam cells and we previously showed that the penetrance of this phenotype can be enhanced when a second let-7 interactor, the let-7 target daf-12, is also knocked down.7 Similarly, while only 53.1% of the lin-41(ma104) animals display precocious alae (±2.4% SEM), the penetrance of this phenotype was significantly (p < 0.05, student’s t-test) increased to 65.0% ± 1.2% for lin-41(ma104); eif-3.E(RNAi) animals and 80.1% ± 0.4% for lin-41(ma104); eif-3.H(RNAi) animals.
Analysis of seam cell fusion and alae formation thus indicate that the translation factors investigated modulate the heterochronic pathway. This shows that depletion of these factors does not simply superficially rescue the let-7(n2853) mutant phenotype but impacts on a pathway known to be regulated by let-7. These data would suggest that translation of one or several heterochronic genes, possibly let-7 target genes, is inefficient and therefore particularly susceptible to further decreases in translation activity. Interestingly, overexpression of eIF3 subunits has been linked to various cancers (reviewed in ref. 35), and our findings indicate that the opposing effects of eIF3 and let-7 on cell differentiation might be a contributing factor (see Discussion).
Discussion
We have identified several novel genetic interaction partners of the let-7 miRNA. Some of these contain predicted let-7 binding sites, and future work may establish them as bona fide let-7 targets. Here, we have focused on the observation that our screen identified several translation initiation factors whose depletion allowed survival of let-7(n2853) worms (Table 1). Systematic depletion of individual translation factors subsequently allowed us to identify suppressors at each step of translation:33 initiation, elongation and termination (Table 2). These suppressors include a subunit of eIF2, which is part of the eIF2/GTP/Met-tRNAiMet ternary complex and all but one subunit of the eIF2B factor, which catalyzes guanine nucleotide exchange on eIF2 bound to GDP. Among the factors required to recruit the 40S ribosomal subunit to the ternary complex to form the 43S pre-initiation complex (PIC), we observed that eIF1A and all but one of the eIF3 subunits that permitted larval growth results are suppressors. The eIF4F complex, which comprises the cap-binding eIF4E, the scaffolding eIF4G, and the ATP-dependent RNA helicase eIF4A, recruits the 43S PIC to mRNA via an interaction between eIF3 and eIF4G. Among these factors, none of the eIF4E homologues (ife-1 to ife-5) individually tested showed suppression, eIF4G and one homologue of eIF4A (F57B9.3) showed developmental arrest, whereas the eIF4A homologue encoded by the F57B9.6 ORF showed potent suppression. Additionally eIF5, eIF5A and eIF5B were also identified as suppressors. Finally, all translation elongation and termination factors tested showed either a developmental block or suppression of the bursting phenotype.
Based on this result, it seems that suppression of the let-7(n2853) bursting phenotype can be rescued by decreasing the activity of virtually any step of the translation initiation process as well as the elongation and termination steps. Indeed, some of the other suppressors found in our screen are predicted, by homology, to be part of the ribosome (rpl-24.2) or function in its biogenesis (the putative snoRNP proteins W01B11.3 and Y48G1A.4 and the putative nucleolar GTPases homologous to yeast Lsg1p and Nog2p) and we also found two poly(A)-tail binding proteins, pab-1 and pabp-2. However, not all translation factors could suppress let-7 lethality. This may be due to redundancy (e.g., ife-1 to ife-5), inefficient depletion by RNAi or a genuine lack of interaction between these two genes.
Unexpectedly, we also observed that depleting eIF6 rescues let-7(n2853) animals and causes precocious heterochronic phenotypes in the presence of wild-type let-7, although this factor was reported to be required for miRNA mediated repression.30 If eIF6 were similarly required for let-7 function, we would have expected to see the opposite phenotypes, i.e., enhancement of weak let-7 alleles and or retarded heterochronic phenotypes. Our data would thus argue against an involvement of eIF6 in let-7 function, consistent with earlier reports from D. melanogaster cells that eIF6 does not seem to be generally involved in promoting miRNA function.31,32
It is also surprising to see that eif-3.D, along with almost all other eIF3 subunits, is found as an efficient suppressor. This observation is in contrast with a recent report indicating that eif-3.D(RNAi) in an RNAi sensitized strain enhanced the weak let-7(mg279) loss-of-function allele, as determined by increased vulval bursting.36 However, we found that eif-3.D(RNAi) can induce vulval bursting even in wild-type animals where ca. 20% of animals die by bursting despite having functional let-7. It is possible that this bursting phenotype may dominate over a weak let-7 allele, particularly when RNAi is performed in an RNAi sensitized strain, as done in the earlier report.36
We are particularly intrigued to see that depletion of eIF3 subunits causes precocious seam cell differentiation in the presence of wild-type let-7. This is because several of the thirteen subunits of human eIF3 have altered expression levels in cancers including lung, breast, cervical, esophageal, prostate and testicular cancers, and this aberrant expression is likely to contribute to oncogenesis.35 For instance, INT6/eIF3e was originally identified as a common integration site of mouse mammary tumor virus,37 and expression of the truncated INT6/eIF3e gene product, but not of the wild-type eIF3e gene, is sufficient to transform a number of cell lines.38,39 Conversely, INT6/eIF3e loss-of-heterozygosity and decreased expression appear to be associated with breast and non-small cell lung cancers,37 suggesting that INT6/eIF3e activity is particularly dosage dependent. Recently, eIF3h overexpression was shown to increase tumorigenic phenotypes in various cell lines,40 and eIF3h was also found in a genome-wide association screen for loci conferring increased risk for colorectal cancer.41 Finally, eIF3a, the largest eIF3 subunit, is overexpressed in human lung, breast, cervical and esophageal cancers,42-45 and reduction of eIF3a levels in two human lung and breast cancer cell lines, respectively, is sufficient to suppress the malignant phenotypes in vitro.46 eIF3a expression is also higher in fetal than in more differentiated tissues,47 and thus reciprocal to let-7 expression.26
As expression of eIF3 subunits in human cells appears to be highly coordinated48 such that forced overexpression of individual subunits leads to increased accumulation of other subunits and incorporation into functional eIF3 complexes, it appears likely that additional eIF3 subunits are deregulated in tumors.
When viewed together with the fact that let-7 functions as a tumor suppressor gene,26 these findings suggest that the opposing roles of eIF3 and let-7 on cell differentiation might be conserved beyond C. elegans. Indeed, increased amounts of eIF3 specifically stimulate translation of mRNAs involved in cell proliferation, in particular MYC and cyclin D1 (reviewed in ref. 48)—mRNAs that are repressed by let-7.26 We might thus speculate that the opposing activities of eIF3 and let-7 on a subset of cellular mRNAs contribute to the oncogenic functions of eIF3.
Taken together, we find widespread suppression of let-7 loss-of-function through decreased cellular translation activity, suggesting that let-7 targets or other heterochronic genes may be translationally regulated to allow proper timing of cell differentiation.
Materials and Methods
let-7(n2853) suppressor screen and RNAi
Wild-type (N2) and let-7(n2853) (MT7626) strains used in this work were provided by the Caenorhabditis Genetics Center (CGC), which is founded by the NIH National Center for Research Resources. The screen was performed using RNAi by feeding with a published RNAi library29 covering ca. 90% of the genes on C. elegans chromosome I. Additional RNAi clones were obtained from RNAi libraries,49,50 or were created in the laboratory as follows by PCR on genomic DNA using the primers listed below. PCR fragments were digested with XbaI and KpnI (pXD10, pXD11 and pXD12), NdeI/XhoI (pHG8) or BamHI/XhoI (pHG9) and ligated into L4440 (reviewed in ref. 51). The resulting constructs were transformed in E. coli HT115 for feeding RNAi experiments.
The screen was performed as illustrated in Figure 1, with every step done in duplicate. Supplements were used at the following concentrations: ampicillin: 100 μg/ml, tetracycline: 12.5 μg/ml, IPTG: 1 mM. Retesting and testing of additional translation factors was done at 20°C and 25°C on 6-cm diameter plates as described.7 In some experiments, carbenicillin was used instead of ampicillin. Suppressor identity was confirmed through plasmid DNA recovery followed by sequencing. In some cases we were unable to stage worms reliably because RNAi caused gonad migration defects in the absence of oocytes. These candidates were discarded.
Enhancement of lin-41(ma104) precocious phenotypes was scored in at least two independent experiments with ≥19 animals per strain and a total of ≥100 animals scored per strain.
Oligonucleotide sequences
Synthetic sequences are in lowercase, restriction sites underlined.
| Name | 5′ to 3′ sequence | Plasmid |
|---|---|---|
| eIF1_F1 | gctctagagcTGAAGTTGCTCACCATCTCG | pXD10 |
| eIF1_R1 | ggggtaccccACTTCCTCGCCACTTCTTCA | (L4440-eIF1) |
| cif-1_F1 | gctctagagcGATGATGTCAAGCAGCTCCA | pXD11 |
| cif-1_R1 | ggggtaccccCATTGTTCCGTTCCGAATCT | (L4440-cif-1) |
| eIF5B_F1 | gctctagagcGAAGAGGATTCGGATGGTGA | pXD12 |
| eIF5B_R1 | ggggtaccccACCTCCTTCCTCTTGGCAAT | (L4440-eIF5B) |
| eif-3.B_F1 | caccatATGGTCGAAATTGACTTTAAT | pHG8 |
| eif-3.B_R1 | tttctcgagTTAGTCTCTCATCTCCTCC | (L4440-eif-3.B) |
| eif-3.C_F1 | tttggatccTGTCTCGCTTCTTCCATGC | pHG9 |
| eif-3.C_R1 | tttctcgagTTAGAAGGCTCGTGGCTTT | (L4440-eif-3.C) |
Precocious seam cells fusion
Precocious seam cell fusion was analyzed using strain wIs79[ajm-1::gfp; scm::gfp] (ref. 9). Microscopy images were acquired using a Zeiss Axioplan microscope equipped with a Zeisscam CCD camera and Axiovision software. Images were cropped and levels adjusted using Adobe Photoshop software.
Acknowledgements
X.D. was supported by a PhD student fellowship from the Boehringer Ingelheim Foundation, F.J.S by NIH R01 grant (GM62594). Part of H.G.’s work on the project was supported by a Human Frontier Science Program Postdoctoral Fellowship. Research in H.G.’s laboratory is supported by the FMI, part of the Novartis Research Foundation.
References
- 1.Bushati N, Cohen SM. microRNA Functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 2.Eulalio A, Huntzinger E, Izaurralde E. Getting to the Root of miRNA-Mediated Gene Silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 3.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 5.Moss EG. Heterochronic genes and the nature of developmental time. Curr Biol. 2007;17:425–34. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 6.Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–37. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 7.Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev Cell. 2005;8:321–30. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 8.Lall S, Grün D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, Kao HL, Gunsalus KC, Pachter L, Piano F, Rajewsky N. A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol. 2006;16:460–71. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 9.Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4:639–50. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 10.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–69. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 11.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 12.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 13.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–30. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayr C, Hemann MT, Bartel DP. Disrupting the Pairing Between let-7 and Hmga2 Enhances Oncogenic Transformation. Science. 2007;315:1576–9. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang T, Zhang X, Obijuru L, Laser J, Aris V, Lee P, Mittal K, Soteropoulos P, Wei JJ. A micro-RNA signature associated with race, tumor size, and target gene activity in human uterine leiomyomas. Genes Chromosomes Cancer. 2007;46:336–47. doi: 10.1002/gcc.20415. [DOI] [PubMed] [Google Scholar]
- 17.Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer. 2007;6:5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, Feig C, Lengyel E, Peter ME. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA. 2007;104:11400–5. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang JC, Babak T, Corson TW, Chua G, Khan S, Gallie BL, Hughes TR, Blencowe BJ, Frey BJ, Morris QD. Using expression profiling data to identify human microRNA targets. Nat Methods. 2007;4:1045–9. doi: 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]
- 20.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ. The let-7 MicroRNA Represses Cell Proliferation Pathways in Human Cells. Cancer Res. 2007;67:7713–22. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 21.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 22.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7 doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 23.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105:3903–8. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, Su TJ, Chiang CC, Li HN, Hong QS, Su HY, Chen CC, Chen WJ, Liu CC, Chan WK, Li KC, Chen JJ, Yang PC. MicroRNA Signature Predicts Survival and Relapse in Lung Cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 Regulates Self Renewal and Tumorigenicity of Breast Cancer Cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 26.Büssing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008 doi: 10.1016/j.molmed.2008.07.001. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–7. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–63. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 29.Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–30. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 30.Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–8. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 31.Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–70. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–53. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 33.Rhoads RE, Dinkova TD, Korneeva NL. Mechanism and regulation of translation in C. elegans. In: The C. elegans Research Community, editor. WormBook. Wormbook; 2006. pp. 1–18.http://www.wormbook. org [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson SM, Lin SY, Slack FJ. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev Biol. 2003;259:364–79. doi: 10.1016/s0012-1606(03)00202-1. [DOI] [PubMed] [Google Scholar]
- 35.Dong Z, Zhang JT. Initiation factor eIF3 and regulation of mRNA translation, cell growth and cancer. Crit Rev Oncol Hematol. 2006;59:169–80. doi: 10.1016/j.critrevonc.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 36.Parry DH, Xu J, Ruvkun G. A Whole-Genome RNAi Screen for C. elegans miRNA Pathway Genes. Curr Biol. 2007;17:2013–22. doi: 10.1016/j.cub.2007.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchetti A, Buttitta F, Pellegrini S, Bertacca G, Callahan R. Reduced expression of INT-6/eIF3-p48 in human tumors. Int J Oncol. 2001;18:175–9. doi: 10.3892/ijo.18.1.175. [DOI] [PubMed] [Google Scholar]
- 38.Rasmussen SB, Kordon E, Callahan R, Smith GH. Evidence for the transforming activity of a truncated Int6 gene, in vitro. Oncogene. 2001;20:5291–301. doi: 10.1038/sj.onc.1204624. [DOI] [PubMed] [Google Scholar]
- 39.Mayeur GL, Hershey JW. Malignant transformation by the eukaryotic translation initiation factor 3 subunit p48 (eIF3e) FEBS Lett. 2002;514:49–54. doi: 10.1016/s0014-5793(02)02307-4. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L, Smit-McBride Z, Pan X, Rheinhardt J, Hershey JW. An oncogenic role for the phosphorylated h-subunit of human translation initiation factor eIF3. J Biol Chem. 2008 doi: 10.1074/jbc.M800956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, Pittman AM, Spain S, Lubbe S, Walther A, Sullivan K, Jaeger E, Fielding S, Rowan A, Vijayakrishnan J, Domingo E, Chandler I, Kemp Z, Qureshi M, Farrington SM, Tenesa A, Prendergast JG, Barnetson RA, Penegar S, Barclay E, Wood W, Martin L, Gorman M, Thomas H, Peto J, Bishop DT, Gray R, Maher ER, Lucassen A, Kerr D, Evans DG, CORGI Consortium, Schafmayer C, Buch S, Völzke H, Hampe J, Schreiber S, John U, Koessler T, Pharoah P, van Wezel T, Morreau H, Wijnen JT, Hopper JL, Southey MC, Giles GG, Severi G, Castellví-Bel S, Ruiz-Ponte C, Carracedo A, Castells A, EPICOLON Consortium, Försti A, Hemminki K, Vodicka P, Naccarati A, Lipton L, Ho JW, Cheng KK, Sham PC, Luk J, Agúndez JA, Ladero JM, de la Hoya M, Caldés T, Niittymäki I, Tuupanen S, Karhu A, Aaltonen L, Cazier JB, Campbell H, Dunlop MG, Houlston RS. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–30. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 42.Bachmann F, Banziger R, Burger MM. Cloning of a novel protein overexpressed in human mammary carcinoma. Cancer Res. 1997;57:988–94. [PubMed] [Google Scholar]
- 43.Dellas A, Torhorst J, Bachmann F, Banziger R, Schultheiss E, Burger MM. Expression of p150 in cervical neoplasia and its potential value in predicting survival. Cancer. 1998;83:1376–83. doi: 10.1002/(sici)1097-0142(19981001)83:7<1376::aid-cncr15>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 44.Chen G, Burger MM. p150 expression and its prognostic value in squamous-cell carcinoma of the esophagus. Int J Cancer. 1999;84:95–100. doi: 10.1002/(sici)1097-0215(19990420)84:2<95::aid-ijc1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 45.Pincheira R, Chen Q, Zhang JT. Identification of a 170-kDa protein overexpressed in lung cancers. Br J Cancer. 2001;84:1520–7. doi: 10.1054/bjoc.2001.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong Z, Liu LH, Han B, Pincheira R, Zhang JT. Role of eIF3 p170 in controlling synthesis of ribonucleotide reductase M2 and cell growth. Oncogene. 2004;23:3790–801. doi: 10.1038/sj.onc.1207465. [DOI] [PubMed] [Google Scholar]
- 47.Liu Z, Dong Z, Yang Z, Chen Q, Pan Y, Yang Y, Cui P, Zhang X, Zhang JT. Role of eIF3a (eIF3 p170) in intestinal cell differentiation and its association with early development. Differentiation. 2007;75:652–61. doi: 10.1111/j.1432-0436.2007.00165.x. [DOI] [PubMed] [Google Scholar]
- 48.Zhang L, Pan X, Hershey JW. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J Biol Chem. 2006;282:5790–800. doi: 10.1074/jbc.M606284200. [DOI] [PubMed] [Google Scholar]
- 49.Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, Vidal M. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–8. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–7. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 51.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998:395–85. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]