Summary
The mammalian Target of Rapamycin Complex 1 (mTORC1) regulates cell growth in response to the nutrient and energy status of the cell, and its deregulation is common in human cancers. Little is known about the overall architecture and subunit organization of this essential signaling complex. We have determined the three-dimensional (3D) structure of the fully assembled human mTORC1 by cryo-electron microscopy (cryo-EM). Our analyses reveal that mTORC1 is an obligate dimer with an overall rhomboid shape and a central cavity. The dimeric interfaces are formed by interlocking interactions between the mTOR and raptor subunits. Extended incubation with FKBP12-rapamycin compromises the structural integrity of mTORC1 in a stepwise manner, leading us to propose a model in which rapamycin inhibits mTORC1-mediated phosphorylation of 4E-BP1 and S6K1 through different mechanisms.
Introduction
The mTOR serine/threonine kinase is a member of the phosphoinositide 3-kinase (PI3K)-related kinase (PIKK) family. This conserved protein integrates diverse upstream signals to regulate growth-related processes, including mRNA translation, ribosome biogenesis, autophagy, and metabolism (Sarbassov et al., 2005a). mTOR nucleates two large, physically and functionally distinct signaling complexes: mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) (Guertin and Sabatini, 2007). mTORC1 consists of mTOR, raptor (regulatory associated protein of mTOR), PRAS40 (proline-rich AKT substrate 40 kDa), and mLST8 (mammalian lethal with sec-13). mTORC2, on the other hand, is composed of mTOR, mLST8, rictor (raptor independent companion of mTOR), mSIN1 (mammalian stress-activated protein kinase interacting protein 1), and Protor-1 (protein observed with rictor-1), and controls cell proliferation and survival by phosphorylating and activating the Akt/PKB kinase (Sarbassov et al., 2005b). The key structural features that differentiate the substrate specificity of mTORC1 and mTORC2 remain unclear.
Unlike mTORC2, mTORC1 appears to play critical roles in cell growth in response to nutrients. The mTOR protein, which consists of multiple HEAT repeats at its N-terminal half followed by the FKBP12-rapamycin binding (FRB) and serine–threonine protein kinase domains near its C-terminal end, has no known enzymatic functions besides its kinase activity. PRAS40 has been characterized as a negative regulator of mTORC1 (Sancak et al., 2007; Vander Haar et al., 2007; Wang et al., 2007), but the functions of other mTOR-interacting proteins in mTORC1 are ambiguous. Previous studies indicate that raptor may have roles in mediating mTORC1 assembly, recruiting substrates, and regulating mTORC1 activity and subcellular localization (Hara et al., 2002; Kim et al., 2002; Sancak et al., 2008). The strength of the interaction between mTOR and raptor can be modified by nutrients and other signals that regulate the mTORC1 pathway, but how this translates into regulation of the mTORC1 pathway remains elusive. The role of mLST8 in mTORC1 function is also unclear, as the chronic loss of this protein does not affect mTORC1 activity in vivo (Guertin et al., 2006). However, the loss of mLST8 can perturb the assembly of mTORC2 and its function. The small GTP-binding protein Rheb (Ras homologue enriched in brain) binds near the mTOR kinase domain (Long et al., 2005) and seems to have a key role in stimulating the kinase activity of mTORC1 (Long et al., 2005; Sancak et al., 2007).
mTORC1 can be hyperactivated by oncogenic phosphoinositide 3-kinase signaling and promotes cellular growth in cancer (Guertin and Sabatini, 2007; Shaw and Cantley, 2006). mTORC1 drives growth through at least two downstream substrates S6 kinase 1 (S6K1) and eIF-4E-binding protein 1 (4E-BP1) (Richter and Sonenberg, 2005; Ma and Blenis, 2009). The regulation of the activity of mTORC1 towards these and yet unidentified substrates appears to be complex and is likely to be dependent on the organization of the various subunits in the mTORC1 complex. The study of mTORC1 phosphorylation of substrate sites has been greatly aided by pharmacological inhibitors of mTORC1, in particular rapamycin. Rapamycin, in complex with its intracellular receptor FKBP12 (FK506-binding protein of 12 kDa), acutely inhibits mTORC1 by binding to the FRB domain of mTOR (Sarbassov et al., 2005a). Yet, the molecular mechanism of how this high affinity interaction perturbs mTOR kinase activity and the fully assembled mTORC1 is currently unknown. Although there have been attempts to model the N-terminal domain of mTOR based on the low-resolution structure of human DNA-PK (Sibanda et al., 2010), these efforts have failed to provide insights into the function and regulation of the mTOR kinase. Thus, a detailed knowledge of mTORC1 structure, including the organization of its components, has the potential to help understand the regulation of its kinase activity and in aiding the development of more effective mTORC1 inhibitors. We report the three-dimensional (3D) structure of human mTORC1 as determined by cryo-EM. This structure together with labeling and biochemical studies reveal the intricate organization of the components within mTORC1 and provide structural insights into the mechanism of its inhibition by FKBP12-rapamycin.
Results and Discussion
Purification of human mTORC1
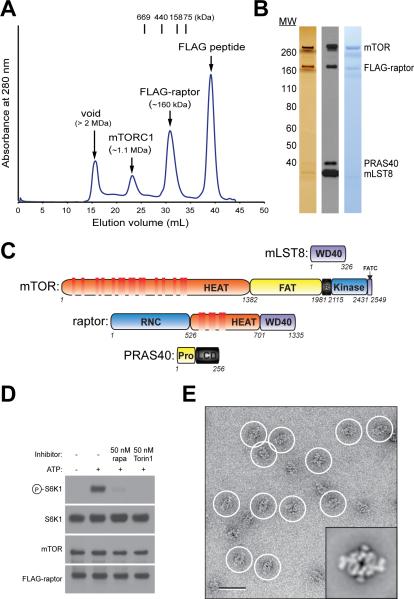
The large size (~1 MDa) and instability of mTORC1 make it difficult to obtain the purified complex for structural analysis. To address this issue, we devised a method to purify microgram quantities of intact and active human mTORC1. Keys to the successful purification of mTORC1 were the development of a human cell line stably expressing a tagged raptor subunit that incorporates into endogenous mTORC1, the identification of buffer conditions that minimize mTORC1 disintegration and/or aggregation during purification, and the implementation of tandem gel filtration chromatography steps to separate mTORC1 from other large contaminants (Figure 1A). Purified mTORC1 consists of equimolar quantities of mTOR, raptor, and mLST8, and of PRAS40 at substoichiometric level (Figure 1B and C). The kinase activity of purified mTORC1 towards S6K1 was sensitive to FKBP12-rapamycin and Torin1, an ATP-competitive inhibitor of mTOR (Thoreen et al., 2009) (Figure 1D). Negative stain EM analysis of the purified complex revealed particles that were homogeneous in size and shape (Figure 1E). Projection averages calculated from the classification of 10,080 particle images illustrated that mTORC1 has an elongated, rhomboid shape with a central, stain-filled cavity and “feet-like” protrusions emanating from both ends of the molecule (Figure 1E, inset, and Figure S1). The averages displayed a two-fold symmetry. This observation, together with the mass estimated from gel filtration, provides evidence for the obligate dimeric organization of mTORC1 that had been previously suggested by genetic and co-immunoprecipitation studies (Zhang et al., 2006; Takahara et al., 2006; Urano et al., 2007; Wang et al., 2006). Although PRAS40 was present in substoichiometric amounts in our purified mTORC1 sample, the fact that all averages show particles with identical overall shape suggests that PRAS40 contributes little to the density of the complex and is not required for proper assembly and stability of mTORC1. Our projection structure does not resemble the monomeric structure obtained from the recent negative stain EM analysis of Saccharomyces cerevisiae TOR in complex with KOG1 (the raptor homolog in yeast) (Adami et al., 2007). These striking differences may be due to the differences in composition and stoichiometry of known components within the two TOR complexes. The sample used in the EM study of the yeast complex may not reflect a fully assembled TORC1 complex but rather a subassembly. For example, LST8, a bona fide yeast TORC1 component, is missing in the analyzed sample of the yeast study. Additionally, human mTOR and raptor are only conserved over limited regions compared to their yeast orthologues, TOR1 and KOG1, respectively. Therefore, it is conceivable that human mTORC1 and yeast TORC1 could adopt different quaternary structures.
Figure 1.
mTORC1 purification. (A) mTORC1 and FLAG-raptor were purified by tandem gel filtration chromatography. Their masses were estimated based on known molecular weight standards as indicated (Thyroglobulin (669 kDa), Ferritin (440 kDa), Aldolase (158 kDa) and Conalbumin (75 kDa)). (B) The gel filtration fraction corresponding to mTORC1 was analyzed by SDS-PAGE followed by silver and Coomassie staining as well as immunoblotting for indicated proteins. (C) Schematics of mTORC1 components illustrating the various predicted domains. (D) In vitro kinase assay showing that purified mTORC1 phosphorylates S6K1 and is inhibited by both rapamycin-FKBP12 (rapa) and Torin1. (E) EM of negatively stained mTORC1. A raw image of mTORC1 particles (circled) and a representative class average from the classification of 10,080 particles (inset). The scale bar represents 50 nm, and the side length of the panel showing the class average is 45 nm. “see also Figure S1”
Cryo-EM structure of mTORC1
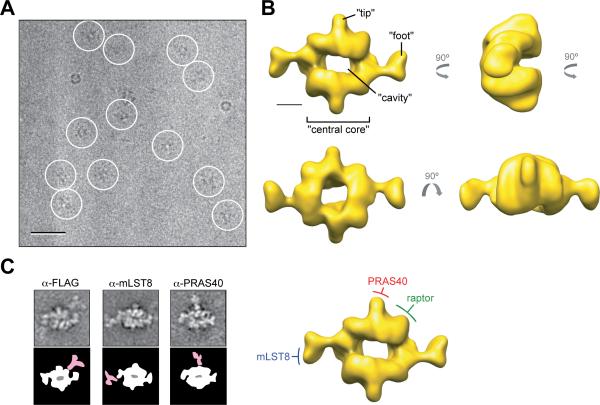
To determine the 3D structure of mTORC1 by cryo-EM, we first produced a reliable initial model by calculating a random conical tilt (RCT) reconstruction with 50°/0° tilt pair images of cryo-negatively stained specimens (Figure S2). Collecting images of vitrified mTORC1 specimens proved difficult due to low protein concentration (attempts to concentrate mTORC1 samples were unsuccessful) and a strong tendency of mTORC1 to dissociate upon contact with the air-water interface. We overcame these difficulties by adsorbing mTORC1 to a thin carbon film prior to vitrification. Even so, only few particles were present (Figure 2A), requiring us to collect many images to obtain a sufficient number of particles for structure determination. The carbon film also induced mTORC1 to adsorb to the grid in a preferred orientation, making it necessary to collect images of tilted specimens to obtain the multiple views needed for 3D reconstruction. The final data set contained 28,325 particle images, including 3,905 from 45° tilted specimens. A 3D reconstruction was calculated by aligning these individual images of the vitrified complex to the initial model produced with the cryo-negatively stained sample, followed by iterative refinement of their orientation parameters. The estimated resolution of the final reconstruction is 26 Å according to the Fourier shell correlation = 0.5 criterion (Figure S2). However, the resolution is clearly anisotropic, with lower resolution in the direction perpendicular to the carbon film, a result of the limited number of views other than the face-on view (Figure S2). mTORC1 has overall dimensions of approximately 290 Å × 210 Å × 135 Å and an estimated volume of 1.4 × 106 Å3 at the contour level of the displayed map, which was chosen to be consistent with the calculated molecular mass of dimeric mTORC1 (Figure 2B).
Figure 2.
Cryo-EM reconstruction of mTORC1 and its molecular organization. (A) Image of a vitrified specimen showing individual mTORC1 particles (circled). The scale bar represents 50 nm. (B) Different views of the 3D reconstruction of mTORC1 filtered to 26 Å, with the main structural features denoted. The scale bar represents 5 nm. (C) Molecular organization of mTORC1. Left panels: Representative class averages from antibody labeling experiments of mTORC1 (top) and schematic representations showing mTORC1 in white and the antibody in pale red (bottom). The side length of each panel is 45 nm. Right panel: Location of raptor, mLST8, and PRAS40 in the cryo-EM density map of mTORC1. “see also Figure S2”
The cryo-EM structure reveals a central cavity that has an oval shape when viewed from one face, but a rectangular shape from the opposite face, with two troughs located at the extensions linking the central core to the “feet-like” structures. While the biological relevance of this cavity remains elusive, its location between the two “monomeric” complexes may enable substrates with multiple phosphorylation sites, such as 4E-BP1 (Gingras et al., 1999), to shuttle between the two mTOR active sites within the complex. Another, albeit less likely, possibility is that the cavity may serve as a docking platform for nucleic acids because its size (~40 Å × 28 Å) is large enough to accommodate double-stranded DNA (dsDNA). While mTORC1 has not yet been shown to interact with dsDNA or other macromolecules, several members of the PIKK family (Keith and Schreiber, 1995), most notably DNA-PK, are known to mediate DNA repair by directly binding to DNA (Gottlieb and Jackson, 1993; Spagnolo et al., 2006).
Subunit organization of mTORC1 and EM structure of raptor
While the cryo-EM structure revealed the overall shape of mTORC1, at the current resolution it was not possible to define intermolecular and intersubunit boundaries. Therefore, we performed antibody labeling experiments to localize individual subunits within mTORC1, including raptor (detected through its FLAG tag), mLST8, and PRAS40. The labeled particles were imaged by negative stain EM and analyzed by classification and image averaging. We discovered that mLST8 localizes to the distal “foot-like” structures, PRAS40 to the small tips in the mid-section of the central core, and the N-terminus of raptor to the corner of the core (Figure 2C). The occasional observation of double-labeled particles provided further assurance for the dimeric organization of mTORC1 (Figure S2).
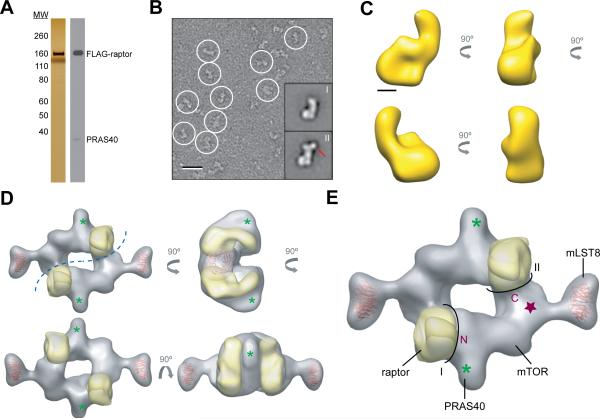
Additionally, we determined the EM structure of raptor. FLAG-tagged raptor that did not incorporate into mTORC1 eluted as a separate peak from gel filtration (Figures 1A and 3A). Purified raptor was homogeneous in size and shape according to negative stain EM analysis (Figures 3B and S3). The 3D reconstruction of raptor, determined by the RCT approach using 60°/0° image pairs of negatively stained specimens (Figure S3), revealed that its overall shape resembles a “comma” with the circular lobe likely representing the predicted C-terminal WD40 domain (Kim et al., 2002) (Figure 3C). Utilizing the antibody labeling data as a guide, the structure of mTORC1 provides an adequate framework in which the EM reconstruction of raptor can be meaningfully fitted (gold surface in Figure 3D). Because mLST8 is solely composed of a seven-bladed beta-propeller (Kim et al., 2003), we next docked two beta-propeller models (PDB code 3EMH) into the “foot” substructures (Figure 3D). By subtracting two copies of raptor and mLST8, the densities occupied by two mTOR subunits can be predicted, while accounting for minor contributions by the two small PRAS40 subunits (Figure 3D). It has been shown that the C-terminal kinase domain of mTOR associates with mLST8 (Kim et al., 2003), suggesting that this domain is likely positioned adjacent to the “foot” (purple star in Figure 3E). From the position of the kinase domain, we deduced that the N-terminus of mTOR interacts with the flat face of one raptor molecule (bottom left view in Figure 3C), forming interface I, whereas the C-terminus interacts with the side of the second raptor molecule (top right view in Figure 3C), forming interface II. The interlocking raptor-mTOR interactions within the central core provide an understanding of the basis of dimerization and illustrate the crucial function of raptor in mediating and maintaining the higher-order organization of mTORC1 (Figure 3E). In contrast, each mLST8 contacts only one mTOR within the complex. Its localization to the distal “foot” structures suggests that it could potentially assist substrate entry into the catalytic site.
Figure 3.
3D reconstruction of raptor and molecular docking. (A) Silver-stained gel and immunoblot of the gel filtration fraction containing free raptor detected the presence of PRAS40. (B) EM image of negatively stained raptor (circled) and two representative class averages from the classification of 12,216 particles (bottom right insets). Class II particles contain an additional density (red arrow) compared to Class I particles, which likely represents PRAS40. The scale bar represents 25 nm, and the side length of the panels showing the class averages is 27 nm. (C) Different views of the raptor 3D reconstruction. The scale bar represents 2.5 nm. (D) Two copies of the raptor 3D reconstruction (gold) and two models of a representative WD40 domain (PDB code 3EMH, red) were placed into the cryo-EM density map (gray). The green asterisk depicts the location of PRAS40 as determined by antibody labeling. The blue dotted line represents the dimer interface. (E) The proposed locations of the N- and C-terminal domains (marked “N” and “C”) and the kinase domain of mTOR (purple star). The black lines labeled “I” and “II” delineate the two interaction interfaces formed by each mTOR molecule with the two raptor subunits. “see also Figure S3”
According to our data and composite model, PRAS40 localizes in close proximity to raptor (asterisk in Figure 3D), which is in agreement with the known binding of PRAS40 to raptor (Sancak et al., 2007; Wang et al., 2007). Interestingly, some class averages of purified raptor showed an additional, small density, which may represent bound PRAS40 (indicated by red arrow in Figures 3B and S3). This interpretation is supported by immunoblots that show PRAS40 to be present in the analyzed raptor fraction (Figure 3A). Thus, our structural data suggest that PRAS40 inhibition is not likely to be achieved through an interaction of PRAS40 with the mTOR kinase domain. Instead, it favors the model that PRAS40 acts as a competitive inhibitor for the binding of mTORC1 substrates to raptor (Wang et al., 2007).
FKBP12-rapamycin destabilizes mTORC1 in a stepwise manner
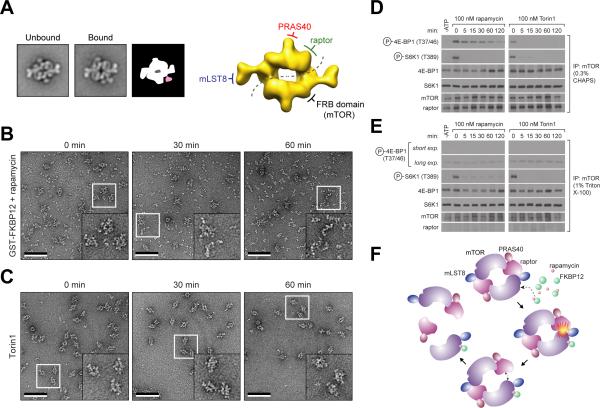
With a more detailed understanding of the subunit organization of mTORC1, we next investigated how rapamycin affects its structure. As an allosteric inhibitor of mTORC1, rapamycin requires the intracellular protein FKBP12 to form a gain-of-function complex, which directly interacts with the FKBP12-rapamycin-binding (FRB) domain of mTOR (Chen et al., 1995; Sabatini et al., 1994). The crystal structure of FKBP12-rapamycin in complex with the FRB domain did not reveal how this interaction prevents phosphorylation of direct mTORC1 substrates (Choi et al., 1996). Previous biochemical studies indicated that binding of FKBP12-rapamycin to mTORC1 induces a conformational change that weakens the mTOR-raptor interaction (Kim et al., 2002). To test this hypothesis, we incubated mTORC1 with N-terminal GST-tagged FKBP12 in the presence of 50 nM rapamycin for 15 minutes, and then visualized the particles by negative stain EM (Figure S4). Although the raw images did not reveal any obvious structural changes (Figure S4), image classification showed that about 10% of the particles featured an additional density, likely constituting FKBP12-rapamycin, tethered to the region we assigned to mTOR and directly opposite of raptor (Figure 4A). Interestingly, we did not observe individual particles or averages of mTORC1 showing two extra densities, suggesting that either mTORC1 cannot accommodate two FKBP12-rapamycin complexes or that this intermediate is short lived.
Figure 4.
Effects of rapamycin-FKBP12 on mTORC1. (A) Representative class averages of untreated mTORC1 (left) and mTORC1 treated with 50 nM of rapamycin and 0.02 μg/μl GST-FKBP12 (middle), and a schematic representation showing the additional density in pale red (right). The side length of each panel is 45 nm. To the right, the location of the FRB domain with respect to the other components in the cryo-EM map of mTORC1 is shown. (B) and (C) Purified mTORC1 was treated with 50 nM of rapamycin and 0.02 μg/μl GST-FKBP12 or 100 nM Torin1. EM images of negatively stained samples were taken at the indicated time points. The inset in each image shows an enlarged view of the area marked by the white square. The scale bars represent 100 nm. (D) and (E) mTOR immunoprecipitates, prepared in lysis buffers containing 0.3% CHAPS or 1% Triton X-100, were subjected to in vitro kinase assays using 4E-BP1 or S6K1 as a substrate in the presence of 100 nM rapamycin and 0.02 μg/μl FKBP12 or 100 nM Torin1. Assays were then analyzed by immunoblotting for the indicated proteins and phosphorylation states. (F) A model depicting a potential mechanism of mTORC1 inhibition by FKBP12-rapamycin. “see also Figure S4”
While relatively short exposure to FKBP12-rapamycin did not affect the structural integrity of mTORC1, extended incubations resulted in a drastic reduction in the total number of intact mTORC1 particles. Many smaller fragments appeared in the background, suggesting that FKBP12-rapamycin may cause disassembly of mTORC1 (Figure 4B). Once initiated, this dissociation appears to be swift, as we were unable to detect intermediates with defined structures during the course of the reaction (data not shown). After a one-hour incubation, virtually no intact mTORC1 particles could be detected, and the sample contained only smaller fragments, likely representing free mTOR or its subcomplexes, and undefined aggregates (Figure 4B). In contrast, Torin1 did not affect mTORC1 stability even after extended incubation (Figure 4C).
The disruption of mTORC1 by FKBP12-rapamycin may play a role in the inhibition by rapamycin of mTORC1 kinase activity towards certain substrates. Consistent with the time-dependent effects of FKBP12-rapamycin on the structural integrity of mTORC1, FKBP12-rapamycin significantly inhibited the in vitro phosphorylation of 4E-BP1 by mTORC1 only after mTORC1 had been incubated with the drug for at least 60 minutes (Figure 4D). In contrast, FKBP12-rapamycin rapidly blocked the phosphorylation of S6K1 by mTORC1, and Torin1 quickly blocked the phosphorylation of both S6K1 and 4E-BP1. In close agreement with in vitro kinase assays, in vivo experiments produced similar time-dependent effects of rapamycin on endogenous 4E-BP1, but not S6K1 (Figure S4). To assess the importance of mTORC1 integrity in the phosphorylation of S6K1 and 4E-BP1, we prepared mTORC1 that lacked raptor (Kim et al., 2002). While raptor-free mTORC1, as predicted (Hara et al., 2002), could not support 4E-BP1 phoshorylation, it was capable of phosphorylating full-length S6K1 (Figure 4E). Furthermore, the phosphorylation of S6K1 by raptor-free mTORC1 was still inhibited by FKBP12-rapamycin. Thus, contrary to previous assumptions, raptor is dispensable for mTORC1 to phosphorylate S6K1 in a rapamycin-sensitive fashion in vitro. Our results suggest an important role for the mTORC1 dimer in 4E-BP1 phosphorylation. Perhaps, 4E-BP1 binds to the raptor in one monomer, but is phosphorylated by the kinase of the adjacent monomer, such that in the absence of dimerization, 4E-BP1 is not in a position to be phosphorylated.
Based on these observations and our knowledge of the molecular organization of mTORC1, we propose the following model for rapamycin-mediated inhibition of mTORC1. The initial binding of one FKBP12-rapamycin to mTORC1 causes a subtle conformational change in mTOR that weakens the mTOR-raptor interaction, but does not suffice to disrupt the dimeric architecture. Moreover, the bound FKBP12-rapamycin likely occludes the binding of or blocks access to the active site for larger-sized substrates, such as S6K1. Over time, either amplified structural strain caused by the first FKBP12-rapamycin or, perhaps, the binding of a second rapamycin complex leads to a fast disintegration of the already “weakened” mTORC1 and the complete abolishment of 4E-BP1 phosphorylation (Figure 4F). Therefore, our work suggests that in vitro rapamycin is an mTORC1 inhibitor that may work through at least two different modes. The fact that within cells rapamycin does not completely inhibit 4E-BP1 phosphorylation (Choo et al., 2008; Thoreen et al., 2009; Feldman et al., 2009) nor mTORC1 stability (Kim et al., 2002) suggests that cells contain buffering mechanisms that counter the effects of rapamycin on mTORC1 and that these are lost when mTORC1 is purified.
Experimental Procedures
Protein expression and purification
mTORC1 was purified from a HEK-293T cell line that stably expresses N-terminally FLAG-tagged raptor by FLAG-M2 monoclonal antibody-agarose and gel filtration. Details of the expression method and purification conditions are described in Supplementary Experimental Procedures.
In vitro kinase assay
Kinase assays were performed using immunoprecipitated mTORC1 and inactive 4E-BP1 or S6K1 as a substrate. Reactions were analyzed by SDS-PAGE and immunoblotting. Details of the assay conditions are described in Supplementary Experimental Procedures.
Electron microscopy
Negatively stained specimens were prepared as described (Ohi et al., 2004). Images were collected with a Tecnai T12 electron microscope (FEI) equipped with a LaB6 filament and operated at an acceleration voltage of 120 kV. Images were recorded on imaging plates at a nominal magnification of 67,000 using a defocus value of –1.5 μm. Cryo-negatively stained specimens were prepared as described (Ohi et al., 2004). Grids used to collect image pairs of 50°/0° tilted specimens, were loaded on an Oxford cryo-transfer holder. Images were taken under low-dose conditions at a nominal magnification of 50,000x and a defocus value of –2.5 μm using a Tecnai F20 electron microscope (FEI) equipped with a field emission electron source operated at an acceleration voltage of 200 kV. For vitrification, Quantifoil R1.2/1.3 400 mesh grids were overlaid with a thin layer of carbon film and glow discharged. 3 μl of mTORC1 (~0.02 mg/ml) was adsorbed to a grid, and the grid was blotted and frozen in liquid ethane using a Vitrobot (FEI). Specimens were examined using a Gatan 626 cryo-holder on a Tecnai F20 electron microscope equipped with a field emission electron source (FEI) operated at 200 kV. Additional details of specimen preparation and data collection are described in Supplementary Experimental Procedures.
Image processing
Details are described in Supplementary Experimental Procedures.
Supplementary Material
Acknowledgments
This work was supported by fellowships from the American Cancer Society and LAM Foundation to S.A.K.; grants from the NIH (AI47389 and CA103866), Department of Defense (W81XWH-07-1-0448), and W.M. Keck Foundation to D.M.S.; fellowships from the Jane Coffin-Childs Memorial Fund and the Canadian Institutes of Health Research to C.K.Y. D.M.S and T.W. are investigators of the Howard Hughes Medical Institute. The molecular electron microscopy facility at Harvard Medical School was established with a generous donation from the Giovanni Armenise Harvard Center for Structural Biology. We thank Yasemin Sancak, Eric Spooner for technical assistance, Zongli Li for assistance in microscopy and image processing, and members of the Sabatini and Walz laboratories for support and discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
- The cryo-EM structure of human mTORC1 was determined at a resolution of 26 Å.
- Interlocking mTOR-raptor interactions maintain the dimeric architecture of mTORC1.
- FKBP12-rapamycin profoundly disrupts the structural integrity of mTORC1.
Supplemental Data
Supplemental Data including 4 figures and Supplemental Experimental Procedures can be found with this article online at XXXXX.
References
- Adami A, Garcia-Alvarez B, Arias-Palomo E, Barford D, Llorca O. Structure of TOR and its complex with KOG1. Mol Cell. 2007;27:509–516. doi: 10.1016/j.molcel.2007.05.040. [DOI] [PubMed] [Google Scholar]
- Chen J, Zheng XF, Brown EJ, Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci USA. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JW, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci USA. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Keith CT, Schreiber SL. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR Interacts with Raptor to Form a Nutrient-Sensitive Complex that Signals to the Cell Growth Machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov dos D, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005a;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005b;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNAPKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010;463:118–121. doi: 10.1038/nature08648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell. 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Takahara T, Hara K, Yonezawa K, Sorimachi H, Maeda T. Nutrient-dependent Multimerization of the Mammalian Target of Rapamycin through the N-terminal HEAT Repeat Region. J Biol Chem. 2006;281:28605–28614. doi: 10.1074/jbc.M606087200. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano J, Sato T, Matsuo T, Otsubo Y, Yamamoto M, Tamanoi F. Point mutations in TOR confer Rheb-independent growth in fission yeast and nutrient-independent mammalian TOR signaling in mammalian cells. Proc Natl Acad Sci U S A. 2007;104:3514–3519. doi: 10.1073/pnas.0608510104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Wang L, Harris TE, Roth RA, Lawrence JC. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem. 2007;282:20036–20044. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- Wang L, Rhodes CJ, Lawrence JC., Jr. Activation of mammalian target of rapamycin (mTOR) by insulin is associated with stimulation of 4EBP1 binding to dimeric mTOR complex 1. J Biol Chem. 2006;281:24293–24303. doi: 10.1074/jbc.M603566200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Billington CJ, Jr., Pan D, Neufeld TP. Drosophila target of rapamycin kinase functions as a multimer. Genetics. 2006;172:355–362. doi: 10.1534/genetics.105.051979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.