Abstract
Purpose
We studied the expression levels of cyclins B1, D1, and E1 and the implications of cyclin overexpression for patient outcomes in distinct breast cancer subtypes defined by clinical variables and transcriptional profiling.
Experimental Design
The expression levels of cyclins B1, D1, and E1 were quantified in 779 breast tumors and 53 cell lines using reverse phase protein arrays and/or transcriptional profiling.
Results
Whereas cyclin E1 overexpression was a specific marker of triple-negative and basal-like tumors, cyclin B1 overexpression occurred in poor prognosis hormone receptor–positive, luminal B and basal-like breast cancers. Cyclin D1 overexpression occurred in luminal and normal-like cancers. Breast cancer subgroups defined by integrated expression of cyclins B1, D1, and E1 correlated significantly (P < 0.000001) with tumor subtypes defined by transcriptional profiling and clinical criteria. Across three hormone receptor–positive data sets, cyclin B1 was the dominant cyclin associated with poor prognosis in univariate and multivariate analyses. Although CCNE1 was present in significantly higher copy numbers in basal-like versus other subtypes (ANOVA P < 0.001), CCNB1 gene copy number did not show gain in breast cancer. Instead, cyclin B1 expression was increased in tumors with co-occurrence of TP53 mutations and MYC amplification, a combination that seems to characterize basal-like and luminal B tumors. CCNB1 gene expression was significantly correlated with PLK, CENPE, and AURKB gene expression.
Conclusion
Cyclins B1, D1, and E1 have distinct expressions in different breast cancer subtypes. Novel PLK, CENPE, and AURKB inhibitors should be assessed for therapeutic utility in poor prognosis cyclin B1–overexpressing breast cancers.
Cyclins are critical to cell cycle progression. Cyclins D1 and E1 are well studied in breast cancer (1–10). There is a strong association between cyclin D1 expression and estrogen receptor (ER) positivity and between cyclin E1 overexpression and ER negativity. Whereas the relationship between cyclin D1 expression and breast cancer outcomes is somewhat controversial, cyclin E1 overexpression has been consistently associated with an increased risk of cancer relapse and death (8, 9). However, cyclin B1 is not nearly as extensively studied in breast cancer as either cyclin D1 or cyclin E1 (11).
Breast cancer represents at least three clinical subtypes based on the expression of the hormone receptors ER and progesterone receptor (PR) and on the presence or absence of HER2 amplification [i.e., hormone receptor (ER and/or PR)–positive, HER2-positive, and triple negative]. The intrinsic gene-based classification is derived using transcriptional profiling and subdivides breast cancer into luminal A and B, HER2-positive, basal-like, and normal-like subtypes (12–14). There is significant overlap between the two classification systems, with triple-negative cancers expressing a basal-like profile and luminal A and B cancers composed largely of hormone receptor–positive tumors (15). The outcomes of patients with luminal B cancers are inferior to those of patients with luminal A cancers.
The comparative expression of cyclins B1, D1, and E1 across these distinct breast cancer subtypes along with the relevance of cyclin overexpression to patient outcomes specifically within these subtypes have not been well reported. Further, the clinical significance of cyclin B1 overexpression in breast cancer is not as well defined as that of cyclin D1 or E1. Thus, we applied reverse phase protein arrays (RPPA; refs. 16–19) and/or transcriptional profiling to quantify the expression of cyclins B1, D1, and E1 in 779 breast tumors and 53 breast cancer cell lines. We show that the expression levels of cyclins B1, D1, and E1 are differentially deregulated in different breast cancer subtypes. Cyclin B1 overexpression is associated with the combination of TP53 mutation and MYC amplification. In three independent tumor sets, cyclin B1 is the dominant cyclin associated with poor prognosis in hormone receptor–positive breast cancer in both univariate and multivariate analyses. Coordinate overexpression of cyclins B1 and D1 and of cyclins B1 and E1 is associated with adverse patient outcomes across all breast cancers and specifically in hormone receptor–positive breast cancers.
Materials and Methods
Human breast tumor samples
Four tumor cohorts were collected under institutional review board–approved protocols. The first cohort (A) was composed of 390 primary breast tumors obtained from the Breast Tissue Frozen Tumor Bank at the M.D. Anderson Cancer Center (Supplementary Table S1). These tumors were subdivided into three clinically relevant categories by immunohistochemistry or RPPA for ER and PR status and by immunohistochemistry, fluorescent in situ hybridization, or RPPA for HER2 status. Transcriptional profiling data were not available for this tumor set.
The second cohort (B) of 168 primary breast tumors was collected from patients enrolled in the Danish DBCG82 b and c breast cancer studies (Supplementary Table S2; ref. 20). These tumors were classified into one of the five intrinsic breast cancer subtypes by transcriptional profiling (12–14).
The third cohort (C) of 51 hormone receptor (ER and/or PR)–positive breast cancers was obtained from a centralized reference laboratory (Quantitative Diagnostic Laboratories; ref. 8). Each patient was diagnosed with breast cancer between 1990 and 1995 at 1 of 12 Chicago area hospitals. Patients in this cohort did not receive adjuvant hormonal therapy or chemotherapy.
The patient characteristics, p53 and Ki67 status (both assessed by immunohistochemistry; ref. 21), gene expression, and gene copy number (aCGH) profiles pertaining to the fourth cohort (D) of 170 breast tumors from the University of California San Francisco Comprehensive Cancer Center Breast Oncology Program Tissue Bank have previously been published (21, 22). Supplementary Fig. S1 summarizes the sample sets used in this study.
Breast cancer cell lines
The lysates of 53 cell lines (Supplementary Table S3) were obtained from the Lawrence Berkeley National Laboratory. The cell lines were cultured in complete medium (RPMI supplemented with 5% fetal bovine serum) before protein extraction.
Reverse phase protein lysate microarray
Protein was extracted from tumors in cohorts A to C and from cell lines and probed for expression of cyclins B1 (1495-1 antibody, Epitomics, Inc.) and D1 and E1 (sc-247 and sc-718 antibodies, Santa Cruz Biotechnology, Inc., respectively) by RPPA as previously described (16–19).
Transcriptional profiling
The microarray system applied to cohort B was the Applied Biosystems Human Genome Survey Microarray version 2.0. Details of the array procedure can be found at the Web site.9 These microarray data were used to apply the 70-gene Amsterdam (Mammaprint; ref. 23) and the 76-gene (24) and genomic grade signatures (25) to cohort B to facilitate a comparison between these genomic signatures and cyclin B1 protein expression levels. However, because of the differences between the Applied Biosystems platform applied to cohort B and the platforms used to develop these signatures, only the 70-gene Amsterdam (Mammaprint; ref. 23) signature could successfully be applied to the transcriptional data for cohort B.
The microarray system applied to breast cancer cell lines and cohort D was the Affymetrix high-density oligonucleotide array human HG-U133A chip (21, 22). These data are available in the CaBIG repository,10 in the Berkeley Lab database,11 and in the ArrayExpress Web site,12 with accession number E-TABM-157/158.
Mass spectroscopy–based approach evaluating PIK3CA mutation status
A mass spectroscopy–based approach evaluating single nucleotide polymorphisms was used to detect 23 known mutations in PIK3CA (PIK3-CA_A1046V, PIK3CA_C420R, PIK3CA_E110K, PIK3CA_E418K, PIK3CA_E453K, PIK3CA_E542K, PIK3CA_E545K, PIK3CA_F909L, PIK3CA_G1049R, PIK3CA_G451L456_V, PIK3CA_H1047L, PIK3-CA_H1047R, PIK3CA_H1047Y, PIK3CA_H701P, PIK3CA_K111N, PIK3CA_M1043V, PIK3CA_N345K, PIK3CA_P539R, PIK3CA_Q060K, PIK3CA_Q546E, PIK3CA_R088Q, PIK3CA_S405F and PIK3-CA_T1025S; ref. 17). PCR and extension primers for PIK3CA were designed using Sequenom, Inc., Assay Design. PCR-amplified DNA was cleaned using EXO-SAP (Sequenom); the primer was extended by IPLEX chemistry, desalted using Clean Resin (Sequenom), and spotted onto Spectrochip matrix chips using a nanodispenser (Samsung). Chips were run in duplicate on a Sequenom MALDI-TOF MassArray system. Sequenom Typer Software and visual inspection were used to interpret mass spectra. Reactions where >15% of the resultant mass ran in the mutant site in both reactions were scored as positive.
Statistical analysis
The RPPA spot signal intensity data from Micro-Vigene (VigeneTech, Inc.) was processed by the R package SuperCurve (version 1.01; ref. 18).13 A fitted curve (called “supercurve”) was plotted with the signal intensities on the Y axis and the relative log 2 concentration of each cyclin protein on the X axis using the nonparametric, monotone-increasing B-spline model (18). The protein concentrations were derived from supercurve for each sample lysate on the slide by curve fitting and then normalized by median polish. Each cyclin protein measurement was subsequently corrected for loading as previously described (16–19).
Means and SDs were generated for expression of each cyclin and compared between groups using two-sample t tests and ANOVA. To examine survival, the time to death or censoring was computed in years since diagnosis for each patient, and recurrence-free survival (RFS) time to event was computed as years to first relapse or death after diagnosis for each patient. Overall survival (OS) time was censored at the date of the last follow-up if death was not observed. RFS time was censored at the date of the last follow-up if no relapses were observed and death was not observed. RFS and OS probabilities were estimated using the Kaplan-Meier product limit method as well as univariable and multivariable Cox models. These statistical analyses were done using the SPSS16.0 (SPSS, Inc.) and NCSS/PASS softwares. Reported P values are two-sided.
For clustering and heat map generation, the softwares X cluster and Treeview were used.
Results
Cyclins display breast cancer subtype–specific expression
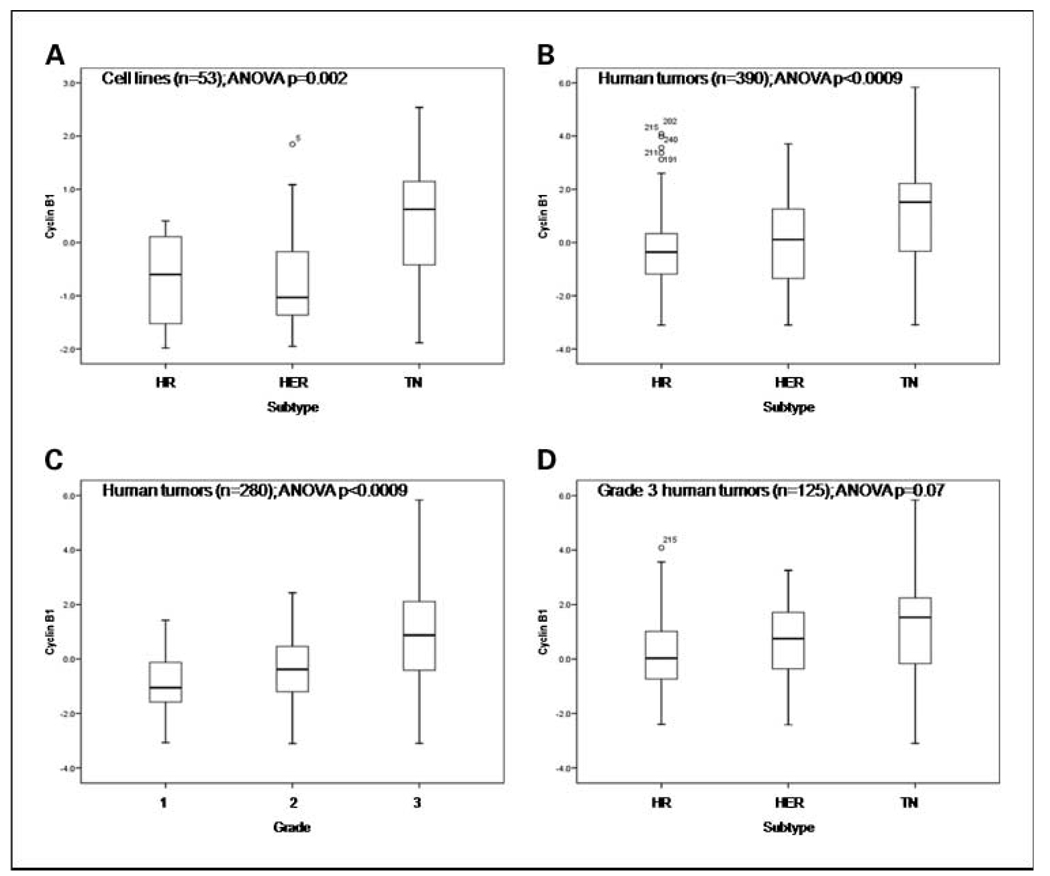
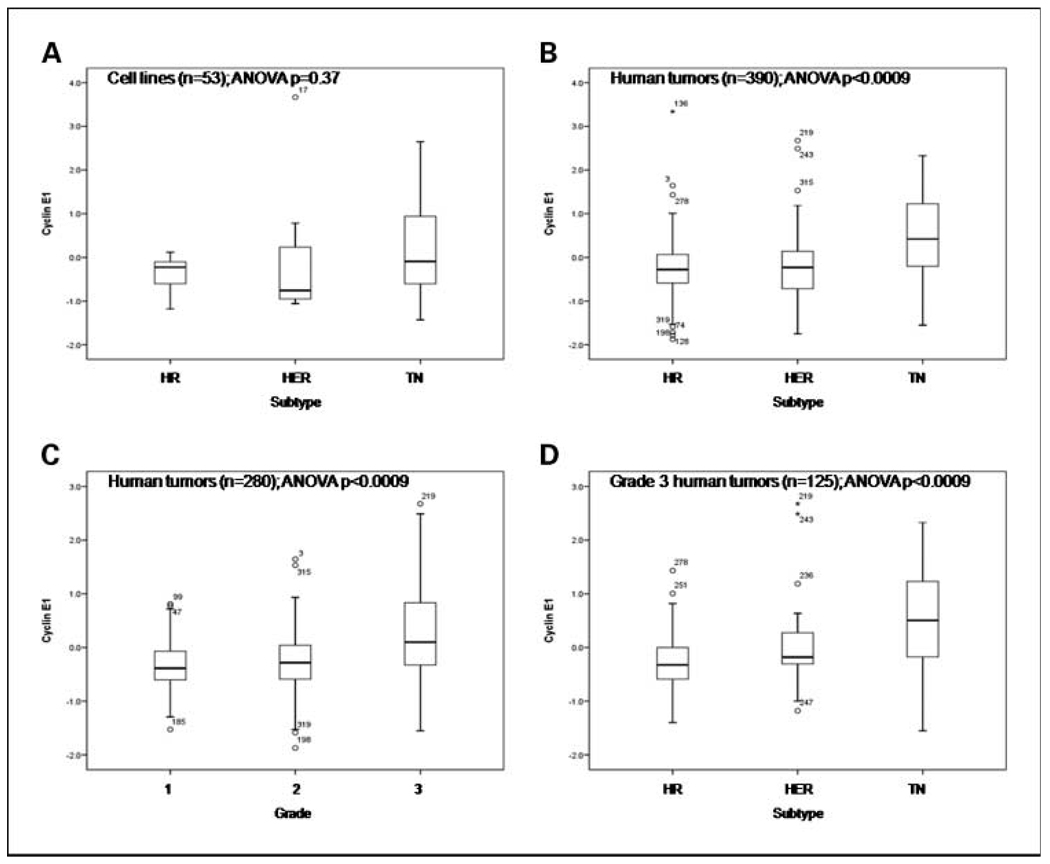
Whereas cyclin B1 (Fig. 1) was expressed at significantly higher levels in triple-negative cell lines compared with other subtypes by RPPA, cyclin E1 (Fig. 2) was not (Fig. 1A and 2A). In human breast cancers (cohort A), both cyclin B1 and E1 proteins were expressed at significantly higher levels in triple-negative tumors compared with hormone receptor–positive and HER2-positive cancers (Fig. 1B and Fig. 2B). Despite their significant and direct associations with tumor grade (Fig. 1C and Fig. 2C), cyclin B1 and E1 expression levels were not significantly different between hormone receptor–positive and HER2-positive breast cancers (P = 0.50 and 0.70, respectively) or cell lines (P = 1.0 and 0.93, respectively). With the analysis confined to 125 high-grade (grade 3) tumors, only cyclin E1 expression remained significantly higher in triple-negative compared with other breast cancers (Fig. 1D and Fig. 2D). These data suggest that cyclins B1 and E1 are markers of breast cancer grade and that cyclin E1 is also a specific marker of triple-negative tumors.
Fig. 1.
Expression of cyclin B1 in distinct subtypes and grade groupings of 390 human breast tumors (cohort A; B–D) and 53 breast cancer cell lines (A). Cyclin B1 was quantified using RPPA and the quantification data were log 2 transformed and mean centered for expression on the Y axis. HER, HER2–positive; HR, hormone receptor–positive; TN, triple-negative.
Fig. 2.
Expression of cyclin E1 in distinct subtypes and grade groupings of 390 human breast tumors (cohort A; B–D) and 53 breast cancer cell lines (A). Cyclin E1 was quantified using RPPA and the quantification data were log 2 transformed and mean centered for expression on the Y axis.
In contrast, cyclin D1 protein expression levels were significantly higher in hormone receptor–positive cell lines and breast cancers and in low-grade breast cancers compared with other subtypes and grades (Supplementary Fig. S2A–C). With the analysis confined to high-grade tumors, cyclin D1 expression remained significantly higher in hormone receptor–positive compared with other cancers (Supplementary Fig. S2D). Thus, cyclin D1 is a marker of low grade in breast cancer and of hormone receptor–positive tumors.
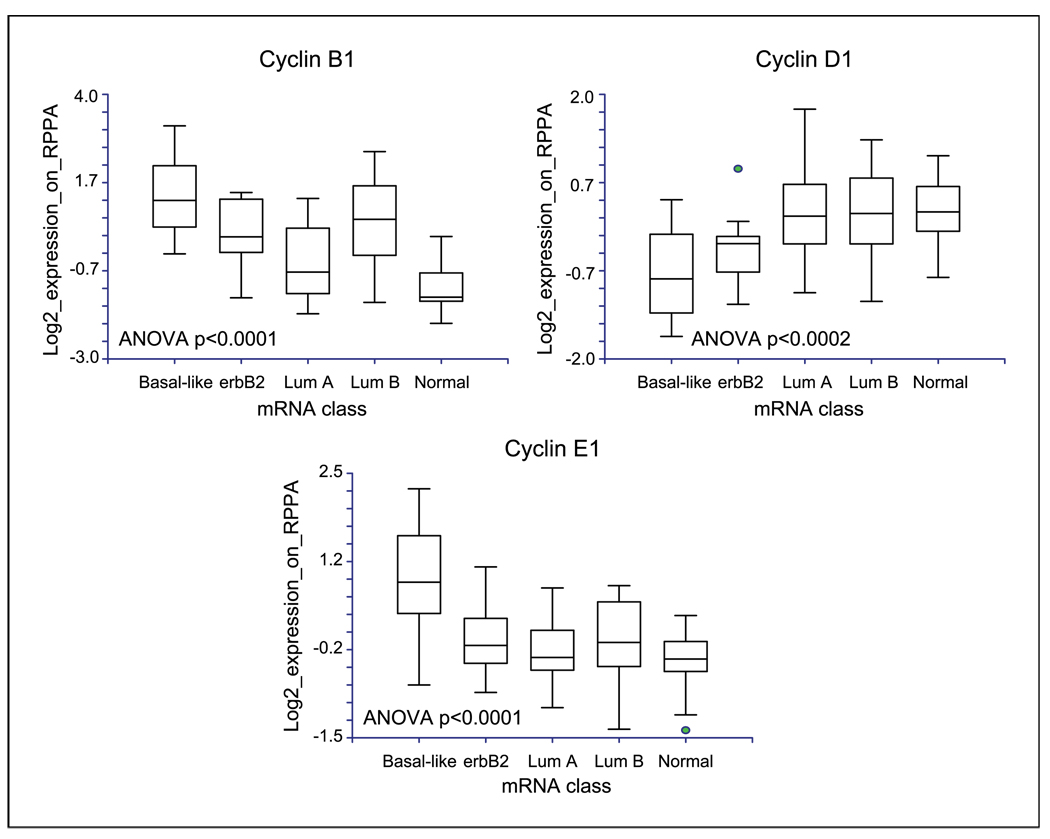
In cohort B, the associations among the expression levels of the three cyclins and the clinically defined breast cancer subgroups were confirmed. In addition, the cyclin B1 protein was expressed at high levels in basal-like and luminal B intrinsic breast cancer subtypes compared with other subtypes; cyclin D1 was expressed at high levels in luminal A, luminal B, and normal-like cancers; and cyclin E1 was expressed at high levels in basal-like cancers (Fig. 3). These associations were confirmed at the transcript level in cohorts B and D (not shown). Thus, breast cancer subtypes defined by transcriptional profiling, as well as subtypes defined using clinical criteria, express cyclins at significantly different levels. Further, the pattern of cyclin expression was consistent in cohorts A, B, and D.
Fig. 3.
Cyclin B1, cyclin D1, and cyclin E1 protein expression levels were compared using RPPA between breast cancer subtypes defined by the intrinsic gene lists in cohort B composed of 168 breast cancers, of which 128 tumors had available transcriptional profiling data (Supplementary Table S2). The cyclins were quantified using RPPA and the log 2 transformed mean centered quantification data were graphed on the Y axis. Erbb2, HER2-positive; Lum A, luminal A; Lum B, luminal B; Normal, normal-like.
There was no significant association between breast cancer stage and the expression level of any assayed cyclin. Unlike cyclin D1, both cyclins B1 and E1 were expressed at significantly higher levels in ductal versus lobular and mixed histology breast cancers (ANOVA P = 0.01, 0.027, respectively).
In general, cyclin B1 was most tightly, and cyclin D1 least tightly, correlated with expression of the known proliferation marker Ki67. In the cell lines and cohorts D and B, respectively, where mRNA expression data were available for all markers, the correlation coefficients between Ki67 mRNA and the cyclin transcripts were as follows: cyclin B1 [0.46 (P = 0.001), 0.63 (P = 0), 0.83 (P = 0)], cyclin D1 [0.06 (P = not significant, NS), −0.10 (P = NS), 0.27 (P = 0)], and cyclin E1 [0.34 (P = 0.01), 0.56 (P = 0), 0.48 (P = 0)]. In cohort D, Ki67 protein expression levels were available. In this data set, the correlation coefficients between Ki67 protein and mRNA expression of the three cyclins were 0.26 (P = 0.03), 0.10 (P = NS), and 0.26 (P = 0.03) for cyclins B1, D1, and E1, respectively.
Cyclin B1 overexpression is associated with poor prognosis in hormone receptor–positive breast cancer
In breast tumor cohorts A and B, high cyclin B1 and E1 but not cyclin D1 expression levels as determined by RPPA were significantly associated with poor RFS and OS times. However, given that breast cancer represents several molecularly and clinically different subtypes, and given that cyclins B1 and E1 are overexpressed in those subtypes that have poor outcomes with current therapies, we examined the effect of cyclin overexpression within the breast cancer subtypes.
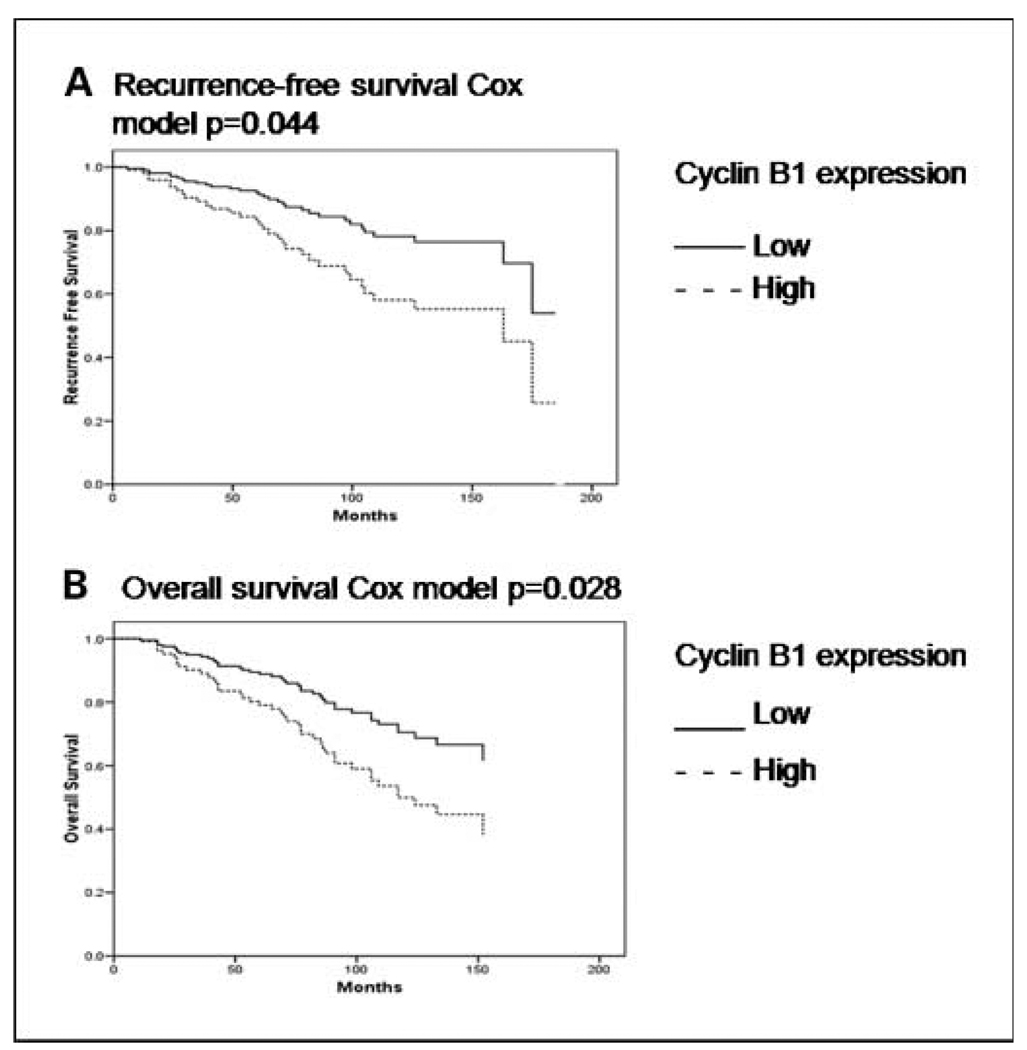
Tumor grade is a major determinant of hormone receptor–positive breast cancer patient outcomes. Given the significant associations between cyclin protein expression levels and tumor grade, we studied the relationship between the expressions of cyclins B1, D1, and E1 and patient outcomes specifically in hormone receptor–positive breast cancer. For this purpose, we used RPPA data derived from 195 early-stage hormone receptor–positive breast tumors in cohort A (Table 1) as a test set and 77 patients with nonmetastatic ER-positive breast cancer from cohort B as a validation set. We specifically selected these tumors because they were derived from patients who were homogeneously treated with adjuvant tamoxifen only after definitive locoregional therapy. In 195 tumors in cohort A, neither cyclin D1 nor cyclin E1 protein expression levels were significant univariate predictors of RFS or of OS, although a borderline association was found between high cyclin D1 expression and impaired RFS (P = 0.08). However, high cyclin B1 protein expression was a significant predictor of tumor recurrence after adjuvant tamoxifen therapy in a univariate analysis (P < 0.001) and in multivariable Cox models (Fig. 4A; Supplementary Table S4). In univariate and multivariate analyses of OS (Fig. 4B; Supplementary Table S4), high tumor cyclin B1 protein expression was also a significant predictor of death.
Table 1.
Association between cyclin protein expression and clinical variables in 195 hormone receptor–positive breast cancers obtained at primary surgery from patients treated subsequently with only adjuvant tamoxifen
| Patient number |
Mean RPPA expression |
95% confidence interval (RPPA) |
P | ||
|---|---|---|---|---|---|
| Lower | Upper | ||||
| Cyclin B1 | |||||
| Stage | 0.82 | ||||
| I | 45 | −0.33 | −0.73 | 0.07 | |
| II | 117 | −0.27 | −0.46 | −0.07 | |
| III | 32 | −0.16 | −0.6 | 0.28 | |
| Histology | 0.08 | ||||
| Ductal | 171 | −0.2 | −0.37 | −0.02 | |
| Lobular | 15 | −0.83 | −1.24 | −0.42 | |
| Mucinous | 8 | −0.63 | −1.29 | 0.03 | |
| Grade | 0.001 | ||||
| 1 | 49 | −0.84 | −1.13 | −0.55 | |
| 2 | 60 | −0.17 | −0.44 | 0.11 | |
| 3 | 9 | 0.13 | −0.55 | 0.82 | |
| PR | 0.72 | ||||
| Negative | 53 | −0.22 | −0.51 | 0.08 | |
| Positive | 142 | −0.28 | −0.48 | −0.09 | |
| Cyclin D1 | |||||
| Stage | 0.07 | ||||
| I | 45 | 0.27 | 0.07 | 0.47 | |
| II | 117 | 0.26 | 0.13 | 0.4 | |
| III | 32 | 0.6 | 0.28 | 0.93 | |
| Histology | 0.87 | ||||
| Ductal | 171 | 0.33 | 0.22 | 0.45 | |
| Lobular | 15 | 0.33 | 0.08 | 0.74 | |
| Mucinous | 8 | 0.19 | −0.14 | 0.51 | |
| Grade | 0.56 | ||||
| 1 | 49 | 0.24 | 0.03 | 0.45 | |
| 2 | 60 | 0.39 | 0.19 | 0.59 | |
| 3 | 9 | 0.24 | −0.1 | 0.58 | |
| PR | 0.58 | ||||
| Negative | 53 | 0.27 | 0.07 | 0.47 | |
| Positive | 142 | 0.34 | 0.21 | 0.47 | |
| Cyclin E1 | |||||
| Stage | 0.18 | ||||
| I | 45 | −0.36 | −0.53 | −0.19 | |
| II | 117 | −0.23 | −0.33 | −0.14 | |
| III | 32 | −0.14 | −0.3 | 0.01 | |
| Histology | 0.33 | ||||
| Ductal | 171 | −0.24 | −0.32 | −0.16 | |
| Lobular | 15 | −0.44 | −0.7 | −0.18 | |
| Mucinous | 8 | −0.16 | −0.37 | 0.04 | |
| Grade | 0.357 | ||||
| 1 | 49 | −0.34 | −0.48 | −0.19 | |
| 2 | 60 | −0.25 | −0.39 | −0.1 | |
| 3 | 9 | −0.5 | −0.85 | −0.15 | |
| PR | |||||
| Negative | 53 | −0.19 | −0.33 | −0.05 | |
| Positive | 142 | −0.27 | −0.36 | −0.18 | 0.35 |
NOTE: The cyclins were quantified using RPPA and the quantification data were expressed in log 2 form for analysis with the clinical variables shown. Note that numbers may not add up to 195 in any particular category because of missing data.
Fig. 4.
In multivariable Cox models (A and B), including cyclin B1 protein expression, tumor stage, grade and patient age, high primary tumor cyclin B1 protein expression assessed as a continuous variable by RPPA was a significant predictor of hormone receptor–positive breast tumor recurrence and patient death in 195 patients treated only with adjuvant tamoxifen after definitive locoregional therapy.
In the independent validation set of 77 patients with nonmetastatic ER-positive breast cancer from cohort B who were treated with adjuvant tamoxifen only after locoregional therapy, there were similar findings with a statistically significant association between high cyclin B1 expression and impaired RFS and OS times. In a multivariable RFS model, including cyclin B1 protein expression, grade, PR status, stage (tumor size and nodal status), and patient age, only cyclin B1 protein expression retained significance as a predictor of RFS (P = 0.004) and OS (P = 0.001). In a two-variable Cox model, high cyclin B1 expression (P = 0.01, 0.02) was a superior predictor, respectively, of poor RFS and OS than intrinsic subtype (P = 0.47, 0.29). Further, cyclin B1 protein expression levels were significantly higher in 31 “high-risk” breast tumors versus 40 “low-risk” breast tumors (P = 0.00006) when risk was assessed using the 70-gene Amsterdam signature (Mammaprint). Both the 70-gene signature (P = 0.04, 0.03) and cyclin B1 protein level (P = 0.01, 0.003) retained prognostic significance in a two-variable multivariate Cox model for RFS and OS, respectively. These data confirm the association between high cyclin B1 protein expression and hormone receptor–positive breast cancer relapse after adjuvant tamoxifen and suggest that cyclin B1 overexpression may be a stronger predictor of adverse hormone receptor–positive breast cancer patient outcomes after hormonal therapy than the intrinsic subgroup. Further, cyclin B1 protein expression adds to the predictive performance of the 70-gene signature.
To investigate the association between cyclin B1 protein expression and prognosis of women with untreated early-stage hormone receptor–positive breast cancer (versus the role of cyclin B1 in prediction of adverse outcomes after adjuvant tamoxifen), we applied RPPA to 51 primary hormone receptor–positive breast tumors derived from women who underwent no adjuvant treatment after locoregional therapy (cohort C). In contrast to cyclins D1 and E1, high cyclin B1 protein expression was a significant predictor of tumor recurrence and death (Supplementary Fig. S3).
Cyclin protein expression in triple-negative breast cancer
In univariate analyses in triple-negative breast cancer patients (cohort A), neither cyclin B1 nor cyclin E1 expression was significantly associated with cancer stage or with RFS or OS. Grade was not analyzed as almost all triple-negative tumors were of high grade.
Integrated cyclin B1, cyclin D1, and cyclin E1 profiling can reproduce breast cancer subtyping
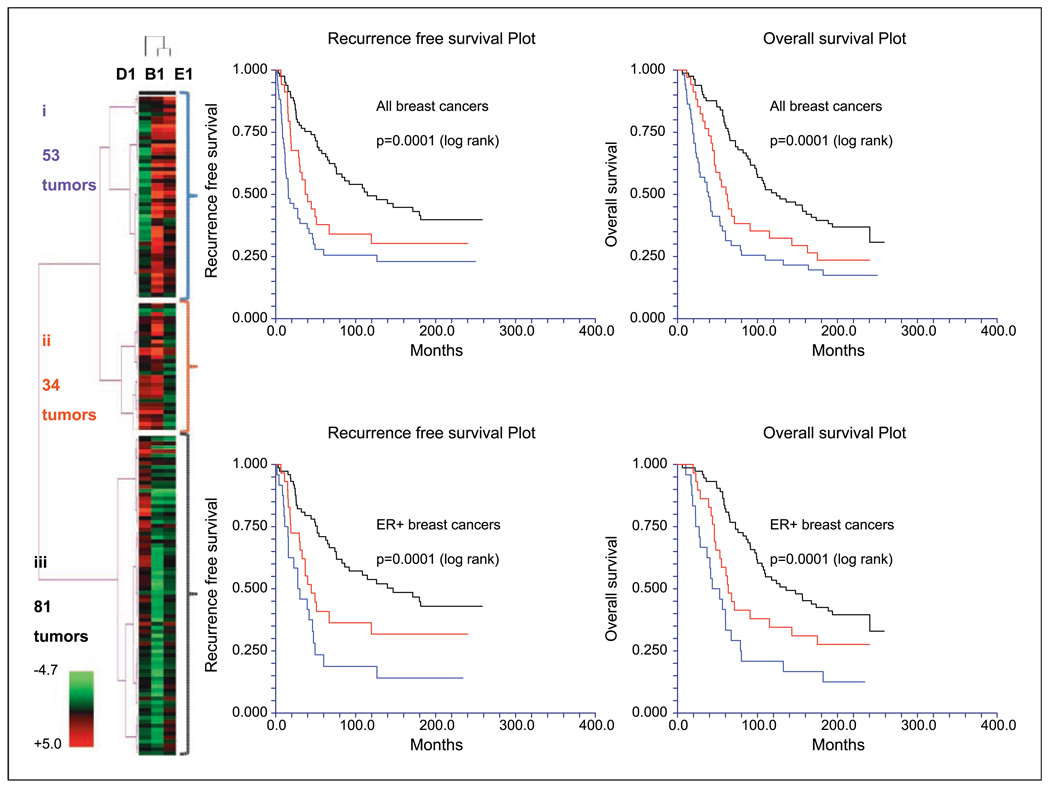
Here, we used all tumors in cohort A as a test set and all tumors in cohort B as a validation set. In cohort A, cyclin B1, D1, and E1 protein expression levels were integrated in a heat map. This analysis defined three tumor subgroups—(i) one with high cyclin B1 and E1 expression levels, (ii) one with high cyclin B1 and D1 expression, and (iii) another subgroup composed largely of tumors with high expression of cyclin D1 only. RFS and OS rates were significantly poorer among patients with breast tumors in subgroups i and ii versus patients with breast cancers in subgroup iii. There was a significant association on cross-tabulation between the three cyclin groups identified by clustering and the three clinically defined breast cancer subgroups. These data were validated in the independent set of breast tumors in cohort B (Fig. 5), in which there was also a statistically significant association between the three-group cyclin clustering and the five-group intrinsic subtyping (Supplementary Table S5; P < 0.000001). The prognostic significance of the cyclin clustering was also retained when this analysis was confined to only patients with ER-positive breast cancer (not shown).
Fig. 5.
An integrated analysis of cyclin B1, cyclin D1, and cyclin E1 expression in breast cancer. In human breast tumor cohort B, cyclin B1, cyclin D1, and cyclin E1 protein expression levels were integrated in the heat map. This analysis defined three large breast cancer subgroups (i, ii and iii) that were associated with significantly different outcomes in all breast tumors and when the analysis was confined to ER-positive (ER+) breast cancers only (color coding matches each subgroup name to the corresponding survival curves). The cyclins were quantified using RPPA and quantification data were log 2 transformed and mean centered for clustering in the heat map.
Cyclin B1 overexpression is associated with the combination of TP53 mutation and MYC amplification
In cohort D, whereas cyclin D1 and cyclin E1 mRNA expression levels were significantly correlated with the copy number of the corresponding genes, this was not the case with cyclin B1. Further, unlike the cyclin E1 gene (CCNE1) that was present in significantly higher copy numbers in basal-like versus other subtypes (ANOVA P < 0.001), CCNB1 gene copy number did not differ significantly between subtypes (ANOVA P = 0.61) and did not show gain in breast cancer. Neither was cyclin B1 (or E1) protein expression level associated with PIK3CA mutation status in cohort A or B. This suggests that the overexpression of cyclin B1 in basal-like and luminal B cancers is a consequence of oncogenic events other than CCNB1 gene amplification or common PIK3CA mutations.
MYC and TP53 genes have been shown mechanistically to regulate the CCNB1 gene promoter in opposing ways (26). Further, in cohort D, MYC amplification was more frequent in basal-like and luminal B tumors than in other breast cancer subtypes. Indeed, >l90% of basal-like and luminal B tumors exhibited amplification of MYC and/or mutation of TP53, compared with only 55% of ERBB2 and 47% of luminal A tumors. We therefore hypothesized that cyclin B1 overexpression in luminal B and basal-like tumors may occur as a consequence of MYC amplification and TP53 mutation.
In cohort D, cyclin B1 mRNA levels were elevated in tumors with MYC amplification compared with tumors possessing a normal MYC gene copy number, and also in TP53-mutant versus TP53-wild type cancers, with the most marked effect being found in tumors harboring both MYC amplification and TP53 mutation (Supplementary Fig. S4). This was validated in cohort B using cyclin B1 protein expression data (RPPA) and MYC gene expression as a surrogate for MYC gene copy number (not shown).
Cyclin B1 overexpression is associated with increased expression of PLK, CENPE, and AURKB
Cyclin B1 protein and mRNA expression levels were tightly correlated in both cohort B (R = 0.68/P = 0) and in breast cancer cell lines (R = 0.57/P = 0.00008). Cyclin B1 mRNA expression was found to be significantly and positively correlated (correlation coefficients >0.5) in each of cohorts B and D and in cell lines with mRNA expression levels of PLK1, PLK4, CENPE, and AURKB. The latter molecules are targets of current drug development programs. These correlations suggest that CCNB1 expression may modulate sensitivity to drugs against these targets.
Discussion
Cyclins D1 and E1 are well studied in breast cancer (1–10). Whereas the reported relationship between cyclin D1 expression and outcome is controversial, cyclin E1 overexpression is consistently associated with an increased risk of breast cancer relapse. Cyclin B1 is less extensively studied in breast cancer (11). The current study is the first to report the differential expression of cyclins B1, D1, and E1 across clinically and molecularly defined breast cancer subgroups. It is also the first study to comprehensively report the clinical relevance of the expression of cyclins B1, D1, and E1 within specific breast cancer subtypes and to conclusively show that cyclin B1 plays as important a role as cyclins D1 and E1 in breast cancer. Indeed, cyclin B1 is the dominant cyclin associated with poor prognosis in hormone receptor–positive breast cancer.
In cohort A, transcriptional profiling data were not available to allow us to subgroup breast cancers into the intrinsic subgroups. However, there is a significant overlap between breast cancer classification using clinical criteria and based on intrinsic gene expression, with triple-negative breast cancers and cell lines expressing a basal-like transcriptional profile, and luminal A and B breast cancers composed largely of hormone receptor–positive tumors (12–14). Further, the differential protein expression levels of cyclins B1, D1, and E1 among different breast cancer subgroups that were defined using the clinical criteria were consistent in both cohorts A and B in this study. We showed that cyclin E1 overexpression occurs specifically in triple-negative and basal-like breast tumors; high cyclin B1 expression occurs in luminal B; poor prognosis hormone receptor–positive and triple-negative/basal-like tumors and high cyclin D1 expression occurs in hormone receptor–positive, luminal B, luminal A, and in normal-like breast tumors. These subtype associations were consistent across three large tumor cohorts (A/B/D).
As reported previously (8, 9), across all breast cancers, cyclin E1 overexpression was associated with high tumor grade and with a significantly increased risk of relapse and death in our present study. However, the effect of cyclin E1 overexpression on patient outcomes is likely due in part to the specific association between high cyclin E1 expression and poor prognosis basal-like and triple receptor–negative breast cancers, an association that persisted even when tumor grade was accounted for. When analyzed within specific breast cancer subtypes, cyclin E1 did not significantly influence RFS or OS times. Indeed, the relatively low expression of cyclin E1 in hormone receptor–positive and luminal breast cancers and its lack of an association with tumor grade or patient outcomes specifically in these breast cancer subtypes suggest that cyclin E1 may not play a major role in hormone receptor–positive breast carcinogenesis.
Cyclin B1 is the dominant cyclin associated with aggressive behavior of hormone receptor–positive breast cancer in both univariate and multivariate analyses across several sample sets. As shown by Cox regression analysis (Supplementary Table S4), the effect of cyclin B1 overexpression on hormone receptor–positive breast cancer patient outcomes may not be entirely attributable to tumor grade. Like cyclin E1, although high cyclin B1 expression occurs in triple-negative/basal-like tumors, the cyclin B1 expression level does not influence the outcomes of patients with this form of breast cancer.
An integrated analysis of cyclins B1, D1, and E1 in breast cancer reveals that different expression patterns are associated with differential patient outcomes (Fig. 5). Coordinate overexpression of cyclins B1 and E1 with low cyclin D1 expression occurs in almost all basal-like breast cancers. Given their distinct expression patterns in breast cancer and the lack of a direct correlation between the expression levels of all three cyclins, cyclins B1, D1, and E1 may serve distinct roles in the pathogenesis of different breast cancer subtypes. Cyclin overexpression is driven at least partly by gene amplification for cyclins D1 and E1 but not for cyclin B1 in breast cancer. Instead, cyclin B1 overexpression is likely driven in part by a combination of TP53 mutation and MYC amplification, a combination that seems pathognomonic of basal-like and luminal B tumors. Cyclin B1 overexpression was tightly correlated with overexpression of PLK, CENPE, and AURKB, all targets of current drug development programs.
Our study has some shortcomings. RPPA is useful for objective quantification of protein expression in cell lines and tumor tissues but does not provide information on the intratumoral or cellular localization of proteins and thus on the effect of different cyclin localization patterns on clinical outcomes. Further, and in particular with regard to cyclin E1 (8), RPPA does not discriminate between the protein products of different splice variants.
In conclusion, the expression levels of cyclins B1, D1, and E1 are differentially deregulated in different breast cancer subtypes. This may suggest that cyclins B1, D1, and E1 may have distinct roles in different breast cancer subtypes. This study is the first to comprehensively examine the relevance of overexpression of cyclins B1, D1, and E1 to the clinical behavior of distinct breast cancer subtypes. Cyclin B1 is the dominant cyclin associated with poor prognosis of hormone receptor–positive breast cancers. The effect of cyclin B1 overexpression on hormone receptor–positive breast cancer patient outcomes is likely not entirely attributable to tumor grade. Novel PLK, CENPE, and AURKB inhibitors may have utility in the treatment of cyclin B1–overexpressing breast cancers that are currently associated with a poor prognosis (basal-like and luminal B cancers).
Translational Relevance
This study is the first to comprehensively examine the relevance of overexpression of cyclins B1, D1, and E1 to the clinical behavior of distinct breast cancer subtypes. The expression levels of cyclins B1, D1, and E1 are differentially deregulated in different breast cancer subtypes, suggesting that these cyclins may have distinct roles in the different subtypes. Cyclin B1 is the dominant cyclin associated with poor prognosis of hormone receptor–positive breast cancers; thus, high cyclin B1 expression identifies breast tumors that are unlikely to be cured with antihormonal therapy alone. Indeed, cyclin B1 overexpression is a stronger predictor of hormone receptor–positive breast cancer recurrence than intrinsic subtype. Given strong correlations between the expression levels of CCNB1 and of PLK, CENPE, and AURKB genes, novel PLK, CENPE, and AURKB inhibitors may have utility in the treatment of aggressive cyclin B1–overexpressing breast cancers (basal-like and luminal B cancers).
Supplementary Material
Acknowledgement
Grant support: Kleberg Center for Molecular Markers at the M. D. Anderson Cancer Center, National Cancer Institute grant PO1CA099031 (G.B. Mills), The Susan G. Komen Foundation Biomarkers Identification and Validation Award FAS0703849 (B.T. Hennessy, A.M. Gonzalez-Angulo, and G.B. Mills), Cancer Research UK Clinician Scientist award (R. Agarwal), the Research Council of Norway grant 175240/S10 (A.L. Borresen-Dale), The MDACC Physician Scientist Program, and the McNair Scholars Program supported by the Robert and Janice McNair Foundation (B.T. Hennessy).
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Gillett C, Smith P, Gregory W, et al. Cyclin D1 and prognosis in human breast cancer. Int J Cancer. 1996;69:92–99. doi: 10.1002/(SICI)1097-0215(19960422)69:2<92::AID-IJC4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 2.Michalides R, Hageman P, van Tinteren H, et al. A clinicopathological study on overexpression of cyclin D1 and of p53 in a series of 248 patients with operable breast cancer. Br J Cancer. 1996;73:728–734. doi: 10.1038/bjc.1996.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McIntosh GG, Anderson JJ, Milton I, et al. Determination of the prognostic value of cyclin D1 overexpression in breast cancer. Oncogene. 1995;11:885–891. [PubMed] [Google Scholar]
- 4.Seshadri R, Lee CS, Hui R, et al. Cyclin DI amplification is not associated with reduced overall survival in primary breast cancer but may predict early relapse in patients with features of good prognosis. Clin Cancer Res. 1996;2:1177–1184. [PubMed] [Google Scholar]
- 5.Han S, Park K, Bae BN, et al. Cyclin D1 expression and patient outcome after tamoxifen therapy in estrogen receptor positive metastatic breast cancer. Oncol Rep. 2003;10:141–144. [PubMed] [Google Scholar]
- 6.Hwang TS, Han HS, Hong YC, et al. Prognostic value of combined analysis of cyclin D1 and estrogen receptor status in breast cancer patients. Pathol Int. 2003;53:74–80. doi: 10.1046/j.1440-1827.2003.01441.x. [DOI] [PubMed] [Google Scholar]
- 7.Bièche I, Olivi M, Noguès C, et al. Prognostic value of CCND1 gene status in sporadic breast tumours, as determined by real-time quantitative PCR assays. Br J Cancer. 2002;86:580–586. doi: 10.1038/sj.bjc.6600109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keyomarsi K, Tucker SL, Buchholz TA, et al. Cyclin E and survival in patients with breast cancer. N Engl J Med. 2002;347:1566–1575. doi: 10.1056/NEJMoa021153. [DOI] [PubMed] [Google Scholar]
- 9.Span PN, Tjan-Heijnen VC, Manders P, et al. Cyclin-E is a strong predictor of endocrine therapy failure in human breast cancer. Oncogene. 2003;22:4898–4904. doi: 10.1038/sj.onc.1206818. [DOI] [PubMed] [Google Scholar]
- 10.Porter PL, Malone KE, Heagerty PJ, et al. Expression of cell-cycle regulators p27Kip1 and cyclin E, alone and in combination, correlate with survival in young breast cancer patients. Nat Med. 1997;3:222–225. doi: 10.1038/nm0297-222. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki T, Urano T, Miki Y, et al. Nuclear cyclin B1 in human breast carcinoma as a potent prognostic factor. Cancer Sci. 2007;98:644–651. doi: 10.1111/j.1349-7006.2007.00444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herschkowitz JI, Simin K, Weigman VJ, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2007;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tibes R, Qiu Y, Lu Y, et al. Reverse phase protein array (RPPA): validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 17.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu J, He X, Baggerly KA, et al. Non-parametric quantification of protein lysate arrays. Bioinformatics. 2007;23:1986–1994. doi: 10.1093/bioinformatics/btm283. [DOI] [PubMed] [Google Scholar]
- 19.Hennessy BT, Lu Y, Poradosu E, et al. Quantified pathway inhibition as a pharmacodynamic marker facilitating optimal targeted therapy dosing: proof of principle with the AKT inhibitor perifosine. Clin Cancer Res. 2007;13:7421–7431. doi: 10.1158/1078-0432.CCR-07-0760. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen HM, Overgaard M, Grau C, Jensen AR, Overgaard J Danish Breast Cancer Cooperative Group. Study of failure pattern among high-risk breast cancer patients with or without postmastectomy radiotherapy in addition to adjuvant systemic therapy: long-term results from the Danish Breast Cancer Cooperative Group DBCG 82 b and c randomized studies. J ClinOncol. 2006;24:2268–2275. doi: 10.1200/JCO.2005.02.8738. [DOI] [PubMed] [Google Scholar]
- 21.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Fridlyand J, Snijders AM, Ylstra B, et al. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer. 2006;6:96. doi: 10.1186/1471-2407-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 25.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 26.Yin XY, Grove L, Datta NS, et al. Inverse regulation of cyclin B1 by c-Myc and p53 and induction of tetraploidy by cyclin b1 overexpression. Cancer Res. 2001;98:6487–6493. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.