Abstract
Dietary long-chain fatty acid (LCFA) uptake across cell membranes is mediated principally by fatty acid transport proteins (FATPs). Six subtypes of this transporter are differentially expressed throughout the human and rodent body. To facilitate drugs discovery against FATP subtypes, we utilized mammalian cell lines stably expressing the recombinant human FATP4 and 5, and developed a high-throughput screening (HTS) assay using a 96-well fluorometric imaging plate reader (FLIPR). LCFA uptake signal-to background ratios were between 3 and 5-fold. Two 4-aryl-dihydropyrimidinones, j3 and j5, produced inhibition of FATP4 with a half-maximal inhibitory concentration (IC50) of 0.21 uM, and 0.63 uM, respectively, and displayed approximately 100-fold selectivity over FATP5. The US Drug Collection library was screened against the FATP5. A hit rate of around 0.4% was observed with a Z’ factor of 0.6 ± 0.2. Two confirmed hits are bile acids, chenodiol and ursodiol with an IC50 of 2.4 and 0.22 uM, respectively. To increase throughput, a single-time-point measurement in 384-well format was developed using the Analyst HT and the results are comparable with 96-well format. In conclusion, the FATP4 and 5 cell-based fluorescence assays are suitable for a primary drug screen, while differentiated cell lines useful for a secondary drug screen.
Keywords: Long-chain fatty acids, Fatty acid transport proteins, High-throughput screening, Fluorometric imaging plate reader (FLIPR), Bile acids
INTRODUCTION
Long-chain fatty acids (LCFAs) are important metabolites and contribute to many cellular structures and functions, including activation of certain protein kinase C isoforms and nuclear transcription factors such as peroxisome proliferator-activated receptors. LCFA uptake across the plasma membrane occurs via both diffusion and protein-mediated processes, but the latter accounts for over 90% of transport1–5. Fatty acid transport proteins (FATPs, i.e., solute carrier family 27) have been identified in human and mouse genomes. In humans, FATPs consist of a family of six homologous proteins, designated as Homo sapiens FATP1–6 (hsFATP1–6). FATP subtypes are differentially expressed throughout the whole body with typically one or two subtypes predominantly expressed in a particular tissue or organ, such as FATP1 and 4 in brain and adipose tissue, FATP1 in muscles, FATP2 in kidneys, FATP3 in lungs, FATP4 in small intestine, FATP5 in liver and FATP6 in heart 4–15.
Growing evidence indicates that LCFA uptake is tightly regulated by a number of plasma membrane-associated proteins, including fatty acid translocase, plasma membrane-bound fatty acid binding protein, and long chain fatty acyl-CoA synthetase 4, 5, 13–15. Furthermore, hormonal regulation of FATP activity may play an important role in energy homeostasis 16. In adipocytes, the adipogenic hormone insulin increases expression of the long chain fatty acyl-CoA synthetase 17 and induces plasma membrane translocation of FATPs from an intracellular perinuclear compartment to the plasma membrane. This process is paralleled by an increase in LCFA uptake 16. Chronic leptin administration decreases fatty acid uptake 18, and acute leptin application has a direct inhibitory effect on insulin-stimulated fatty acid uptake. Furthermore, treatment with tumor necrosis factor-alpha inhibited basal and insulin-induced LCFA uptake, and reduced FATP1 and FATP4 expression levels 16. Disturbed fatty acid metabolism and homeostasis are associated with insulin resistance. In obesity and other metabolic syndromes, dysfunction of these FATPs may contribute to elevations in serum free fatty acids and the genesis of type 2 diabetes 4, 19–20.
Lacking biologic probes for each subtype of the FATP family, studies of the physiologic role of an individual subtype principally have relied on gene knockout mouse models in vivo and over-expression or antisense knockdown in vitro 4, 12, 20, 22. FATP1 knockout mice displayed less insulin resistance following lipid infusion or a high-fat diet compared to wild-type mice 20. In contrast, homozygote deletion of the FATP4 gene led to early embryonic lethality. While this result illustrates an essential role for FATP4 in embryonic development and survival, the knockout model failed to give insight onto a potential role for FATP4 in lipid absorption or insulin resistance 12, 22. Nevertheless, it was speculated that a selective inhibitor against FATP4 could specifically reduce LCFA absorption with minimal interfering with other nutrient intakes 4. FATP5 is exclusively expressed in the liver and localized to the basal plasma membrane of hepatocytes, congruent with a role in LCFA uptake from the circulation. Overexpression of FATP5 in mammalian cells increased the uptake of 14C-oleate and fluorescence-labeled LCFA 4. Conversely, LCFA uptake was reduced in vitro in hepatocytes isolated from FATP5 knockout mice. The FATP5 knockout mice have lower hepatic triglyceride and free fatty acid content in the liver compared to wildtype mice 23. Furthermore, detailed phenotypic analysis unexpectedly revealed that FATP5 deletion mice did not become overweight under a high-fat diet. Since FATP5 does not express in the gastrointestinal, FATP5 null mice had normal fat absorption. This phenotype likely resulted from both a decrease in general food intake and an increase in energy expenditure 23. These findings not only indicate an important role for FATP5 in regulation of liver fat content, but also suggest FATP5 possibly being involved in body weight homeostasis. Therefore, a selective and potent FATP5 inhibitor could provide a powerful biologic probe to further study the FATP5 function. Furthermore, it could provide a basis for the development of a specific therapeutic for treatment of fatty liver disease, where abnormal fat metabolism is primarily responsible for pathogenesis.
High-throughput screening (HTS) of a large compound library against an FATP subtype could be the initial step toward identifying novel subtype-selective compounds that specifically inhibit LCFA absorption or transport. Conventional methods for the assessment of LCFA uptake using radiolabeled fatty acids and fluorescent fatty acid analogs with fluorescence activated cell sorters (FACS) are very slow and are not suitable for HTS in drug discovery. Recently an LCFA uptake measurement method using the QBT™ Fatty Acid Uptake Assay Kit (QBT kit) has been used to determine real-time LCFA uptake kinetics in 3T3-L1 adipocytes using Gemini or Flexsation fluorescence plate readers 24, 25. The 3T3-L1 adipocytes require differentiation from fibroblast-like progenitors, a process that has been inconsistent and time-consuming. Moreover, 3T3-L1 adipocytes are murine in origin and express several proteins involved in LCFA uptake in addition to FATP1 and 4 14, 26. Thus, a defined target inhibitor is not easily identifiable. To overcome this problem, HTS assays using live yeast cells expressing murine or human FATP2 have been developed and used either 96- or 384-well format to identify small compounds that reduced the import of a fluorescent fatty acid analog, 4,4-difluoro-5-methyl-4-bora-3a,4a-diaza-s-indacene-3-dodecanoic acid (C(1)-BODIPY-C(12)) by FATP2 27,28. In the present study, we have developed an HTS assay (96- or 384-well format) against recombinant hsFAPT4 or FATP5 stably expressed in mammalian cell lines for the screening of subtype-specific inhibitors using a fluorometric imaging plate reader (FLIPR) or the Analyst HT with the QBT assay reagent.
MATERIALS AND METHODS
HsFATP4 and 5 stably expressing cell lines and reagents
Creation of human kidney embryonic (HKE) 293 cell lines stably expressing human recombinant HsFATP1, 4 and 5 has been described previously 4. Cell culture media and supplements were purchased from Gibco Invitrogen (Invitrogen, Carlsbad, CA). The QBT™ Fatty Acid Assay Kits were purchased from MDS analytical Technologies (formerly Molecular Devices Co. Sunnyvale, CA). Chenodeoxycholic acid, oleic acid and palmitic acid were purchased from Sigma-Aldrich (St. Louis, MO). The two FATP4 inhibitors j3 and j5 were synthesized based on a recent publication 29. US Drug Collection (USDC) library consisting of 1040 compounds with known pharmacology was purchased from Microsource (Gaylordsville, CT). All compounds were dissolved in pure DMSO at 10 mM as stock solution.
Cell cultures
The hsFATP1, hsFATP4 or hsFATP5 cells and vector control HEK293 cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) containing 2,250 mg/L D-glucose, pyroxidine hydrochloride, and 0.5 mM CaCl2, supplemented with 10% fetal bovine serum (FBS), and 100 units penicillin-100 ug streptomycin/ml, and supplemented with 1 mg/ml G418 (for genetic selection). Cells were seeded into PDL coated black-walled, clear-bottom microtiter plates (Becton-Dickinson) at a cell density of 80,000 cells/well for 96-well format and 20,000/well for 384-well format. Plates were used for assay 12–16 hours after seeding.
3T3-L1 preadipocytes (American Type Culture Collection, No. CCL92.1) was differentiated following the published protocol 24,25. Cell differentiation was assessed by visual inspection of lipid droplet accumulation under a microscope and cells were seeded into 96-well plates at 100,000 cells/well 12–16 hours before the assay.
The QBT kit and fluorescence intensity measurement
The QBT mixture was prepared as instructed in the product insert (R8133, MDS analytical Technologies), and consisted of a proprietary quenching agent and 2 uM of the fluorescently labeled fatty acid BODIPY-FA. The cell-impermeable quenching agent effectively quenched the fluorescence of BODIPY-FA in solution. Cells took up the BODIPY-FA and intracellular fluorescence was measured using a bottom-read detector FLIPR or Analyst HT (both from MDS analytical Technologies). All synthetic small molecules were dissolved in DMSO as stock solutions and then diluted with 1× loading buffer (1× Hank’s balanced salt solution with 20 mM HEPES buffer and 0.2% fatty acid free bovine serum albumin (BSA)). For kinetic assays in 96-well format, culture media in the plate were replaced with 150 ul 1× loading buffer plus 50 ul diluted compounds at 5× concentration (or 175 ul 1× loading buffer plus 25ul diluted compounds at 10× concentration), and incubated for 30 min at 37°C in a 5% CO2 incubator. At the end of the incubation time, 50 ul of the QBT mixture was added to the well. To examine the inhibitory effect of fatty acid derivatives, they were dissolved in ethanol as stock solutions and diluted with 1× loading buffer. The dilution was then equilibrated with the QBT mix for 24 hours in 4° C before being added to the plate for the uptake assay. Immediately after the addition of QBT mix to cells, fluorescence readings were started using the FLIPR with an excitation filter of λex = 488 nm and an emission filter of λem = 540 nm at room temperature. For single-time-point assays in 384-well format, culture media in the plate were replaced with 35 ul 1× loading buffer plus 5 ul diluted compounds at 10× concentration, and incubated for 30 min at 37°C in a 5% CO2 incubator. At the end of the incubation, 10 ul of the QBT mixture was added to the well. One min and 1 hour following the dye incubation, fluorescence intensity of each well was measured using the Analyst HT with an excitation filter of λex = 488 nm and an emission filter of λem = 540 nm at room temperature. The pilot drug screens were conducted in a blinded manner and the identities of the hits were not decoded until the completion of the verification using concentration-response tests.
Data analysis
For kinetic assays, the uptake of BODIPY-FA was observed continuously for one hour on the FLIPR. The relative fluorescence unit (RFU) level of each well was normalized by subtracting the fluorescence level in the first data point. The end point RFU after one hour observation was used as the maximum fatty acid uptake. For single-time-point assay in 384-well format, the absolute fluorescence intensity of each well was obtained as the maximum fatty acid uptake. Percentage changes in fluorescence signals (inhibition or enhancement) compared with experimental controls were calculated from the mean of duplicate measurements in the primary assay development. Since we cannot measure the final free fatty acid concentrations in the wells, the potency of inhibition by fatty acids were based on the starting concentrations with the assumption that they bind to proteins in the growth medium to a similar degree. Similarly, the small molecule potency was estimated based on their apparent molar concentrations since we did not determine their protein binding profiles. Concentration-response curves were constructed and fitted to a four-parameter sigmoidal function to yield a half-maximal inhibitory concentration (IC50). As appropriate, data were represented as mean ± sem of at least three measurements.
The Z’ factor reflects both the assay signal dynamic range and the control data variation associated with the signal measurement 30 The Z’ factors from each plate were calculated based on the following equation:
To identify initial hits (inhibitory compounds) from the pilot screen at a single measurement, we used the statistical z-score, i.e. it is the ratio of the difference between the sample value and the mean (X) of all wells in the entire plate over the standard deviation (SD) of the mean. In this case, z-score was calculated using the following formula:
The inhibitory compounds were defined in our pilot screen with a z score = −3 z, i.e. with 3 SD from the mean of compounds in the entire plate.
RESULTS
Signal-to-background ratio for the assay using hsFATPs-expressing cells
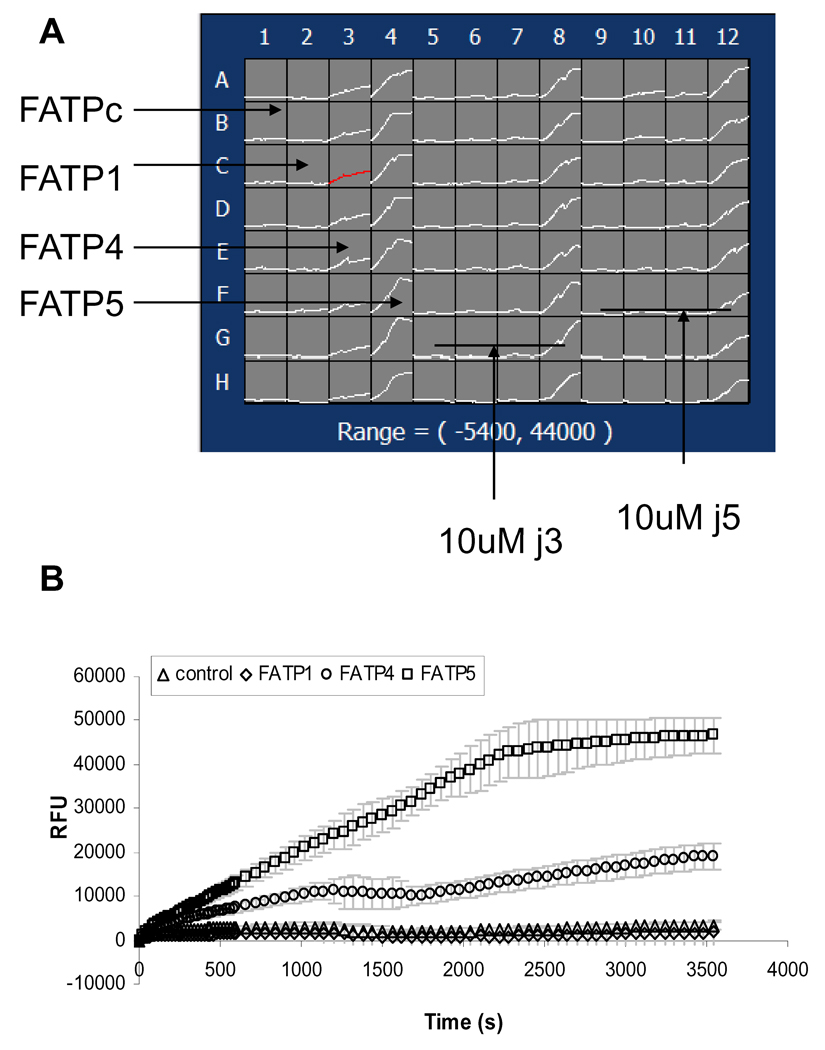
We started with determining the “signal-to--background ratio” by comparing active LCFA uptake in hsFATP1, 4 or 5 expressing cells to vector-control cells (HEK293 cells transfected with an empty expression vector and selected under the same conditions as FATPs-expressing cells) to evaluate the specific signal over nonspecific uptakes for the HTS assay. As Figure 1 shows, real-time uptake kinetics of fluorescently labeled fatty acid BODIPY-FA by the three different FATP subtypes of cells and vector-control cells were measured by the FLIPR within 1 min after the addition of the QBT mix solution (50 ul/well) using a multi-pipette. Approximately 3 and 15 fold increases were observed in LCFA uptake by the hsFATP 4- or 5- expressing cells compared to the vector-control, respectively. The low signals of hsFATP1 might be due to low surface expression of the transporter in the stable cell line tested. For HTS assay development purpose, in the consequent studies, we concentrated on hsFATP4 and hsFATP5.
Figure 1.
Kinetic signals of fluorescently labeled long-chain fatty Acid (BODIPY-FA) uptake in four stable cell lines expressing recombinant hsFATP1, FATP4, FATP5 and FATPc (vector-control) using a FLIPR. A. Screen shot of a FLIPR signals in 96-well plate. Column1,5,9: FATPc; Column 2, 6, 10: FATP1; Column 3, 7, 11: FATP4, Column 4, 8, 12, FATP5; Column 1–4: control; Column 5–8 10 uM j3; Column 9–12: 10 uM j5. B. Averaged relative fluorescence unit (RFU) (mean ± sem, n = 8 wells) taken from the plate shown in A plotted against recording time.
Competitive inhibition of BODIPY-FA uptake by fatty acid derivatives
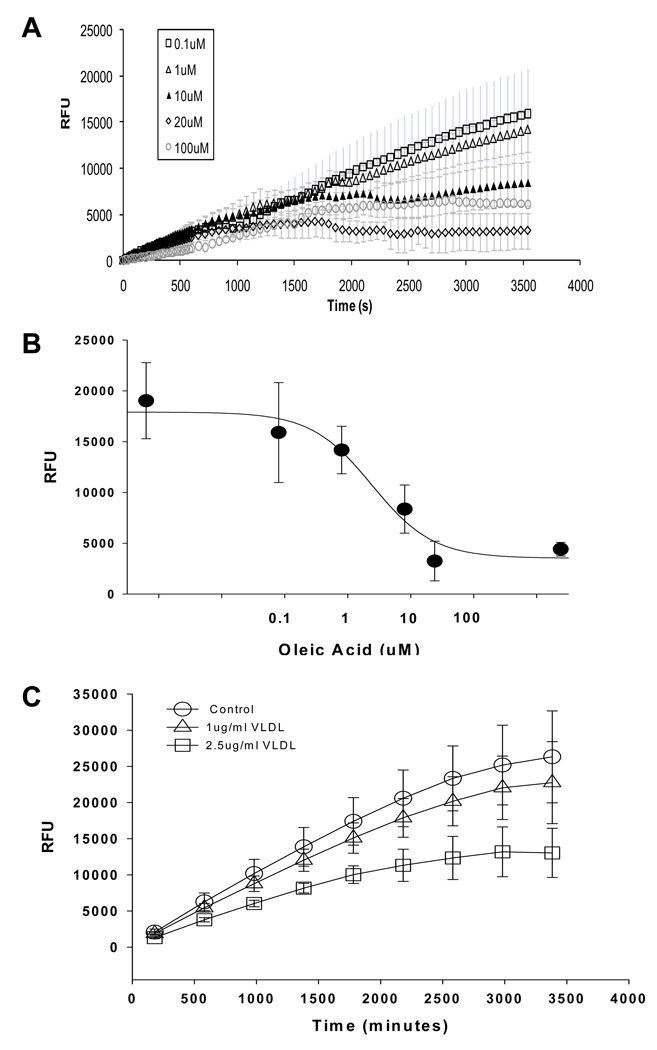
The fluorescently labeled fatty acid BODIPY-FA is actively transported through the cell membrane by the FATPs. Therefore, unlabeled fatty acid derivatives should act as competitive inhibitors to decrease the intracellular fluorescence. Natural substrate oleic acid competitively inhibited the FATP4 uptake with an IC50 of approximately 3.1 uM with 2 uM BODIPY-FA in the QBT mix solution (Figure 2A and B). Similarly, very-low-density lipoprotein (VLDL) cholesterol also decreaseed the uptake in a concentration-related manner (Figure 2C). However, oleic acid must be pre-equilibrated with the BODIPY-FA in the QBT mix and BSA for at least 24 hours before the assay. Without pre-equilibration, oleic acid would compete with BODIPY-FA for the BSA binding first. This would initially cause an increase of fluorescence because more free BODIPY-FA (without BSA binding) is available for the uptake. This process might be a reason for a great variability of the IC50 tested in separated experiments. Moreover, palmitic acid, another natural LCFA apparently did not inhibit fatty acid uptake by FATP4 or FATP5 (data not shown).
Figure 2.
Inhibition of fluorescently labeled BODIPY-FA uptake by natural long-chain fatty acids in the hsFATP4-expressing cells. A. Averaged signals (n=4 wells) in the presence of different oleic acid concentrations were plotted against recording time. Note: oleic acid was equilibrated with the BODIPY-FA in the QBT mix for 24 hours before placing onto the cells in wells. B. Concentration-response curve of the oleic acid in inhibition of BODIPY-FA uptake by FATP4 with an estimated IC50=3.1 uM. Data were taken from A. C. Averaged signals (n=4 wells) in the presence of different very-low-density lipoproteins (VLDL) concentrations were plotted against recording time. VLDL at 2.5 ug/ml inhibits 42% of the maximum uptake.
Selective inhibition of FATP4 by 4-aryl-dihydropyrimidinones
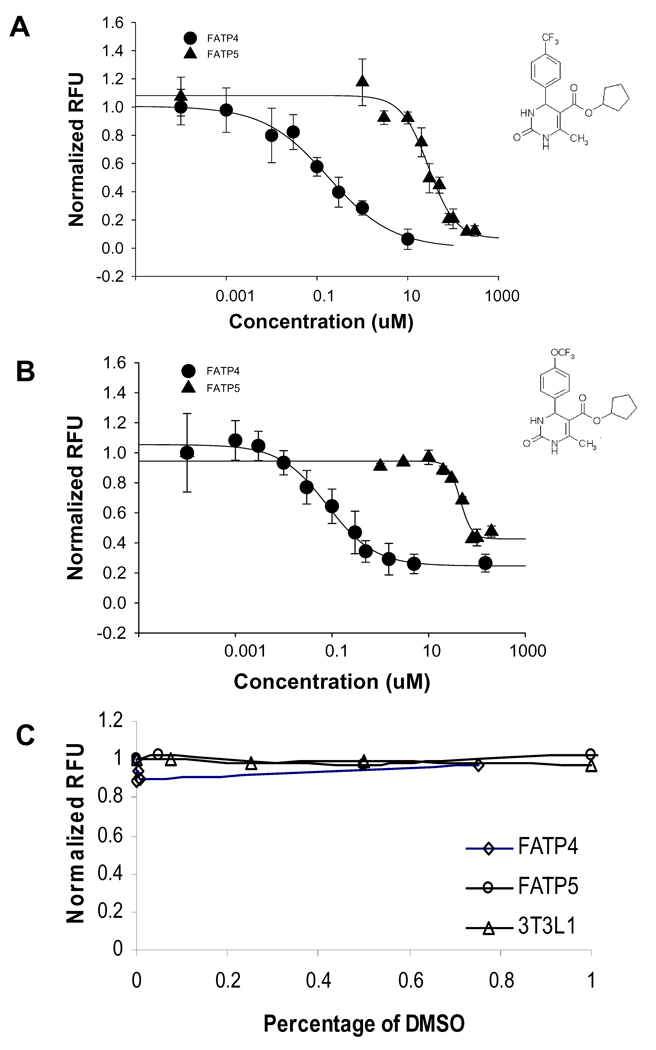
During development of the HTS assay, a report described, for the first time, a class of small molecules (4-aryl-dihydropyrimidinones) that produce potent inhibition of hsFATP4 29. Two selected compounds in the published chemical class, j3 and j5, were synthesized and tested. As shown in Figure 3A and B, compounds j3 and j5 produced potent inhibition of FATP4-mediated uptake, with IC50 values of 0.21 ± 0.22 uM (n=3 separate measurements) and 0.63 ± 0.33 uM (n=3 separate measurements), respectively. Furthermore, both compounds displayed approximately 100-fold greater selectivity for hsFATP4 over hsFATP5 with IC50 of 32 ± 5.2 uM for j3 (n=3 separate measurements) and 36 ± 9.2 uM for j5 (n=3 separate measurements). Neither compound blocked the baseline uptake by the vector-control cells (data not shown). The inhibition onset of j3 and j5 was rapid compared to the inhibition kinetics of the natural substrates tested (data not shown), possibly due to different binding kinetics of the compounds to the transporter. To eliminate potential binding rate differences that led to apparent selectivity, the drug incubation time was varied from 1 to 90 min with an incremental time at 15 min. The inhibitory effect of j5 proved consistent regardless of the compound incubation time (data not shown).
Figure 3.
Two small molecules j3 and j5 selectively inhibit fatty acid uptake through FATP4 over FATP5. A. Compound j3 is more potent against FATP4 (filled circle, IC50 = 0.21 ± 0.39 uM (n=3 plates) than against FATP5 (filled triangle IC50 =32.3 ± 8.7 uM). The insert is the j3 chemical structures. B. Compound j5 is more potent against FATP4 (filled circle, IC50 =0.63 ± 0.58 uM) than against FATP5 (filled triangle IC50 =36.3 ± 16.2 uM). The insert is the j5 chemical structures. Note: each IC50 value was averaged from n=3 separate measurements (different plates), while the curves plotting normalized RFU against compound concentrations shown here are from one representative plate with multiple wells measurements (n≥3 wells). C. Effects of different concentrations of DMSO on normalized RFU in FATP4 (circle), FATP5 (square) and activated 3T3L1 adipocytes (triangles). DMSO was added 30 minutes prior to the QBT mixture addition.
The highest concentration of dimethylsulfoxide (DMSO) contained in the j3 and j5 testing wells was 0.1%. Since in primary screening most compounds are dissolved in DMSO as stock solutions, we examined assay tolerance to this most commonly used solvent. Varying concentrations of DMSO (0.05 – 1%) were added to hsFATP4- or 5-cells, vector-control cells and 3T3 L1 adipocytes, and incubated for 30 min at 37°C prior to LCFA uptake measurement by the FLIPR. DMSO at ≤1 % at the preincubation time used here apparently did not cause significant signal changes in hsFATP4, hsFATP5 and 3T3L-1 cells (Figure 3C).
Pilot drug screen against hsFATP5
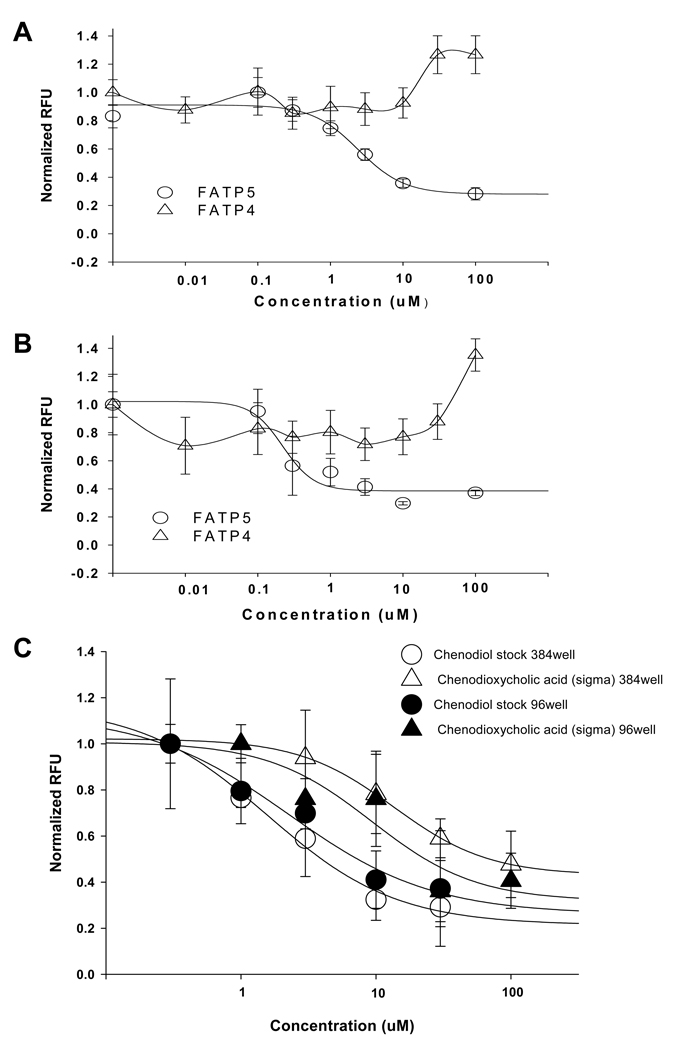
We further evaluated the quality and reproducibility of the assay by conducting a pilot screening of the USDC library consisting of 1,040 compounds. By the time we developed this assay, selective FATP4 inhibitors (such as j3 and j5) just became available 29, but no FATP5 specific inhibitors or HTS assay against FATP5 target have been reported thus far. We therefore concentrated on our initial screening against hsFATP5. All test compounds were dissolved in DMSO at 10 mM as stock solutions and screened at a final single concentration of 10 uM in a single well measurement. For each plate, the identification of active compounds (hits) was based the z score of each compound as described in the section of Material and Methods. Compounds with z scores ≤ −3 indicate potential inhibiting, while ≥ +3 enhancing BODIPY-FA uptake. Using this selection criterion, 9 hits out of 1040 were identified. In the following concentration-response tests using the same assay in a duplicated measurement, 4 inhibitory compounds were verified among the initial hits, indicating a hit rate at 0.4% for the USDC library. Two of the confirmed hits were bile acids, chenodiol (chenodeoxycholic acid) and ursodiol (ursodeoxycholic acid) 31. Further tests revealed that chenodiol and ursodiol caused concentration-dependent inhibition of hsFATP5 with an IC50 of 2.4 uM and 0.22 uM, respectively (Figure 4A and B). Interestingly, both compounds, up to 30 uM, did not inhibit, while at 100 uM appeared to increase the fatty acid uptake by the hsFATP4, suggesting these two bile acids have apparent selectivity between these two subtypes of hsFATPs. A screening window coefficient, termed the “Z’ factor” 30 was determined for the assay ranging from 0.5 to 0.7, with a mean of 0.6 ± 0.2 (n = 7 plates). These values indicate that the assay is consistent and has good reproducibility.
Figure 4.
A. Concentration-dependent inhibition curves of chenodiol (USDC, i.e., chenodeoxycholic acid, Sigma) against FATP5 (open circle, IC50 = 2.4 uM) and FATP4 (open triangle; no inhibition, but with an apparent signal increase at 30 and 100 uM). B. Concentration-dependent inhibition curves of ursodiol (USDC, i.e., ursodeoxycholic acid, Sigma) against FATP5 (open circle, IC50 = 0.22 uM) and FATP4 (open triangle, no inhibition but with an apparent signal increase at 100 uM). C. Concentration-dependent inhibition curves of chenodiol (USDC) and chenodeoxycholic acid (Sigma) measured by Analyst HT in 384-well format (open triangle) or by a FLIPR in 96-well format (close circle). The IC50 of chenodiol was estimated at 2.3 uM obtained by 384-well assay while the values of 1.6 – 2.4 uM obtained from the 96-well assay. The IC50 value of chenodioxycholic acid was estimated at 13.5 uM in 384-well (open triangle) while 9.3 uM in 96-well format (close triangle).
Single-time-point fluorescence intensity measurement in 384-well plate format
To demonstrate this fluorescence-based hsFATP-cell assay can be run in an HTS format, the assay was modified into a 384-well format with a single-time-point measurement using a conventional plate reader (Analyst HT). Cells expressing hsFATP5 were plated in 20,000/well 12–16 hours before the assay. After loading of dye, the fluorescence intensity of each well was measured immediately (as the baseline) and at 1 hour time-point following dye incubation. Seven plates were tested with Z’ = 0.6 ± 0.1, similar to that observed in a 96-well assay. The IC50 of chenodiol (2.3 uM) obtained in 384-well is consistent with the value (1.6 – 2.4 uM) obtained from 96-well assay. Chenodeoxycholic acid (Sigma) is the same chemical as chenodiol (US Drug Collection). The IC50 of chenodeoxycholic acid from Sigma obtained in 384-well 13.5 uM is also similar to its potency (9.3 uM) determined by a FLIPR in 96 well. These results indicate the single-time-point measurement in 384-well format is suitable for primary HTS.
LCFA uptake assay in 3T3 L1 adipocytes
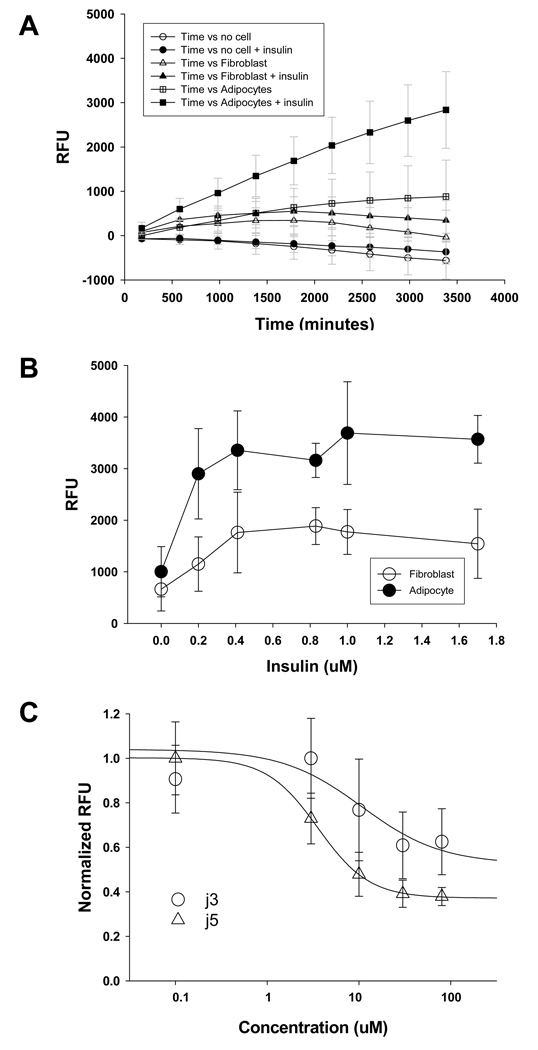
LCFA uptake by native cells is likely to be more complex than by a recombinant cloned FATP expressed in a mammalian cell line. Moreover, this process is hormone-regulated. We therefore compared stimulation and inhibition of fatty acid uptake by cells endogenously expressing FATPs. To this end we started with 3T3 L1 adipocytes, which have been shown to express both FATP1 and 4. Differentiation protocol for 3T3-L1 cells followed the published procedure 24,25 and the differentiated cells were assessed by visual inspection of lipid droplet accumulation under a microscope. 3T3-L1 adipocytes or undifferentiated fibroblasts were seeded into 96-well plates at 100,000 cells/well and treated with insulin (0.2 – 1.7 uM) as indicated in the Figure 5. Uptake kinetics of fluorescently labeled fatty acid BODIPY-FA by the 3T3-L1 cells were measured by the FLIPR within 1 min after the addition of the QBT mix solution (100 ul/well). Insulin greatly enhanced uptake in a concentration-dependent manner in the differentiated adipocytes, but had much weaker effects in their precursor’s fibroblasts or preadipocytes (Figure 5A and B). The maximal insulin-induced uptake over the baseline of adipocytes was about two- to three-fold, consistent with the results obtained in the previous report using Flexsation fluorescence plate readers 24. The two FAPT4 inhibitors j3 and j5 partially blocked fatty acid uptake with IC50 of 9.6 uM and 3.6 uM, respectively in the 3T3-L1 adipocytes (Figure 5C). The inhibition potency against 3T3-L1 adipocytes fell between the potency against FATP4 (IC50 of 0.21 ul and 0.63 ul for j3 and j5, respectively) and FATP5 (IC50 of 32 and 36 uM for j3 and j5, respectively). 3T3-L1 adipocytes have been shown to express both FATP1 and 4. Our data suggests that j3 and j5 might have weaker effect on FATP1 compared to FATP4. Alternatively, some cellular components in native cells may interact with the compounds and alter the drug potency. The two compounds had no effects on undifferentiated 3T3-LI preadipocytes (data not shown) suggesting specific drug action on the expressed FATPs in the differentiated adipocytes.
Figure 5.
Fatty acid dye uptake in differentiated 3T3L1 adipocytes. A. Incubation of 1 uM insulin substantially increases the fatty acid dye uptake in differentiated 3T3-L1 adipocytes (filled square n=20 wells vs. open square n = 15), while it has a minimal effect on the uptake of non-differentiated 3T3-L1 fibroblast cells (filled triangle, n = 16 vs. open triangle n = 12). B. The fatty acid uptake in adipocytes (filled circle) potentiated by insulin was greater than that in fibroblast cells (open circle). C. Compounds j3 and j5 inhibit the uptake by 3T3L1 adipocytes with IC50 9.8 uM for j3 and 3.6 uM for j5, respectively.
Inhibition of LCFA uptake by FATP4 inhibitors in mouse enterocytes
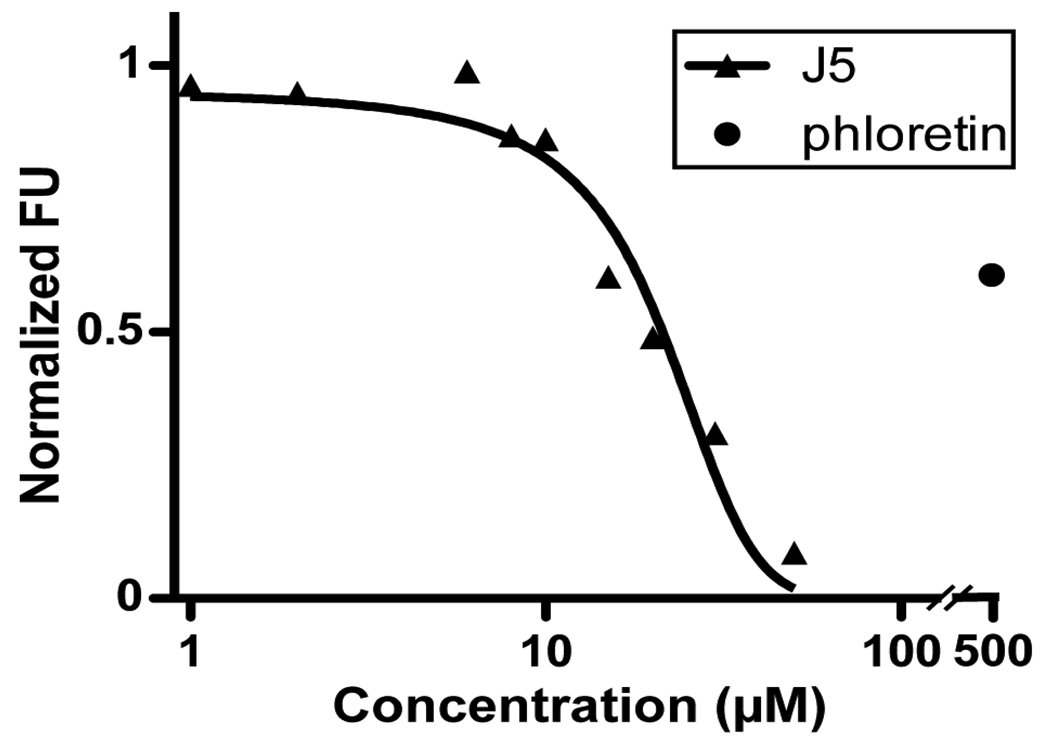
FATP4 is the primary intestinal LCFA transporter and should be inhibited by the FATP4 inhibitors, such as j5. Thus it is physiologically relevant to determine the effect of j5 on LCFA uptake by isolated primary mouse enterocytes. Using primary enterocytes for the FLIPR assay would require large number of cells and will make it time consuming and costly. Moreover, the non-adherent nature of enterocytes would affect the consistency of the assay. As an alternative we utilized a flow cytometer based assay in conjunction with BODIPY-fatty acid to measure short-term LCFA uptake following isolation of duodenal enterocytes. Enterocytes were exposed for 60 s to a 2 uM BODIPY-FA solution containing several different concentrations of j5 as indicated in Figure 3. The cell viability was assessed with propidium iodide and LCFA uptake by viable cells was determined by FACS. Addition of j5 robustly inhibited LCFA uptake by enterocytes with an apparent IC50 of 21 uM. Maximal reduction of enterocyte LCFA uptake by j5 surpassed inhibition by the widely used nonspecific inhibitor phloretin (500 uM; Figure 6). FATP4 is the only FATP expressed in the small intestine. The different inhibitory potency of j5 observed between the FATP4 stable cell line and the primary enterocytes might result from different assay sensitivities or from different environmental factors in the native enterocytes and mammalian cell line.
Figure 6.
FACS based LCFA uptake assay using acutely dissociated mouse enterocytes. Cells were incubated with j5 (1 – 30 uM) or phloretin (500 uM), a non-specific FATP inhibitor. FACS was used to gate on viable cells and determine LCFA uptake expressed as % of control (containing 0.5% DMSO). Curve represents a non-linear regression (R2 = 0.96) with an estimated IC50 of 21 uM.
DISCUSSION
HTS assay viability using the stable cell lines expressing recombinant hsFATPs
Our study shows that the hsFATP4 and 5 cell-based fluorescence assays using a FLIPR or a conventional fluorescence plate reader are suitable for primary drug screening. Cellular LCFA uptake involves both passive diffusion and FATP-mediated transport. The background of fluorescence fatty acid uptake could be due to the passive diffusion or transportation by endogenous FATPs in HEK293 cells. The established hsFATP4 and 5 stable cell lines have substantial and significant signal-to-background ratio compared to vector-control cell line (Figure 1). The current hsFATP1 cell line had poor signal, indicating further dilution cloning or optimization of expression conditions is required in future assay development for this subtype. The fluorescence-labeled LCFA uptake can be inhibited by certain natural LCFAs (Figure 2). Furthermore, examinations of compounds j3 and j5 not only confirmed the previously reported hsFATP4 inhibitory potencies 29, but also found that these two inhibitors have more than 100-fold selectivity over hsFAPT5 (Figure 3). The assay has good tolerance to DMSO up to 1% with 30 minutes incubation prior to the measurement (Figure 3C).
In the pilot screen using a small compound library of approximately 1,000 (USDC) against the hsFATP5 to evaluate assay reproducibility, a Z’ factor ranging from 0.5 to 0.7 was obtained, indicating good assay reproducibility. Interestingly, two apparent hsFATP5 inhibitors identified in random screening of the USDC library were bile acids (Figure 4).
Our assay targeted two human fatty acid transporters, hsFATP4 and 5 expressed in a human mammalian cell line, which differs from the studies published by Li et al 27,28 where a different fluorescent quencher, trypan blue, and C1-BODIPY-C12 were combined to develop a yeast cell-based assay against murine or human FATP2 27,28. This assay relies on the functional expression of mammalian FATPs in a yeast strain lacking the endogenous fatty acid transporter. While the use of yeast cells allows for rapid cell proliferation it has the disadvantage of reconstituting the function of a mammalian transporter. Indeed, half of the six mammalian FATPs, including FATP5, do not enhance C1-BODIPY-C12 uptake when expressed in yeast, while these molecules are clearly able to enhance free fatty acids uptake when expressed in mammalian cells. The HEK cell-based assay presented here can be more easily adopted into typical primary drug screening pipelines along with other mammalian cells expressing different receptors or ion channels using common instruments such as a FLIPR 33 or a conventional plate reader, for example, Analyst HT.
Differential effects of FATP4 inhibitors and bile acids against hsFATP4 and hsFATP5
The two confirmed hits chenodiol (i.e., chenodeoxycholic acid) is primary bile acid formed by synthesis in the liver; while ursodiol (also known as ursodeoxycholic acid), is one of the secondary bile acids, which are metabolic byproducts of intestinal bacteria. Bile acids are known as substrates of FATP5 in the liver. FATP5 has been shown not only be able to transport fatty acid through the cell membrane but also possesses bile acid-CoA ligase activity 21. It is unclear whether the bile acids act as inhibitors directly on the transport protein, or as substrates for bile acid-CoA ligase and indirectly inhibiting the uptake. The two bile acids have no inhibitory effect on the fatty acid uptake through FATP4, indicating that the bile acids cannot bind to the hsFATP4.
Secondary screens using native cells expressing FATPs
It is important to verify the hits identified from primary screening using recombinant cell lines with native cells that endogenously express the transporter of interests. 3T3-L1 adipocytes are murine in origin and express several proteins involved in LCFA uptake with the FATP1 and 4 as the predominant subtypes 25. The reduced potency of the two FATP4 inhibitors j3 and j5 in 3T3-L1 adipocytes compared to the recombinant hsFATP4 cell line suggests that the compounds may also have higher selectivity with FATP4 than FATP1. However, we also found a reduced efficiency of j5 in murine enterocytes, which do not express FATP1. This may indicate that in primary cells FATPs, the compound may interact with other accessory proteins such as CD36. Alternatively, the accessibility of the compounds to the native FATP proteins is limited. Unlike primary enterocytes, 3T3-L1 adipocytes could be used for secondary screening with a FLIPR. However, we found that the differentiation process was not only time-consuming (8–12 days from initiation of differentiation to the assay), but also inconsistent, yielding great variability in signal-to-background ratios among different batches of differentiated cells. Other native cell lines such as Caco-2 and HepG2 cells and three endothelial cell lines (b-END3, HAEC, and HMEC) express different FATPs could potentially be used in secondary screening 28,32.
Potential drug classes acting as inhibitors of FATPs
A family of tricyclic, phenothiazine-derived drugs was shown to inhibit the hsFATP2 expressed in yeast and such effects were linked to the possible cause of metabolic side effects, including hypertriglyceridemia in patients chronically taking this class of antipsychotic drugs 28. There are 21 compounds labeled as “antipsychotic” drugs in the USDC library. In our pilot primary screening, these drugs did not produce any inhibition on the hsFATP5, suggesting these antipsychotic drugs might have selectivity against FATP2 over FATP5, or resulted from the different expression systems. In contrast, our finding for the first time shows that bile acids cause concentration-dependent inhibition of LCFA uptake by FATP5 with apparent selectivity over FATP4. Bile acids have long been viewed as digestive detergents to facilitate fat absorption in small intestine, and the primary role in regulation of cholesterol catabolism. However, bile acids are now recognized as hormones involved in the regulation of various metabolic processes 31. Our preliminary finding that bile acids can selectively inhibit the liver-specific FATP5, without effects on the intestine-predominant FATP4 further supports the notion of bile acids functioning as hormones, and might have clinical implications. In conclusion, we have developed an HTS assay employing fluorescent fatty acid uptake by the recombinant human FATP4 and FATP5 stably expressed in HEK cells. This assay can be used for primary HTS for discovering novel compounds against hsFATP4 or 5, while the 3T3-L1 adipocytes or primary cells (e.g., enterocytes or hepatocytes) could be suitable in secondary screening for hit confirmation, lead identification and optimization.
Acknowledgements
This work was supported by NIH R21NS57052 (Xie), and R01 MH078194 (Xie). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Neurological Disorders and Stroke and the National Institute of Mental Health. We are grateful to Dr. Ling Jong for synthesis of compounds j3 and j5 for our research.
REFERENCE
- 1.Schaffer JE. A novel adipocyte long chain fatty acid transport protein. Eur J Med Res. 1996 Jan 19;1(4):176–180. [PubMed] [Google Scholar]
- 2.Stahl A, Gimeno RE, Tartaglia LA, Lodish HF. Fatty acid transport proteins: a current view of a growing family. Trends Endocrinol Metab. 2001 Aug;12(6):266–273. doi: 10.1016/s1043-2760(01)00427-1. [DOI] [PubMed] [Google Scholar]
- 3.Stremmel W, Pohl L, Ring A, Herrmann T. A new concept of cellular uptake and intracellular trafficking of long-chain fatty acids. Lipids. 2001 Sep;36(9):981–989. doi: 10.1007/s11745-001-0809-2. [DOI] [PubMed] [Google Scholar]
- 4.Stahl A. A current review of fatty acid transport proteins (SLC27) Pflugers Arch. 2004 Feb;447(5):722–727. doi: 10.1007/s00424-003-1106-z. [DOI] [PubMed] [Google Scholar]
- 5.Ehehalt R, Fullekrug J, Pohl J, Ring A, Herrmann T, Stremmel W. Translocation of long chain fatty acids across the plasma membrane--lipid rafts and fatty acid transport proteins. Mol Cell Biochem. 2006 Mar;284(1–2):135–140. doi: 10.1007/s11010-005-9034-1. [DOI] [PubMed] [Google Scholar]
- 6.Hui TY, Bernlohr DA. Fatty acid transporters in animal cells. Front Biosci. 1997;2:d222–d231. doi: 10.2741/a185. [DOI] [PubMed] [Google Scholar]
- 7.Fitscher BA, Riedel HD, Young KC, Stremmel W. Tissue distribution and cDNA cloning of a human fatty acid transport protein (hsFATP4) Biochim Biophys Acta. 1998 Dec 22;1443(3):381–385. doi: 10.1016/s0167-4781(98)00231-0. [DOI] [PubMed] [Google Scholar]
- 8.Pohl J, Ring A, Ehehalt R, Herrmann T, Stremmel W. New concepts of cellular fatty acid uptake: role of fatty acid transport proteins and of caveolae. Proc Nutr Soc. 2004 May;63(2):259–262. doi: 10.1079/PNS2004341. [DOI] [PubMed] [Google Scholar]
- 9.Pohl J, Ring A, Hermann T, Stremmel W. Role of FATP in parenchymal cell fatty acid uptake. Biochim Biophys Acta. 2004 Nov 8;1686(1–2):1–6. doi: 10.1016/j.bbalip.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Gimeno RE. Fatty acid transport proteins. Curr Opin Lipidol. 2007 Jun;18(3):271–276. doi: 10.1097/MOL.0b013e3281338558. [DOI] [PubMed] [Google Scholar]
- 11.Gimeno RE, Hirsch DJ, Punreddy S, et al. Targeted deletion of fatty acid transport protein-4 results in early embryonic lethality. J Biol Chem. 2003 Dec 5;278(49):49512–49516. doi: 10.1074/jbc.M309759200. [DOI] [PubMed] [Google Scholar]
- 12.Gimeno RE, Ortegon AM, Patel S, et al. Characterization of a heart-specific fatty acid transport protein. J Biol Chem. 2003 May 2;278(18):16039–16044. doi: 10.1074/jbc.M211412200. [DOI] [PubMed] [Google Scholar]
- 13.Koonen DP, Glatz JF, Bonen A, Luiken JJ. Long-chain fatty acid uptake and FAT/CD36 translocation in heart and skeletal muscle. Biochim Biophys Acta. 2005 Oct 1;1736(3):163–180. doi: 10.1016/j.bbalip.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 14.Richards MR, Harp JD, Ory DS, Schaffer JE. Fatty acid transport protein 1 and long-chain acyl coenzyme A synthetase 1 interact in adipocytes. J Lipid Res. 2006 Mar;47(3):665–672. doi: 10.1194/jlr.M500514-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Duttaroy AK. Transport of fatty acids across the human placenta: a review. Prog Lipid Res. 2009 Jan;48(1):52–61. doi: 10.1016/j.plipres.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Stahl A, Evans JG, Pattel S, Hirsch D, Lodish HF. Insulin causes fatty acid transport protein translocation and enhanced fatty acid uptake in adipocytes. Dev Cell. 2002 Apr;2(4):477–488. doi: 10.1016/s1534-5807(02)00143-0. [DOI] [PubMed] [Google Scholar]
- 17.Kansara MS, Mehra AK, Von Hagen J, Kabotyansky E, Smith PJ. Physiological concentrations of insulin and T3 stimulate 3T3-L1 adipocyte acyl-CoA synthetase gene transcription. Am J Physiol. 1996 May;270(5 Pt 1):E873–E881. doi: 10.1152/ajpendo.1996.270.5.E873. [DOI] [PubMed] [Google Scholar]
- 18.Steinberg GR, Bonen A, Dyck DJ. Fatty acid oxidation and triacylglycerol hydrolysis are enhanced after chronic leptin treatment in rats. Am J Physiol Endocrinol Metab. 2002 Mar;282(3):E593–E600. doi: 10.1152/ajpendo.00303.2001. [DOI] [PubMed] [Google Scholar]
- 19.Basu A, Basu R, Shah P, Vella A, Rizza RA, Jensen MD. Systemic and regional free fatty acid metabolism in type 2 diabetes. Am J Physiol Endocrinol Metab. 2001 Jun;280(6):E1000–E1006. doi: 10.1152/ajpendo.2001.280.6.E1000. [DOI] [PubMed] [Google Scholar]
- 20.Fisher RM, Gertow K. Fatty acid transport proteins and insulin resistance. Curr Opin Lipidol. 2005 Apr;16(2):173–178. doi: 10.1097/01.mol.0000162322.39548.b1. [DOI] [PubMed] [Google Scholar]
- 21.Doege H, Baillie RA, Ortegon AM, et al. Targeted deletion of FATP5 reveals multiple functions in liver metabolism: alterations in hepatic lipid homeostasis. Gastroenterology. 2006 Apr;130(4):1245–1258. doi: 10.1053/j.gastro.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Herrmann T, van der Hoeven F, Grone HJ, et al. Mice with targeted disruption of the fatty acid transport protein 4 (FATP 4, Slc27a4) gene show features of lethal restrictive dermopathy. J Cell Biol. 2003 Jun 23;161(6):1105–1115. doi: 10.1083/jcb.200207080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hubbard B, Doege H, Punreddy S, et al. Mice deleted for fatty acid transport protein 5 have defective bile acid conjugation and are protected from obesity. Gastroenterology. 2006 Apr;130(4):1259–1269. doi: 10.1053/j.gastro.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 24.Liao J, Sportsman R, Harris J, Stahl A. Real-time quantification of fatty acid uptake using a novel fluorescence assay. J Lipid Res. 2005 Mar;46(3):597–602. doi: 10.1194/jlr.D400023-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Ho M, Foxall S, Higginbottom M, et al. Leptin-mediated inhibition of the insulin-stimulated increase in fatty acid uptake in differentiated 3T3-L1 adipocytes. Metabolism. 2006 Jan;55(1):8–12. doi: 10.1016/j.metabol.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 26.Stahl A, Hirsch DJ, Gimeno RE, et al. Identification of the major intestinal fatty acid transport protein. Mol Cell. 1999 Sep;4(3):299–308. doi: 10.1016/s1097-2765(00)80332-9. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Black PN, DiRusso CC. A live-cell high-throughput screening assay for identification of fatty acid uptake inhibitors. Anal. Biochem. 2005 Jan 1;336(1):11–19. doi: 10.1016/j.ab.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Black PN, Chokshi A, et al. High-throughput screening for fatty acid uptake inhibitors in humanized yeast identifies atypical antipsychotic drugs that cause dyslipidemias. J Lipid Res. 2008 Jan;49(1):230–244. doi: 10.1194/jlr.D700015-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Blackburn C, Guan B, Brown J, et al. Identification and characterization of 4-aryl-3,4-dihydropyrimidin-2(1H)-ones as inhibitors of the fatty acid transporter FATP4. Bioorg Med Chem Lett. 2006 Jul 1;16(13):3504–3509. doi: 10.1016/j.bmcl.2006.03.102. [DOI] [PubMed] [Google Scholar]
- 30.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 31.Staels B, Fonseca VA. Bile acids and metabolic regulation: mechanisms and clinical responses to bile acid sequestration. Diabetes Care. 2009 Suppl 2:S237–S245. doi: 10.2337/dc09-S355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandoval A, Fraisl P, Arias-Barrau E, et al. Fatty acid transport and activation and the expression patterns of genes involved in fatty acid trafficking. Arch Biochem Biophys. 2008 Sep 15;477(2):363–371. doi: 10.1016/j.abb.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Xie XM, Van Deusen AL, Vitko I, Barrett PQ, Huynh N, Cheng H, Yang N, Edward Perez-Reyes E. Validation of high throughput screening assays against three subtypes of Cav3 T-type channels using molecular and pharmacologic approaches. Assay and Drug Development Technologies. 2007;5(2):191–203. doi: 10.1089/adt.2006.054. [DOI] [PubMed] [Google Scholar]