Abstract
Objective
Wegener's granulomatosis (WG) is a systemic inflammatory disease causing substantial morbidity. This study seeks to understand the biology underlying WG, and to discover markers of disease activity useful in prognosis and treatment guidance.
Methods
Gene expression profiling was performed using total RNA from PBMC and granulocyte fractions from 41 WG patients and 23 healthy controls. Gene set enrichment analysis (GSEA) was performed to search for candidate WG-associated molecular pathways and disease activity biomarkers. Principal component analysis (PCA) was used to visualize relationships between subgroups of WG patients and controls. Longitudinal changes in PR3 expression were evaluated using RT-PCR, and clinical outcomes including remission status and disease activity were determined using the BVAS-WG.
Results
We identified 86 genes significantly up-regulated in WG PBMCs and 40 in WG PMNs relative to controls. Genes up-regulated in WG PBMCs were involved in myeloid differentiation, and included the WG autoantigen, PR3. The coordinated regulation of myeloid differentiation genes was confirmed by gene set analysis. Median expression values of the 86 WG PBMC genes were associated with disease activity (p=1.3 × 10−4), and patients expressing these genes at a lower level were only modestly different from healthy controls (p=0.07). PR3 transcription was significantly up-regulated in the PBMCs (p=1.3 ×10−5, FDR=0.002), but not in the PMNs (p=0.03, FDR=0.28) of WG patients, and changes in BVAS-WG tracked with PBMC PR3 RNA levels in a preliminary longitudinal analysis.
Conclusion
Transcription of PR3 and related myeloid differentiation genes in PBMCs may represent novel markers of disease activity in WG.
Introduction
Wegener's Granulomatosis (WG) is a systemic inflammatory disease characterized by granulomatous inflammation of the upper and lower respiratory tracts and necrotizing arteritis affecting small and medium sized arteries. Though significant improvement in patient outcomes have been realized over the past two decades, the longitudinal clinical assessment and management of WG remains complicated by difficulties in differentiating WG-related disease activity from disease and/or treatment-related damage (1-3). The discoveries of anti-neutrophilic cytoplasmic antibodies (ANCA) (4) and the highly specific targeting of the neutrophil serine protease proteinase 3 (PR3, myeloblastin) in WG (5) suggested the use of PR3-ANCA as a potential biomarker that could mitigate some of the clinical assessment difficulties described above. Indeed, ANCA are found in over 90% of patients with WG during the course of their illness (6), and several reports over the past two decades have suggested that elevated antibody titers are associated with more severe disease manifestations, increased risk of flare, and poorer prognosis (4, 7, 8). Further, a mechanistic role for PR3-ANCA in the pathogenesis of WG has been postulated in numerous in vitro studies (9-11).
However, recent longitudinal data from the Wegener's Granulomatosis Etanercept Trial (WGET), demonstrate that though anti-PR3 antibodies are highly specific for the diagnosis of WG, their use as biomarkers for assessing disease activity, determining risk of flare, and gauging remission status is actually quite limited (12). As a result, the current gold-standard methodology for defining these endpoints in WG utilizes consensus-derived clinical indices (13, 14), which may underestimate low and subclinical disease activity in some cases, and overestimate clinical activity in others. Thus, the search for more discriminant biomarkers of disease activity in WG remains a top investigative priority.
Microarray techniques have been used in recent years to identify putative pathways of mechanistic and prognostic relevance in the systemic rheumatic diseases (15, 16), and have also been employed with increasing success to discover new prognostic biomarkers in several forms of cancer (17, 18). Newer quantitative analytical strategies such as gene set enrichment analysis (GSEA) (19, 20) have recently been employed to systematically analyze pathway regulation in gene expression datasets permitting the evaluation of coordinately regulated but only moderately over-expressed sets of genes within a dataset. Whole blood-based gene expression studies have previously been conducted in patients with several forms of ANCA-associated diseases including WG (21, 22); however, no systematic expression profiling study specifically in WG has been performed to date.
In this study, we employed quantitative signature analysis to study gene expression profiles and pathway enrichment in both PBMC and granulocyte fractions from a large and carefully defined cohort of patients with WG. Using this approach, we identified the coordinated expression of genes involved in myeloid differentiation in patients with WG. Strikingly, this gene expression signature (which included the primary WG autoantigen PR3) was expressed primarily in the PBMC, and not the neutrophil peripheral blood fraction. High expression of both the gene expression signature and the PR3 (PRTN3) gene itself was noted in the PBMCs of patients with active disease (BVAS-WG > 0), and similar low signature expression levels were seen in both patients in remission and in healthy controls. The level of PR3 expression in PBMCs tracked disease activity status in a limited longitudinal analysis. Taken together, these findings suggest that gene expression analysis of WG PBMCs may be a powerful tool to help gauge disease activity in WG.
Patients and Methods
Patients
Forty one WG patients seen for clinical care in the Johns Hopkins Vasculitis Center and twenty-three healthy controls were included in this study. All patients met the 1990 American College of Rheumatology classification criteria for Wegener's granulomatosis (23), and all but four of the patients in the cohort had a history of ANCA and/or PR3/MPO positivity as determined by immunofluorescence and/or ELISA in the context of routine clinical care. Patients with other ANCA-associated diseases such as microscopic polyangiitis, the Churg-Strauss syndrome, and drug-induced ANCA-associated vasculitis were excluded from the study. All patients gave informed consent, and all samples were obtained under the auspices of a human subject internal review board-approved protocol. No identifying demographic or clinical information accompanied any of the samples during processing or analysis.
Disease activity scores were measured using a modification of the Birmingham Vasculitis Activity Score-Wegener's Granulomatosis (BVAS-WG) (13). Only factors that were present the day of sample collection were included in the score calculations. Factors present in the 28 previous days, included in standard BVAS-WG calculations, were excluded in order to most accurately correlate the transcriptional profiles from the day of the blood draw with the clinical findings present on that day. Absent disease activity was defined as a BVAS-WG of 0, and remission as maintaining a BVAS-WG score of 0 for 6 months or more.
Isolation of Peripheral Blood Mononuclear cells and Neutrophils
Venous blood was collected by simple venipuncture under aseptic conditions. All samples were processed within one hour of collection to minimize gene expression variations associated with longer sample incubation times (24). PBMCs were separated by Ficoll density gradient, and PMNs isolated following 1-3 rounds of hypotonic RBC lysis. Cells were assessed for viability by trypan blue dye exclusion, were immediately lysed in Trizol reagent (Invitrogen, Carlsbad, California) and stored at −80°C. The purity of the resultant cell populations were assessed by flow cytometry on a FACS caliber flow cytometer, and analyzed using CellQuest Pro software. Neutrophils were identified by co-staining with the following fluorophore-conjugated antibodies: CD15-PE/CD16-FITC/CD11b-APC (BD Bioscience, San Jose CA).
DNA Microarray Analysis
Total RNA was extracted from either the PBMC or PMN fractions using the Trizol reagent method (Invitrogen, Carlsbad, California). Additional purification was performed on RNeasy columns (Qiagen, Valencia, CA), and the quality of total RNA samples was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Biotin-labeled, complementary RNA (cRNA) was prepared from total RNA according to the chip manufacturer's protocol (Illumina, San Diego, CA). cRNA was hybridized to Illumina HumanRef-8 v2 Expression BeadChips, and signal was detected with streptavidin-Cy3. All signal intensity quantification was performed using an Illumina BeadStation 500GX Genetic Analysis Systems scanner. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (25) and are accessible through GEO Series accession number GSE18885 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE18885).
Quantitative RT-PCR (QRT-PCR) Analysis
cDNA was obtained from total RNA using the Archive Kit according to the manufacturer's protocol (Applied Biosystems, Foster City, CA). Probes and primers were designed and synthesized by Applied Biosystems. All PCR amplifications were carried out in duplicate on an ABI Prism® 7300 Sequence Detection System, using a fluorogenic 5’ nuclease assay (TaqMan® probes). Relative gene expressions were calculated by using the 2-ΔΔCt method as described in (26). The ΔCt value of each sample was calculated using 3 endogenous control genes (GAPDH, ACTB, and PGK1).
Analytical Methods and Statistical Analysis
A single intensity (expression) value for each Illumina probe on the array was obtained using Illumina BeadStudio software with standard settings and no background correction. The expression values for all the probes for each sample were scaled to have median 256 (28) and then log (base 2) transformed. Probes that did not have at least 8 samples with Illumina detection p-values < 0.01 were eliminated from further consideration. Genes (i.e., Illumina probes) considered to be significantly differentially expressed between two groups of samples were those satisfying the three criteria: (i) Two-sided Welch t-test p-values less than or equal to 0.01 (10−2) (27); (ii) a Benjamini-Hochberg false discovery rate (FDR) less than or equal to 0.1 (28); and (iii) a fold change above 1.5 or below 1/1.5 (calculated using geometric means). The two-sided t-test was used to obtain p-values for differences between groups unless otherwise noted.
Heat maps (and the ordering by hierarchical clustering of the samples and the genes in heat maps) were based on normalization of the expression values for each sample using z-transformation (27, 29, 30) (utilizing only the probes which had an Illumina detection p-value < 0.01 for least one sample), followed by z-transformation of the normalized expression values for each Illumina probe across all samples. Hierarchical clustering was performed using the Cluster and TreeView software programs (31). The clustering algorithm was set to complete linkage clustering using uncentered correlation.
Differential expression of gene sets was analyzed using GSEA (19). The Broad Institute JAVA Desktop software Version 2.0 of GSEA was utilized with the Preranked option, and with the Molecular Signatures Database C2 curated gene sets file c2.all.v2.5.symbols.gmt containing 1892 gene sets from known pathways and from published studies (http://www.broadinstitute.org/gsea/msigdb/index.jsp). The ranking metric was the Gaussian z-value corresponding to the 1-sided p-value from the Welch t-test of differential expression between 41 WG samples and 23 Controls (z>0 for upregulated in WG) (Suppl. Table 2). Gene sets with an estimated false discovery rate less than 0.05 were considered significant, following the recommendation on page 22 of the GSEA user guide (available at http://www.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html). Default parameters were used with the following exceptions: 2000 permutations were performed and the random number generator seed was set to 159265. When more than one Illumina probe existed for a given gene, the ranking metric with the largest magnitude was used for GSEA.
Principal Component Analysis (PCA) (32, 33) was performed with custom software written in IDL (ITT Visual Information Solutions, Boulder, CO) using the scaled log transformed expression values for the Illumina probes, which were ranked by Welch t-test p-values. Kinemage files of the results of the PCA were prepared via IDL and plotted using the MAGE molecular graphics 3-D software (http://kinemage.biochem.duke.edu).
Results
Identification of a myeloid differentiation signature in WG
Microarray analysis was performed on RNA extracted from separated PBMC and neutrophil fractions from 41 patients with confirmed WG and 23 healthy adult controls. The demographic and clinical characteristics of both patients and controls are shown in Table 1. No significant differences in gender (p=1) or mean age (p=0.2) were noted between patients and controls. The majority of patients were ANCA (+) (88%), had systemic disease (73%), and were on immunosuppression (68%) at the time of blood draw. More than half of the cohort had mild to moderately active disease as measured by the day-of collection BVAS-WG.
Table 1.
Demographic and Clinical Characteristics
| Characteristic | Patients (N=41) | Controls (N=23) | p value | ||
|---|---|---|---|---|---|
| Age (Mean +/− S.D.) | 50 +/− 15 | 45 +/− 14 | 0.2 | ||
| N | % | N | % | ||
| Gender | |||||
| M | 20 | 49 | 11 | 48 | 1 |
| F | 21 | 51 | 12 | 52 | |
| Wegener's Type | |||||
| Limited | 11 | 27 | |||
| Systemic | 30 | 73 | |||
| Renal Disease | |||||
| None | 21 | 51 | |||
| Inactive | 16 | 39 | |||
| Active | 4 | 10 | |||
| ANCA Status | |||||
| Negative | 4 | 10 | |||
| Pr3-ANCA | 31 | 76 | |||
| MPO-ANCA | 5 | 12 | |||
| Unknown | 1 | 2 | |||
| Corticosteroid Usage | |||||
| No | 13 | 32 | |||
| Yes | 28 | 68 | |||
| Immunosuppression | |||||
| None | 13 | 32 | |||
| Methotrexate | 13 | 32 | |||
| Azathioprine | 8 | 20 | |||
| Cyclophosphamide | 4 | 10 | |||
| Mycophenolate | 2 | 4 | |||
| Rituximab | 1 | 2 | |||
| BVAS-WG | |||||
| 0 | 19 | 46 | |||
| 1 to 2 | 10 | 25 | |||
| 3 to 5 | 11 | 27 | |||
| >5 | 1 | 2 | |||
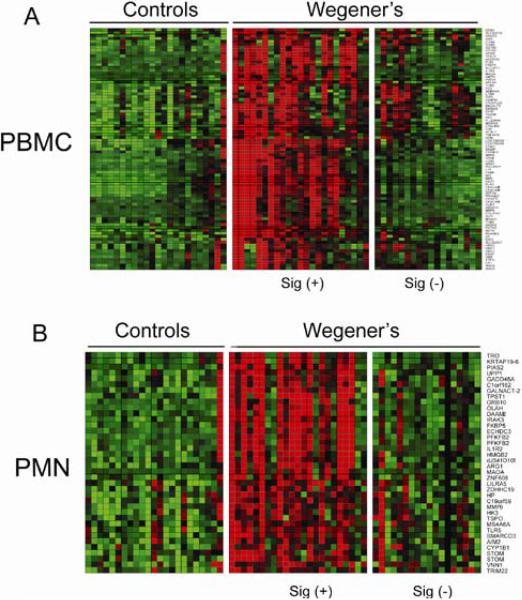
Using the combination of statistical significance and fold change criteria described in Methods, we identified 86 significantly up-regulated genes in WG PBMCs and 40 in WG PMNs relative to controls (Figure 1A & B). The genes up-regulated in both PBMCs and PMNs (N=9) included arginase (ARG1), hexokinase (HK3), monoamine oxidase (MAOA), haptoglobin (HP), and the interleukin 1 receptor IL1R2. Indeed, the majority of genes significantly up-regulated in WG PMNs reflected general cellular activation pathways (Supplemental Table 1). Notably, neutrophil granule constituent genes were markedly up-regulated in the WG PBMC fraction alone, including those encoding many of the major and minor ANCA antigens such as LCN2 (#6), ELA2 (#8), MPO (#15), BPI (#18), CTSG (#19), AZU1 (#22), and PRTN3 (#45) (Supplemental Table 2). Other “neutrophilic” innate immune response genes, including defensins α1 and α4, and cathelicidin (CAMP) were similarly up-regulated exclusively in the PBMC fraction.
Figure 1.
Identification of discrete sets of up-regulated genes in Wegener's PBMCs and PMNs. Heat maps of the expression levels of the genes significantly differentially expressed between WG patients and normal controls in PBMCs (n=86) (A), and in PMNs (n=40) (B).
Though the mature proteins encoded by these genes are stored in mature neutrophil granules, their transcription occurs primarily in earlier granulocytic precursors (34). Thus, the increased transcription of these genes in the WG PBMC fraction is unlikely to arise from mature neutrophils; indeed, the percentage of CD15+/CD16+/CD11b+ neutrophils in both the WG and control PBMC fractions was low (< 3%), and did not differ between the two groups (p=0.26, data not shown).
These data suggested the coordinated up-regulation of genes expressed during early myeloid development in the PBMC fraction of WG patients. To examine this possibility in greater depth, gene set analysis using GSEA was performed. As seen in Table 2, six of the top ten most highly regulated gene sets between WG PBMCs and controls pertained to normal and abnormal myeloid development (all with p-values < 0.001 and FDR < 4.2 × 10−4). In contrast, no enrichment of interferon, TNF, IL-1, or other cytokine-associated gene sets was noted in the top 50 returns (for GSEA summaries, see Supplemental Tables 3 and 4). Interestingly, only two of the top ten returns in WG PMNs related to myeloid development. These data confirm that the enrichment of genes and gene sets involved in early myeloid development is a defining characteristic of WG PBMCs.
Table 2.
Gene set enrichment analysis of changes in gene expression between WG patients and controls
| Gene Set Name | Major Pathway | Gene Set Size | GSEA Normalized Enrichment Score | GSEA nominal p-value | Estimated FDR q-value |
|---|---|---|---|---|---|
| VERHAAK_AML_NPM1_MUT_VS_WT_UP | Leukemia | 154 | 2.80 | < 0.001 | 0 |
| APPEL_IMATINIB_UP | Leukemia | 28 | 2.76 | < 0.001 | 0 |
| ROSS_MLL_FUSION | Leukemia | 70 | 2.73 | < 0.001 | 0 |
| LIAN_MYELOID_DIFF_GRANULE | Myeloid Differentiation | 19 | 2.61 | < 0.001 | 0 |
| HSA01032_GLYCAN_STRUCTURES_DEGRADATION | N/A | 25 | 2.58 | < 0.001 | 0 |
| PARK_RARALPHA_MOD | Myeloid Differentiation | 54 | 2.55 | < 0.001 | 0 |
| MARTINELLI_IFNS_DIFF | Myeloid Differentiation | 22 | 2.54 | < 0.001 | 0 |
| LAL_KO_3MO_UP | N/A | 38 | 2.43 | < 0.001 | 0 |
| HSA04610_COMPLEMENT_AND_COAGULATION_CASCADES | N/A | 36 | 2.38 | < 0.001 | 3.85E-04 |
| CARIES_PULP_UP | N/A | 174 | 2.37 | < 0.001 | 4.17E-04 |
Expression of the WG signature is associated with active disease
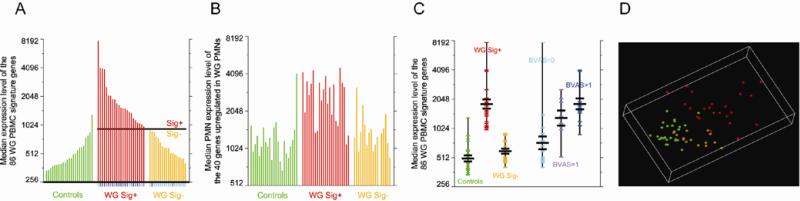
Given the significant up-regulation of pathophysiologically relevant constituent genes in the WG PBMC fractions, we defined a “WG signature” as the median of the expression values for the 86 up-regulated PBMC genes in each subject. This signature was significantly up-regulated in patients vs. controls (p= 7.6 × 10−8), and in patients with active disease (BVAS-WG>0) vs. patients with inactive disease (BVAS-WG=0) (p=1.3 × 10−4). As seen in Figure 1A, WG patients could be clustered into two distinct groups based on their expression of signature genes. WG signature expression values were significantly higher in one group (“signature positive” (sig +), n=23) vs. the other (“signature negative (sig -), n=18) (p=2.2 × 10−10). The median PBMC expression level for the signature genes determining the separation line between the Sig + and Sig - groups was defined to be the second highest median value for the controls plus the difference between that and the third highest median for the controls; the control sample with the highest median was considered to be an outlier (Figure 2A). Using this cutoff value, signature status was tightly associated with disease activity; 16 of the 18 Sig- WG patients had a BVAS-WG = 0, while 20 of the 23 Sig+ WG patients had a BVAS-WG > 0 (p = 1.13 × 10−6 by the one-tailed Fisher's exact test) (Figure 2A). Though signature positivity was associated with disease activity in general (BVAS >0), the magnitude of signature expression was only modestly correlated with the degree of BVAS elevation (Pearson product-moment correlation coefficient r= 0.49, p=0.001). Using the median expression level in PMNs of the 40 genes significantly up-regulated in WG vs. control PMNs gives a considerably less distinct separation between the Sig+ and Sig- WG samples (Figure 2B).
Figure 2.
Association of the WG signature with disease activity. (A), Bar charts of the median expression levels of the significantly up-regulated genes in WG vs. control PBMCs, and (B), WG vs. control PMNs. The samples in (B) are in the same order as in (A). The short vertical lines below the x-axis in (A) indicate the BVAS score for each sample; cyan for BVAS=0, lavender for BVAS=1, and blue for BVAS>1. The single gap in the WG Sig- section in (B) represents a PMN sample that did not pass stringent quality control. (C), Medians of the expression levels in PBMCs of the 86 signature genes, grouped as indicated. The mean and the mean ± the standard error of the mean for each group are designated by black lines (D), PCA visualization of all samples using the top 400 most significantly regulated genes (up or down) in WG vs. control PBMCs, ranked by p-value, viewed using the top three PCA axes. The fraction of the total variance captured in the top three PCA axes was 76%. Controls are in green, and the WG samples are colored by positive (red) and negative (gold) signature status.
Expression of the WG signature differed only modestly between controls and Sig - patients (p=0.07); extending the analysis to include the full set of gene expression profiles in the PBMCs of the Sig- group did not yield any genes that were significantly differentially expressed from controls (Supplemental Table 5). This was an unexpected finding, given that 15/18 (83%) of the Sig- WG patients were on chronic immunosuppressive therapy that could have altered the gene expression profiles of their circulating immune cells. Indeed, no consistent or predictable patterns of gene expression were seen in the patient group when we clustered the samples by immunosuppressive regimen or dosing schedule, concomitant corticosteroid use, or corticosteroid dose (data not shown).
As a group, patients with very low disease activity scores (BVAS-WG=1) were indistinguishable from those with more active disease (BVAS-WG >1) (p=0.15), but differed significantly from patients with a BVAS of 0 (p=0.02), Sig- patients (p=0.003), and healthy controls (p=0.0007) (Figure 2C). Since a BVAS-WG of 1 could arise from a variety of clinical scenarios, we asked whether these patients had any distinguishing characteristics that could account for their gene expression similarities with patients who were more clinically active. Remarkably, all but two of these patients were scored based on the presence of persistent granulomatous disease alone (2 with nasal crusting, 1 with subglottic stenosis, and 2 with pulmonary nodules). These data suggest that signature status may be a useful complementary tool to help assess incipiently active disease in patients with persistent granulomatous manifestations.
The preceding analysis examined median expression levels of a signature consisting of the most highly up-regulated genes between WG and control PBMCs. To determine if these changes were representative of the total variation in gene expression between the groups, PCA was performed using the 400 most differentially (up or down) expressed genes in PBMCs between all WG patients and controls. As seen in the PCA plot in Figure 2D, controls (green) and Sig- WG patients (gold) cluster together in space, indicating similar aggregate expression among the 400 genes in these subjects. We noted with interest that all ten patients with a BVAS-WG=0 who clustered with the controls in the PCA plot remained in remission after 6 months of follow-up. Though no firm conclusions can be drawn from these data due to the small sample size and limited longitudinal follow-up time, these data raise the possibility that signature expression may be a useful tool to gauge remission status over time, a concept that can be directly tested in future longitudinal studies.
PR3 transcription in PBMCs tracks disease activity over time
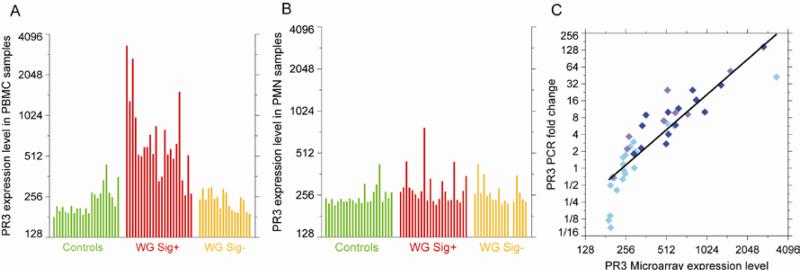
Having noted the association of the WG signature with disease activity, we asked whether transcriptional changes in PR3, the primary WG autoantigen, were similarly associated. PR3 transcription in PBMCs was highly correlated with the WG signature over all 64 samples (R2=0.78, data not shown), and was up-regulated uniquely in the PBMCs (p=1.3 ×10−5, FDR=0.002), and not the PMNs (p=0.03, FDR=0.28), of signature positive WG patients (Figures 3A & B). RT-PCR was used as a surrogate for microarray analysis in nine WG patients seen in longitudinal follow-up to evaluate whether changes in PR3 transcription tracked disease activity scores over time. As seen in Figure 3C, there was a high degree of correlation between gene expression values obtained by microarray and RT-PCR (R2=0.79). Similar findings were obtained with AZU and BPI, both component genes of the WG signature.
Figure 3.
PR3 transcription occurs primarily in PBMCs and not in PMNs in WG. (A), Bar chart of PR3 expression levels in WG and control PBMCs and (B), PMNs. Samples are displayed in the same order as in Figure 2A. (C), Comparison of PR3 expression by RT-PCR and microarray for all 41 Wegener's PBMC samples. For RT-PCR, fold changes are relative to a pool of RNA from healthy controls. The points are color coded by patient BVAS score as in Figure 2A; cyan for BVAS=0, lavender for BVAS=1, and blue for BVAS>1.
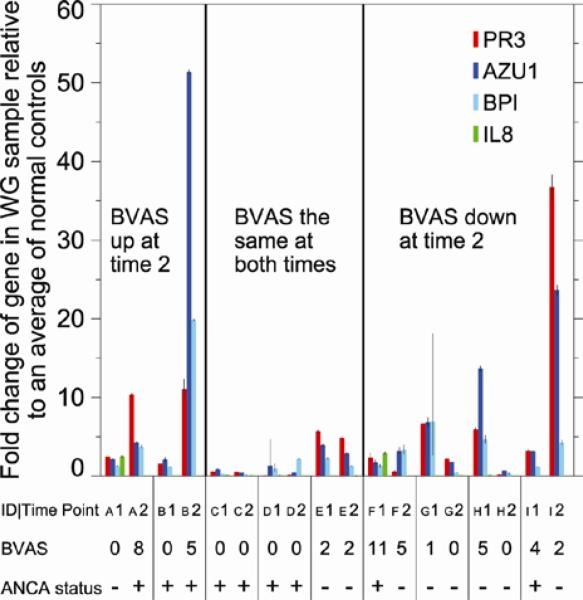
In this analysis, increased PR3 expression was seen in patients with active disease, though as in the previous signature correlation analysis, the magnitudes of the expression changes did not parallel the size of the changes in BVAS-WG scores (Figure 4). However, changes to and from remission tracked with corresponding decreases or increases in PR3 RNA levels in all but one patient (Patient “I”), and persistently low transcription was seen in patients in remission. In contrast, changes in ANCA titer tracked with disease activity less than half the time (3/9 patients); three had persistently high ANCA titers regardless of disease status, and 3 patients with active disease had undetectable serologies (Figure 4). Taken together, these cross-sectional and preliminary longitudinal clinical data suggest that PR3 and related myeloid differentiation gene transcription in the peripheral blood of WG patients might represent a novel marker for assessing disease activity in WG, especially in patients with mild systemic and/or chronic granulomatous manifestations. .
Figure 4.
Longitudinal expression of PR3 and other WG signature genes in WG patients. RT-PCR results for selected signature genes including PR3 were performed on paired samples (PBMCs) taken from patients at two time points at least 3 months apart. Fold change values are presented as average fold change = 2-(average ΔΔCt) for genes in the patient samples relative to a pool of control samples. Error bars for fold changes from PCR correspond to 2-(average ΔΔCt ± sem) where sem is the standard error of the mean for each gene.
Discussion
In this study, we used quantitative signature analysis of gene expression data from separated PBMC and PMN fractions to identify a gene expression signature that both differentiated WG patients from controls and associated with active disease. Strikingly, the WG signature, and in particular neutrophil granule component genes involved in myeloid differentiation (including the primary WG autoantigen PR3) were significantly up-regulated in WG vs. controls in the PBMC, and not the PMN leukocyte fraction. Further, expression in PBMCs of interferon-induced genes, though previously described in a variety of autoimmune rheumatic diseases such as lupus, dermatomyositis, rheumatoid arthritis, and scleroderma (35-38), was not observed in this study. Although the “granulopoiesis” signature previously described in SLE PBMCs (15) partially overlaps with the WG signature described here (see GSEA summary, Supplemental Table 3), the combination of a myeloid differentiation signature without an interferon or other inflammatory cytokine fingerprint has not previously been reported. In this regard, our study both supports the concept of shared inflammation-induced pathways and gene expression “modules” in the systemic rheumatic diseases (39), and highlights the important, and perhaps unique, role that myeloid differentiation may play in mediating both phenotype and disease activity in WG.
The highly specific immune response to PR3 has focused attention on the role of the mature neutrophil in the pathogenesis of WG for nearly two decades. Our data demonstrates that while neutrophils are clearly activated in patients with active WG (Figure 1B), the activation-induced expression of genes encoding both the major and minor ANCA antigens known to be involved in the immunopathogenesis of WG occurs exclusively in the PBMC, and not PMN leukocyte fraction. While mature neutrophils and PBMCs separate during ultracentrifugation in a density gradient, immature myeloid precursor cells sediment with the mononuclear fraction (40). As such, myeloid precursors mobilized during the inflammatory process represent one potential source of the WG PBMC signature. Indeed, low-density granulocytes (LDGs) sedimenting in the PBMC fraction and exhibiting promyelocytic differentiation characteristics in vitro have been demonstrated in patients with active SLE (41), and inflammation-induced mobilization of myeloid precursors from the bone marrow and transcription of myeloid differentiation genes in the peripheral blood has also been noted (42). These mechanisms might explain both the increased PR3 transcription in the peripheral blood of patients with active cystic fibrosis (43), and the increased transcription of genes involved in neutrophil function and activation demonstrated in patients with staphylococcal and pneumococcal infections (39, 44).
Given these data and the clinical difficulty in distinguishing early signs of respiratory infection from a WG flare, one possible confounding explanation for our findings is that the WG signature may be comprised of genes up-regulated during concurrent infection. While we did not directly evaluate gene expression in patients with infections in this study, we did evaluate a published list of 10 discrete gene expression modules (consisting of > 1100 genes) up-regulated in 14 patients infected with S. pneumoniae compared to 12 healthy controls (39) for overlap with the 86 gene WG PBMC signature defined here. Interestingly, although several genes were found in both lists (including defensin α4, MMP9, ELA2, and BPI), the major WG autoantigen genes PR3 and MPO and nearly 50 others were uniquely up-regulated in active WG (data not shown). These findings are supported by additional recent data demonstrating that gene expression profiling of peripheral blood samples could be used to distinguish patients with Kawasaki's disease from those with acute adenoviral or group A streptococcal infections (45). These data reveal that while overlap in gene expression profiles does indeed exist and likely represents activation of common inflammatory pathways, it cannot not fully explain the gene expression signature noted in WG PBMCs. This is further underscored by the highly significant association of signature and disease activity status, and the lack of signature positivity amongst control group members that we observed in this study.
Our study was not designed to analyze phenotype-specific differences in gene expression profiles in patients with different rheumatic diseases, or between patients with different ANCA-associated vasculitides. We instead focused our attention on WG because of its unique clinical (granulomatous vasculitis) and immunologic (highly specific anti-PR3 immunity) phenotype, which distinguishes it from both the other systemic rheumatic diseases and related vasculitides such as the Churg-Strauss syndrome and microscopic polyarteritis. However, the WG signature defined here partially overlapped with the up-regulated gene lists generated in earlier whole blood-based studies in ANCA-associated vasculitis (21, 22) suggesting the presence of shared peripheral inflammatory effector pathways among these conditions. Whether gene expression profiling in PBMCs could be used to discriminate between patients with phenotypically distinct ANCA-associated vasculitides is an important question that is currently being investigated.
Although gene expression profiling has been quite useful in the assessment of disease activity in multiple cross-sectional studies (46), its role in the longitudinal assessment of the systemic rheumatic diseases continues to be defined. Transcriptional profile data was recently shown to correlate well with activity scores in the longitudinal follow-up of lupus patients, and in some cases transcription of relevant genes preceded increases in observed clinical activity (39). However, in a larger study, changes in interferon signature expression and disease activity indices did not correlate well longitudinally, and in many cases the signature remained quite stable despite documented changes in clinical activity (47). In this study we have presented preliminary longitudinal data suggesting that WG signature status tracks with disease activity over time. Defining whether this WG PBMC signature, consisting of myeloid differentiation and not cytokine-induced genes, will ultimately correlate with longitudinal changes in disease status will require further investigation.
Given the lack of correlation of both circulating (48) and in situ PR3 protein levels with disease activity in WG (49), and the controversial role of the use of ANCA in this regard (12), our identification of PR3 and related gene transcription in PBMCs as a potential marker of disease activity in the longitudinal assessment of WG is exciting. Further, our finding that healthy controls and Sig - WG patients are only modestly different by PBMC gene expression even in the presence of immunosuppressive medications suggests that the suppression of signature genes may be a marker of a return to immunologic homeostasis. If confirmed in rigorous longitudinal studies, these data raise the intriguing possibility that changes in signature/PR3 status, used in conjunction with clinical evaluation and activity score determination, could help more accurately predict clinical outcomes such as risk of flare and maintenance of remission in WG in the future.
Supplementary Material
Supplemental Material
Supplemental Table 1. The 40 genes significantly upregulated in WG vs. control PMNs, sorted by fold-change.
Supplemental Table 2. The 86 genes significantly upregulated in WG vs. control PBMCs, sorted by fold-change.
Supplemental Table 3. GSEA summary for WG vs. control PBMCs using genes ranked by the Gaussian z-value corresponding to their Welch t-test one-sided p-value.
Supplemental Table 4. GSEA summary for WG vs. control PMNs using genes ranked by the Gaussian z-value corresponding to their Welch t-test one-sided p-value.
Supplemental Table 5. Welch t-test results and Benjamini-Hochberg FDR for signature negative WG samples vs. controls, PBMC data.
Acknowledgements
We thank Dr. Antony Rosen for his important insights and thoughtful review of the manuscript, and Dr. David Hellmann for thoughtful discussions and continued support of the Johns Hopkins Vasculitis Center.
Dr. Levine's work was supported by the NIH (grants K08 AR50892 and P30 AR053503), and by Brian & Bettina Finn and Family. Kristen Johnson's work was supported by a Doris Duke Charitable Foundation Student Fellowship. Dr. Barnes was supported in part by the Mary Beryl Patch Turnbull Scholar Program. Dr. Andrade and Dr. Levine are Lowe Family Scholars in the Johns Hopkins Bayview Center for Innovative Medicine.
Footnotes
Study design: Levine, Andrade, Cheadle, Berger, Barnes
Acquisition of data: Levine, Andrade, Cheadle, Berger, James, Johnson, Park, Mullins, Chrest, Ehrlich
Analysis and interpretation of data: Cheadle, Berger, Andrade, Johnson, Park, Watkins, Chen, Ehrlich, Chrest, Levine
Manuscript preparation: Levine, Cheadle, Berger, Andrade, Barnes
Statistical analysis: Cheadle, Berger
References
- 1.Seo P, Min YI, Holbrook JT, Hoffman GS, Merkel PA, Spiera R, et al. Damage caused by Wegener's granulomatosis and its treatment: prospective data from the Wegener's Granulomatosis Etanercept Trial (WGET). Arthritis Rheum. 2005 Jul;52(7):2168–78. doi: 10.1002/art.21117. [DOI] [PubMed] [Google Scholar]
- 2.Seo P. Wegener's granulomatosis: managing more than inflammation. Curr Opin Rheumatol. 2008 Jan;20(1):10–6. doi: 10.1097/BOR.0b013e3282f18bef. [DOI] [PubMed] [Google Scholar]
- 3.Merkel PA, Seo P, Aries P, Neogi T, Villa-Forte A, Boers M, et al. Current status of outcome measures in vasculitis: focus on Wegener's granulomatosis and microscopic polyangiitis. Report from OMERACT 7. J Rheumatol. 2005 Dec;32(12):2488–95. [PubMed] [Google Scholar]
- 4.van der Woude FJ, Rasmussen N, Lobatto S, Wiik A, Permin H, van Es LA, et al. Autoantibodies against neutrophils and monocytes: tool for diagnosis and marker of disease activity in Wegener's granulomatosis. Lancet. 1985 Feb 23;1(0140-6736; 8426):425–9. doi: 10.1016/s0140-6736(85)91147-x. [DOI] [PubMed] [Google Scholar]
- 5.Gupta SK, Niles JL, McCluskey RT, Arnaout MA. Identity of Wegener's autoantigen (p29) with proteinase 3 and myeloblastin. Blood. 1990 Nov 15;76(0006-4971; 10):2162. [PubMed] [Google Scholar]
- 6.Finkielman JD, Lee AS, Hummel AM, Viss MA, Jacob GL, Homburger HA, et al. ANCA are detectable in nearly all patients with active severe Wegener's granulomatosis. Am J Med. 2007 Jul;120(1555-7162; 7):643–14. doi: 10.1016/j.amjmed.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Boomsma MM, Stegeman CA, van der Leij MJ, Oost W, Hermans J, Kallenberg CG, et al. Prediction of relapses in Wegener's granulomatosis by measurement of antineutrophil cytoplasmic antibody levels: a prospective study. Arthritis Rheum. 2000 Sep;43(0004-3591; 9):2025–33. doi: 10.1002/1529-0131(200009)43:9<2025::AID-ANR13>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 8.Slot MC, Tervaert JW, Boomsma MM, Stegeman CA. Positive classic antineutrophil cytoplasmic antibody (C-ANCA) titer at switch to azathioprine therapy associated with relapse in proteinase 3-related vasculitis. Arthritis Rheum. 2004 Apr 15;51(2):269–73. doi: 10.1002/art.20234. [DOI] [PubMed] [Google Scholar]
- 9.Yang JJ, Tuttle RH, Hogan SL, Taylor JG, Phillips BD, Falk RJ, et al. Target antigens for anti-neutrophil cytoplasmic autoantibodies (ANCA) are on the surface of primed and apoptotic but not unstimulated neutrophils. Clin Exp Immunol. 2000 Jul;121(1):165–72. doi: 10.1046/j.1365-2249.2000.01228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Csernok E, Ai M, Gross WL, Wicklein D, Petersen A, Lindner B, et al. Wegener autoantigen induces maturation of dendritic cells and licenses them for Th1 priming via the protease-activated receptor-2 pathway. Blood. 2006 Jun 01;107(0006-4971; 11):4440–8. doi: 10.1182/blood-2005-05-1875. [DOI] [PubMed] [Google Scholar]
- 11.Csernok E, Moosig F, Gross WL. Pathways to ANCA production: from differentiation of dendritic cells by proteinase 3 to B lymphocyte maturation in Wegener's granuloma. Clin Rev Allergy Immunol. 2008 Jun;34(3):300–6. doi: 10.1007/s12016-007-8056-8. [DOI] [PubMed] [Google Scholar]
- 12.Finkielman JD, Merkel PA, Schroeder D, Hoffman GS, Spiera R, St Clair EW, et al. Antiproteinase 3 antineutrophil cytoplasmic antibodies and disease activity in Wegener granulomatosis. Ann Intern Med. 2007 Nov 06;147(1539-3704; 9):611–9. doi: 10.7326/0003-4819-147-9-200711060-00005. [DOI] [PubMed] [Google Scholar]
- 13.Stone JH, Hoffman GS, Merkel PA, Min YI, Uhlfelder ML, Hellmann DB, et al. A disease-specific activity index for Wegener's granulomatosis: modification of the Birmingham Vasculitis Activity Score. International Network for the Study of the Systemic Vasculitides (INSSYS). Arthritis Rheum. 2001 Apr;44(4):912–20. doi: 10.1002/1529-0131(200104)44:4<912::AID-ANR148>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 14.Merkel PA, Curthbertson D, Hellmich B, Hoffman GS, Jayne D, Kallenberg CG, et al. Comparison of disease activity measures for ANCA-associated vasculitis. Ann Rheum Dis. 2008 Jul 29;(1468-2060) doi: 10.1136/ard.2008.097758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003 Mar 17;197(0022-1007; 6):711–23. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakou M, Knowlton N, Frank MB, Bertsias G, Osban J, Sandel CE, et al. Gene expression in systemic lupus erythematosus: Bone marrow analysis differentiates active from inactive disease and reveals apoptosis and granulopoiesis signatures. Arthritis Rheum. 2008 Nov;58(0004-3591; 11):3541–9. doi: 10.1002/art.23961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009 Feb 19;360(8):790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 18.Auman JT, McLeod HL. Applications of genomic tools to colorectal cancer therapeutics. Curr Opin Mol Ther. 2008 Dec;10(6):548–54. [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005 Oct 25;102(0027-8424; 43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003 Jul;34(3):267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 21.Yang JJ, Pendergraft WF, Alcorta DA, Nachman PH, Hogan SL, Thomas RP, et al. Circumvention of normal constraints on granule protein gene expression in peripheral blood neutrophils and monocytes of patients with antineutrophil cytoplasmic autoantibody-associated glomerulonephritis. J Am Soc Nephrol. 2004 Aug;15(1046-6673; 8):2103–14. doi: 10.1097/01.ASN.0000135058.46193.72. [DOI] [PubMed] [Google Scholar]
- 22.Alcorta DA, Barnes DA, Dooley MA, Sullivan P, Jonas B, Liu Y, et al. Leukocyte gene expression signatures in antineutrophil cytoplasmic autoantibody and lupus glomerulonephritis. Kidney Int. 2007 Oct;72(0085-2538; 7):853–64. doi: 10.1038/sj.ki.5002371. [DOI] [PubMed] [Google Scholar]
- 23.Leavitt RY, Fauci AS, Bloch DA, Michel BA, Hunder GG, Arend WP, et al. The American College of Rheumatology 1990 criteria for the classification of Wegener's granulomatosis. Arthritis Rheum. 1990 Aug;33(8):1101–7. doi: 10.1002/art.1780330807. [DOI] [PubMed] [Google Scholar]
- 24.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Moser K, Ortmann WA, et al. Expression levels for many genes in human peripheral blood cells are highly sensitive to ex vivo incubation. Genes Immun. 2004 Aug;5(5):347–53. doi: 10.1038/sj.gene.6364098. [DOI] [PubMed] [Google Scholar]
- 25.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002 Jan 1;30(1):207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan JS, Reed A, Chen F, Stewart CN., Jr Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006 Feb 22;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan W. A comparative review of statistical methods for discovering differentially expressed genes in replicated microarray experiments. Bioinformatics. 2002 Apr;18(4):546–54. doi: 10.1093/bioinformatics/18.4.546. [DOI] [PubMed] [Google Scholar]
- 28.Benjamini H, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. JRSS-B. 1995;57:289. [Google Scholar]
- 29.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003 May;5(2):73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nadon R, Woody E, Shi P, Rghei N, Hubschle H, Susko E, et al. Statistical inference in array genomics. In: Geschwind DGJ, editor. Microarrays for the Neurosciences Cambridge. MIT Press; 2002. [Google Scholar]
- 31.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998 Dec 8;95(25):14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jolliffe I. Principal Component Analysis. 2nd Edition Springer-Verlag; New York: 2002. [Google Scholar]
- 33.Ringnér M. What is principal component analysis? . Nat Biotechnol. 2008;26(3):303. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 34.Cowland JB, Borregaard N. The individual regulation of granule protein mRNA levels during neutrophil maturation explains the heterogeneity of neutrophil granules. J Leukoc Biol. 1999 Dec;66(0741-5400; 6):989–95. doi: 10.1002/jlb.66.6.989. [DOI] [PubMed] [Google Scholar]
- 35.Pascual V, Farkas L, Banchereau J. Systemic lupus erythematosus: all roads lead to type I interferons. Curr Opin Immunol. 2006 Dec;18(0952-7915; 6):676–82. doi: 10.1016/j.coi.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 36.Baechler EC, Bauer JW, Slattery CA, Ortmann WA, Espe KJ, Novitzke J, et al. An interferon signature in the peripheral blood of dermatomyositis patients is associated with disease activity. Mol Med. 2007 Jan-Feb;13(1-2):59–68. doi: 10.2119/2006-00085.Baechler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duan H, Fleming J, Pritchard DK, Amon LM, Xue J, Arnett HA, et al. Combined analysis of monocyte and lymphocyte messenger RNA expression with serum protein profiles in patients with scleroderma. Arthritis Rheum. 2008 May;58(5):1465–74. doi: 10.1002/art.23451. [DOI] [PubMed] [Google Scholar]
- 38.van der Pouw Kraan TC, Wijbrandts CA, van Baarsen LG, Voskuyl AE, Rustenburg F, Baggen JM, et al. Rheumatoid arthritis subtypes identified by genomic profiling of peripheral blood cells: assignment of a type I interferon signature in a subpopulation of patients. Ann Rheum Dis. 2007 Aug;66(8):1008–14. doi: 10.1136/ard.2006.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008 Jul;29(1):150–64. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fritsch G, Buchinger P, Printz D. Use of flow cytometric CD34 analysis to quantify hematopoietic progenitor cells. Leuk Lymphoma. 1993 Aug;10(6):443–51. doi: 10.3109/10428199309148201. [DOI] [PubMed] [Google Scholar]
- 41.Lu H, Denny M, Anderson M, McCune W, Kaplan M. The Role Of Low Density Granulocytes In Endothelial Damage in Systemic Lupus Erythematosus. Arthritis and Rheumatism. 2008;(9 (Suppl)):58, S949. [Google Scholar]
- 42.Wang D, Paz-Priel I, Friedman AD. NF-kappa B p50 regulates C/EBP alpha expression and inflammatory cytokine-induced neutrophil production. J Immunol. 2009 May 1;182(9):5757–62. doi: 10.4049/jimmunol.0803861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Just J, Moog-Lutz C, Houzel-Charavel A, Canteloup S, Grimfeld A, Witko-Sarsat V, et al. Proteinase 3 mRNA expression is induced in monocytes but not in neutrophils of patients with cystic fibrosis. FEBS Lett. 1999 Sep 03;457(0014-5793; 3):437–40. doi: 10.1016/s0014-5793(99)01098-4. [DOI] [PubMed] [Google Scholar]
- 44.Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007 Mar 1;109(5):2066–77. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popper SJ, Watson VE, Shimizu C, Kanegaye JT, Burns JC, Relman DA. Gene transcript abundance profiles distinguish Kawasaki disease from adenovirus infection. J Infect Dis. 2009 Aug 15;200(4):657–66. doi: 10.1086/603538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bauer JW, Bilgic H, Baechler EC. Gene-expression profiling in rheumatic disease: tools and therapeutic potential. Nat Rev Rheumatol. 2009 May;5(5):257–65. doi: 10.1038/nrrheum.2009.50. [DOI] [PubMed] [Google Scholar]
- 47.Petri M, Singh S, Tesfasyone H, Dedrick R, Fry K, Lal P, et al. Longitudinal expression of type I interferon responsive genes in systemic lupus erythematosus. Lupus. 2009 Oct;18(11):980–9. doi: 10.1177/0961203309105529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohlsson S, Wieslander J, Segelmark M. Increased circulating levels of proteinase 3 in patients with anti-neutrophilic cytoplasmic autoantibodies-associated systemic vasculitis in remission. Clin Exp Immunol. 2003 Mar;131(3):528–35. doi: 10.1046/j.1365-2249.2003.02083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schnabel A, Csernok E, Braun J, Gross WL. Activation of neutrophils, eosinophils, and lymphocytes in the lower respiratory tract in Wegener's granulomatosis. Am J Respir Crit Care Med. 2000 Feb;161(2 Pt 1):399–405. doi: 10.1164/ajrccm.161.2.9904076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material
Supplemental Table 1. The 40 genes significantly upregulated in WG vs. control PMNs, sorted by fold-change.
Supplemental Table 2. The 86 genes significantly upregulated in WG vs. control PBMCs, sorted by fold-change.
Supplemental Table 3. GSEA summary for WG vs. control PBMCs using genes ranked by the Gaussian z-value corresponding to their Welch t-test one-sided p-value.
Supplemental Table 4. GSEA summary for WG vs. control PMNs using genes ranked by the Gaussian z-value corresponding to their Welch t-test one-sided p-value.
Supplemental Table 5. Welch t-test results and Benjamini-Hochberg FDR for signature negative WG samples vs. controls, PBMC data.