Abstract
Purpose of review
The current rise in diet-related diseases continues to be one of the most significant health problems facing both the developed and the developing world. The use of metabolomics – the accurate and comprehensive measurement of a significant fraction of important metabolites in accessible biological fluids – for the assessment of nutritional status, is a promising way forward. The basic toolset, targets, and knowledge are all being developed in the emerging field of metabolomics, yet important knowledge and technology gaps will need to be addressed in order to bring such assessment to practice.
Recent findings
Dysregulation within the principal metabolic organs (e.g. intestine, adipose, skeletal muscle, liver) are at the center of a diet-disease paradigm that includes metabolic syndrome, type 2 diabetes, and obesity. The assessment of both essential nutrient status, and the more comprehensive systemic metabolic response to dietary, lifestyle, and environmental influences (e.g. metabolic phenotype) are necessary for the evaluation of status in individuals that can identify the multiple targets of intervention needed to address metabolic disease.
Summary
The first proofs of principle building the knowledge to bring actionable metabolic diagnostics to practice through metabolomics are now appearing.
Keywords: Health Assessment, Metabolic Assessment, Nutritional Assessment, Metabolic Phenotype, Metabolomics
INTRODUCTION
The human genome initiative established a new vision for biological research and its translation to health [1], and metabolomics has emerged as one of the toolsets for bringing it to practice [2]. One goal is to apply its combination of analytics, informatics and biochemistry to identify indicators of health that could guide individuals to improve it with nutrition [3]. Analyzing metabolites in blood is already the basis of diagnosing and monitoring heart disease and diabetes risk (e.g. cholesterol, triglycerides and glucose), for example. Metabolomics promises to expand this inherent capability of plasma and urine metabolites to assess health status.
The challenge of applying metabolomics to nutrition is to increase not just the number of metabolites measured but also the knowledge of how variations in the different metabolites–as absolute and relative concentrations-correlate with nutrition and health. Although we have established the levels of the essential nutrients that are adequate to prevent overt deficiency, there is still much to be learned about optimal levels in different individuals, which do more than just prevent deficiency and instead optimize specific metabolic functions and processes.
The systemic response of metabolism to the combined effects of nutrient status, genetic background, epigenetic changes, lifestyle choices, and environmental fluctuations within an individual at a specific point in time (e.g. metabolic phenotype) (Figure 1) is potentially a more sensitive and actionable reflection of nutritional and metabolic status. With such nutritional and metabolic status indicators in place, intelligent interventions, including foods, supplements, and lifestyle modifications, would be recommended to guide an individual’s metabolic phenotype in a more beneficial or personally desired direction. Such a vision would bring a fundamentally different perspective to human diet management and empower a much more detailed, interactive and ultimately valuable industrial engine to delivering health. At this point in time, it is appropriate to ask, where are we in the scientific development of the needed tools and knowledge?
Figure 1. Assessment of metabolic phenotype.
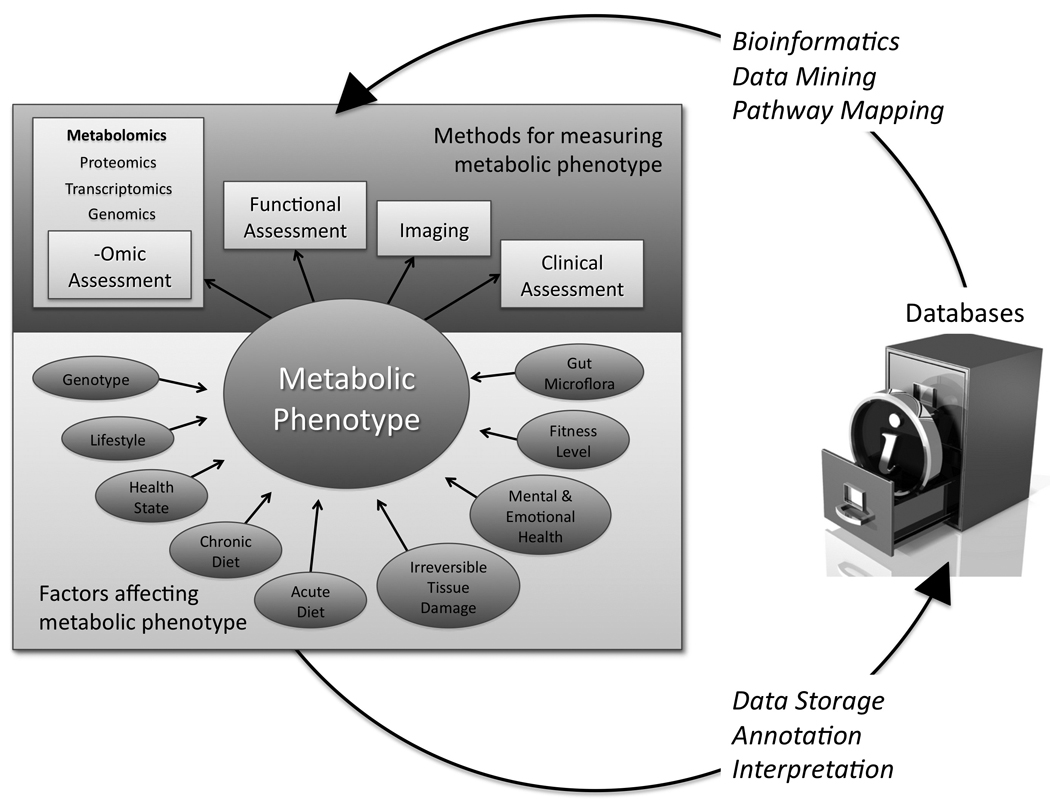
Genotype, the acute and chronic effects of diet, lifestyle, health state, gut microflora, the extent of irreversible tissue damage, mental and emotional health, fitness level and other factors influence the metabolic phenotype of an individual at any point in time. Metabolic phenotype can be measured in a number of ways including imaging, clinical assessment, functional assessment, and –omic assessment (which includes genomics, transcriptomics, proteomics, and metabolomics). Data produced by the various measurement and assessment techniques are annotated and interpreted, then stored in electronic databases as public data repositories, which are in turn used for bioinformatics, data mining, and pathway mapping to both inform future research and extract further information out of the collected data [13].
THE TOOLSET
Metabolomics has been variously defined, and controversy still exists as to the specifics of the nomenclature proposed. At this point, the Metabolomics Society (metabolomicssociety.org) is adhering to a convention consistent with the other 'omic sciences: gene–genome–genomics, protein–proteome– proteomics, and metabolite–metabolome–metabolomics. For the purposes of this review, metabolomics is defined as the global analysis of metabolites—small molecules generated in the process of metabolism—that represent the sum total of all the metabolic pathways in an organism, with a focus on the identification of each pathway and its role in an organism’s function. Although various techniques have been proposed that create highly complex signatures based on either mass, magnetic resonance or chemical signals, if these signatures are not assigned to the identity and quantity of specific metabolites they will not be discussed in this review.
Metabolomics has been included in the NIH roadmap as a core technology in the overall initiative to guide the development of and assist in delivering the solutions to human metabolic diseases [4]. Application of metabolomics specifically to the field of nutrition faces a number of challenges. The metabolome is not definable in the same sense as the genome. Metabolites change constantly in every cell and body fluid, for example in response to food intake (Figure 2), hence the metabolome will always be a biological concept that is only pragmatically definable. Nonetheless, all of our cells and biofluids contain a finite number of key metabolites and homeostasis is generally maintained so that the actual variations in any given metabolite ‘pool’ are typically minor relative to the abundance of the metabolites. These basic molecules of human metabolism, i.e. those that all humans have in relatively constant amounts and without which life is not possible can be considered the endogenous metabolome. Defining the molecules that constitute this core metabolite pool is an immediate priority of the field of metabolomics. The Human Metabolome Project [5*] has established the first draft of the metabolome. In addition to the endogenous metabolome, there are perhaps hundreds of thousands of molecules in the flora and fauna consumed as food that are absorbed into blood, metabolized, and released into body fluids like urine and saliva. Our resident microflora also produce metabolites that can contribute to and alter the metabolome of human biofluids. Ongoing metabolomics research will continue the process of chronicling and annotating these molecules especially as they affect the endogenous metabolome.
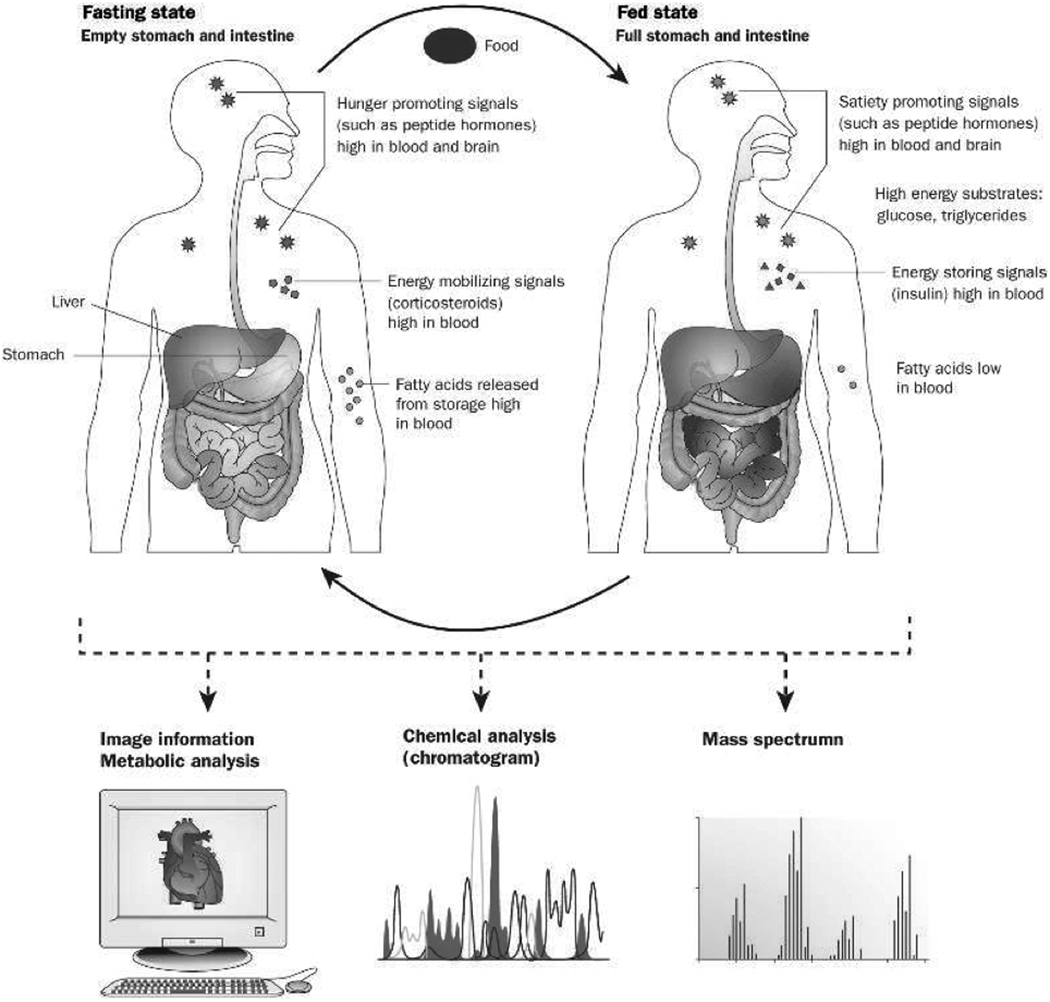
Figure 2. Dynamics of fasted and fed states and their metabolic assessment.
Metabolic regulation is a dynamic process varying from fasted to fed states. Levels of metabolites rise and fall in response to the influx of nutrients from foods, as do the signaling systems that stimulate appetite and mobilize energy in times of need, and inhibit eating and direct energy-rich molecules into storage in times of surplus. The combined molecular-imaging and metabolic-profiling capabilities available today as a result of revolutionary advances in analytical sciences provide an unprecedented opportunity to accurately monitor these processes. If brought to practice as part of routine health care, these tools would similarly revolutionize the diagnosis, treatment and prevention of metabolic diseases.
Reproduced with permission from: Macé K, Corthésy-Theulaz I, Fay LB et al; Effects of food on metabolic regulation and disorders; Nature Insight: Obesity and Diabetes 2006; 444 (7121): A1–A5. © Nature Publishing Group 2006.
THE SAMPLES
There remain critical needs for sampling protocols and analytical platforms for metabolomics. No single analytical technology is capable of measuring all metabolites in a biological sample. The diversity in chemical structures, abundances and reactivities has to date frustrated all attempts to simultaneously identify and quantify all metabolites in a single, high throughput platform. The problem of identification is being addressed in part through increasingly accurate libraries of metabolite spectra (mass NMR etc.) and chemical properties of diverse biological samples [5*]. The problem of quantitation is more daunting. Nonetheless, for an individual’s nutritional status, the amount of metabolites, not their simple presence or absence is necessary. For quantitation of metabolites in biological samples both the numerator, i.e. the absolute mass or molar abundance of each metabolite, and the denominator, the absolute volume, mass or integrated molar ensemble of the biological sample are needed. NIH established the metabolomics standard reference material project in which scientists and methods development can be unified around a single sample [6, 7]. The ongoing development of analytical platforms is beyond the scope of this review [8–12].
THE TARGETS
The goal is to measure nutritional status. Given the scope of this challenge, the scientific community is assembling electronic databases to combine and compare information (Figure 1). The storage of annotated data from diverse studies into public sites will enable the mining of existing data for new discoveries, will drive hypothesis generation and inform the design of new studies [13].
Essential Nutrient Status
The knowledge of essential nutrients is mature. However, subsets of the population still suffer from deficiencies or suboptimal intakes of essential nutrients due to overt poverty, food choices, genetic polymorphisms that increase or modify needs, conditions or medications that alter nutrient utilization or metabolism, or malabsorption syndromes such as celiac disease. In some cases, e.g. vitamin D, diet and lifestyle changes that minimize alternative sources (e.g. sun exposure), are causing inadequacies and even deficiencies. In each of these cases, accurate measures of nutritional status of individuals in the developed world or specific groups in developing countries would identify those involved and ideally indicate the nature of the deficiencies. For such nutritional status measures, metabolic profiling holds considerable promise.
Metabolomic approaches have been used to pursue the signatures of nutrient-related diseases. NMR based analyses identified metabolites whose abundance in urine were diagnostic in celiac patients [14]. Priego et al. [15] demonstrated that the fat soluble nutrients and their metabolites could be measured in serum by LC-MS, though these methods have not yet been brought to diagnostics of human nutrient status. The pathways associated with vitamin D metabolism are particularly attractive as measures of nutrient status since they represent a proof of principle of how examining the quantitative representation of intermediates within a pathway can provide information beyond a single nutrient. For example, as vitamin D metabolism is activated by low calcium status, metabolomics of the vitamin D pathway identifies both vitamin D and calcium status [16].
Metabolic Status: beyond essential nutrients
Nutritional health depends on more than essential nutrient intake. In fact, the global epidemic of non-communicative diseases is largely driven by diets in which essential nutrition is adequate, but chronic imbalances of diet in a background of varying lifestyles are causing metabolic diseases. Diagnosing such varying health states is expected to require simultaneously the measurement of nutritional and metabolic status across a wide range of physiologic, metabolic and nutritional conditions in a range of human genotypes, lifestages and lifestyles (Figures 1 and 2). Building extensive knowledge across such a complex healthscape will require studies from a range of disciplines and targets.
The Intestine: postprandial metabolism meets the microbiome
Research focusing on the intestine is extending 3 variables: time, genetics and microbial ecology. Scientists are pursuing the dynamics of diet (postprandial metabolism), the diversity of diet responses in humans, and the complex interplay between diet and the ‘other genome,’ the microbiota within each human. Although complex, the research to date using metabolomics has established some fascinating new issues for nutrition.
The concept that differences in metabolism resolve healthy and diseased populations has been extended to apparently healthy individuals in a study examining postprandial lipid responses to the same oral challenge [17]. This study found that as many as 50% of measured lipid species were discriminators of metabolic phenotype. Whereas the inter-individual variation among the subjects was high, the intra-individual variation within each subject in the dynamic responses in particular metabolites was low in free-living subjects even over 3 months.
The intestinal microbiome is a new dimension of human health in which diet, its composition and dynamics, plays a key role. The bacteria within us constitute a parallel genome (actually thousands of microbial genomes) with their assembled metabolic activities. Studies have indeed begun to build the tools to characterize the contribution of the microbiome to overall metabolism [18*]. A decidedly evolutionary perspective was taken to explore the interaction between diet and the microbiome. The study showed that the basis of human milk’s guidance of the human infant’s microbiome is metabolic [19**]. The genomic sequence and metabolic characterization of Bifidobacterium longum subsp. Infantis from the intestine of breast fed infants revealed multiple genes in 4 discrete clusters that are required to metabolize the human oligosaccharides in milk [20] and thus support its ecological niche in the infant.
Metabolomics as a strategy was in turn used to great advantage by complex protocols to simultaneously examine the discrete contributions of dietary carbohydrate, explicitly consumed bacteria, and the host [21]. All 3 were critical inputs to the net metabolism of the ‘panorganism’ [21]. This is a landmark study from a combined academic and industrial collaboration examining multiple strategies to probe the diverse nutritional contributions of the microbiota to metabolic health. The importance of the microflora to the abundance of a diverse array of metabolites in human plasma was confirmed separately [11*]. It can now truly be stated that a complete examination of nutritional health must consider the health of the microbiome.
Metabolomics and Adipose: the storage tissue for fuel
The tissue specific annotation of metabolism of adipose is not a new theme of nutrition-related research. However, the importance of this dynamic tissue to metabolic health and disease has brought adipose to modern celebrity status. Yet, metabolomics studies have only begun to be designed to illuminate the activities and functions of adipose to overall health. Mattilla et al., [22] described an integrative approach to the methods required for metabolomics of adipose focusing logically on lipids and amino acids.
Metabolomics and Skeletal Muscle: the sink for fuel
Epidemiologic data show profound benefits of exercise to health. Asking why, skeletal muscle has been studied for its impact on multiple aspects of metabolism, physiology and immunology. The mechanistic understanding of cellular responses to exercise represents one of the great achievements of cell biology over the past few years [23**, 24**]. While these mechanisms have yet to be annotated into metabolomics protocols, studies have documented the importance of altered muscle metabolism to whole body energy regulation. Choi et al. [25*], showed the effects of exercise on cellular products of metabolism and protection from metabolic disease. Koves et al. [26], examined fuel metabolites in a genetic model of insulin resistance demonstrating that the acyl carnitines, metabolites previously thought to be solely cellular effectors, vary significantly associated with the success of fuel disposal. Finally, examining humans, exposed to exercise as a standardized input variable, using metabolomics to measure outcomes, revealed a wide array of metabolic pathways that are quantitatively responsive to exercise consistent with cellular mechanistic predictions [27], and to nutritional modulation in the immediate post-exercise period [28].
Metabolomics and the Liver
The suite of metabolic disorders associated with excess calorie intake is epidemic. The liver’s failure in processing this excess is as challenging as measuring it. Diet assessment is imprecise, with individuals underreporting energy intake by as much as 600 Calories/d [29]. Probing the liver’s status with measures of its metabolic products has been a hallmark of diagnostics for decades.
Lipidomic studies have revealed an unexpected signaling role for specific fatty acids. In an elegant study combining lipid analysis with molecular cell biology, Cao et al. identified a novel function in what they termed a lipokine, or lipid hormone, in mice [30**]. Increased lipogenesis in the adipose tissue of knockout mice resulted in both higher adipose tissue levels and higher circulating levels of palmitoleate (16:1n7), which suppressed lipogenesis in the liver, resisted steatosis, and increased muscle insulin sensitivity and glucose uptake.
In humans, analysis using stable isotopes and mass isotopomer distribution continues to be the gold standard for analyzing the quantitative flux of metabolites through pathways. Studies though few have documented one inescapable reality. individuals vary in their metabolism and its regulation. For example, some women maintained consistently low fractional lipogenesis rates of <10% regardless of diet, whereas in others, 96 h of controlled glucose or sucrose overfeeding caused a spike in lipogenesis rates as high as 4-fold from baseline to account for 50% of plasma lipids [31]. Parks et al. found that acute consumption of fructose stimulated lipogenesis and triglyceride storage in the liver [32*]. This exciting study showed that increasing the proportion of fructose relative to glucose from 0 to 50% to 75% resulted in higher fractional lipogenesis and hyperlipidemia after the second meal, regardless of an 80% decrease in circulating free fatty acids.
With the rising incidence of non-alcoholic steatohepatitis (NASH), liver health has become a dietary target. Lipidomic profiles were taken of 9 patients with simple steatosis, 9 patients with NASH and 9 age- and BMI-matched controls [33]. As expected, total lipids were higher in fatty liver and NASH, with concomitant increases in lipid species indicative of higher turnover of lipids and a reduced synthesis or increased utilization of phospholipids. NASH and fatty liver patients also had decreased conversion of linoleic acid (18:2n6) to its long-chain product arachidonic acid (20:4n6).
Animal models continue to provide information not available with humans. Lipidomic profiles of fatty livers of alcoholic pigs were compared with controls [34]. Chronic alcohol consumption increased lipogenesis and steatosis whereas, phospholipid production and VLDL secretion decreased. Taken together, metabolomic analysis revealed specific pathways involved in metabolic disease, enabling both the assessment of status, and targeted interventions that address the underlying metabolic dysfunction rather than just treating symptoms.
CONCLUSION
The nutritional status of humans remains a major challenge for public health agencies, clinical practice and the food industry, as people around the world are suffering diet related illness due to inappropriate food choices and lifestyles. Routine assessment could provide the means to recognize individual variations in nutritional status but considerable knowledge will be needed to bring such assessment to practice. Metabolomics as a science is actively developing analytical platforms capable of accurately measuring a significant fraction of important metabolites in accessible biological fluids. In parallel the scientific community is beginning to apply these tools to annotating how these metabolic profiles differ in individuals according to their health. Electronic databases are being designed to house the results of this initiative as a public knowledge resource. The dysregulations in hepatic lipid metabolism are at the center of a new diet-disease paradigm (metabolic syndrome, type 2 diabetes, obesity) and are amenable to a first generation of personalizing health through metabolic assessment. The first proofs of principle building the knowledge to bring actionable metabolic diagnostics to practice through metabolomics are now appearing.
Acknowledgments
Funding sources:
The University of California Discovery Program (05GEB01NHB), the National Institute of Environmental Health Sciences (P42ES004699), the California Dairy Research Foundation and the CHARGE study (P01 ES11269).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Collins FS, Green ED, Guttmacher AE, Guyer MS. A vision for the future of genomics research. Nature. 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- 2.German JB, Watkins SM, Fay LB. Metabolomics in practice: emerging knowledge to guide future dietetic advice toward individualized health. J Am Diet Assoc. 2005;105:1425–1432. doi: 10.1016/j.jada.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Zeisel SH. Nutrigenomics and metabolomics will change clinical nutrition and public health practice: insights from studies on dietary requirements for choline. Am J Clin Nutr. 2007;86:542–548. doi: 10.1093/ajcn/86.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zerhouni E. Medicine. The NIH Roadmap. Science. 2003;302:63–72. doi: 10.1126/science.1091867. [DOI] [PubMed] [Google Scholar]
- 5. Forsythe IJ, Wishart DS. Exploring human metabolites using the human metabolome database. Curr Protoc Bioinformatics. 2009;Chapter 14(Unit14 8) doi: 10.1002/0471250953.bi1408s25. Article describing the data structure, resources and mining tools of the first publically accessible, web based, electronic metabolome database established in Canada.
- 6.Phinney KW, Sander LC, Sharpless KE, Wise SA. NIST Develops Serum-Based Standard Reference Materials to Assess Nutritional Status. National Institute of Standards and Technologies. 2006 http://www.cstl.nist.gov/projects/fy06/food0683904.pdf.
- 7.Phinney KW, Sander LC, Sharpless KE, Turk GC, Wise SA. Innovation in Metabolomics: A New Standard Reference Material for Metabolites in Human Plasma. National Institute of Standards and Technology. 2006 http://www.cstl.nist.gov/projects/fy06/health0683905.pdf.
- 8.Buscher JM, Czernik D, Ewald JC, et al. Cross-Platform Comparison of Methods for Quantitative Metabolomics of Primary Metabolism. Anal Chem. 2009;81:2135–2143. doi: 10.1021/ac8022857. [DOI] [PubMed] [Google Scholar]
- 9.Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Issaq HJ, Abbott E, Veenstra TD. Utility of separation science in metabolomic studies. J Sep Sci. 2008;31:1936–1947. doi: 10.1002/jssc.200700601. [DOI] [PubMed] [Google Scholar]
- 11. Wikoff WR, Anfora AT, Liu J, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. This article uses metabolomics as the means to gain a more detailed and comprehensive description of the contribution of gut bacteria to the metabolites present in plasma. Although the study in mice addressed the secondary metabolism of drugs by gut bacteria, the definitive results have broad implications to diet.
- 12.Zhang NR, Yu S, Tiller P, et al. Quantitation of small molecules using high-resolution accurate mass spectrometers - a different approach for analysis of biological samples. Rapid Commun Mass Spectrom. 2009;23:1085–1094. doi: 10.1002/rcm.3975. [DOI] [PubMed] [Google Scholar]
- 13.Lemay D, Zivkovic AM, German JB. Building the bridges to bioinformatics in nutrition research. Am J Clin Nutr. 2007;86:1261–1269. doi: 10.1093/ajcn/86.5.1261. [DOI] [PubMed] [Google Scholar]
- 14.Bertini I, Calabro A, De Carli V, et al. The metabonomic signature of celiac disease. J Proteome Res. 2009;8:170–177. doi: 10.1021/pr800548z. [DOI] [PubMed] [Google Scholar]
- 15.Priego Capote F, Jimenez JR, Granados JM, de Castro MD. Identification and determination of fat-soluble vitamins and metabolites in human serum by liquid chromatography/triple quadrupole mass spectrometry with multiple reaction monitoring. Rapid Commun Mass Spectrom. 2007;21:1745–1754. doi: 10.1002/rcm.3014. [DOI] [PubMed] [Google Scholar]
- 16.Aronov PA, Hall LM, Dettmer K, et al. Metabolic profiling of major vitamin D metabolites using Diels-Alder derivatization and ultra-performance liquid chromatography-tandem mass spectrometry. Anal Bioanal Chem. 2008;391:1917–1930. doi: 10.1007/s00216-008-2095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zivkovic AM, Wiest MM, Nguyen U, Nording ML, Watkins SM, German JB. Assessing Individual Metabolic Responsiveness to a Lipid Challenge Using a Targeted Metabolomic Approach. Metabolomics. 2009;5:209–218. doi: 10.1007/s11306-008-0136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin FP, Dumas ME, Wang Y, et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol. 2007;3:112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sela DA, Chapman J, Adeuya A, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. Study drew a direct line between the human milk glycome (oligosaccharides of human milk) the genome of bacterium B. infantis and the unique genes encoding the metabolic activities digesting these oligosaccharides that promote the competitive success of this microorganism within the breast fed infant intestine.
- 20.LoCascio RG, Ninonuevo MR, Freeman SL, et al. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem. 2007;55:8914–8919. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- 21. Martin FP, Sprenger N, Yap IK, et al. Panorganismal Gut Microbiome-Host Metabolic Crosstalk. J Proteome Res. 2009;8:2090–2105. doi: 10.1021/pr801068x. An ambitious study combining multi-dimensional interventions (dietary prebiotics, probiotics and combinations) with multiple tissue and fluid generalized and targeted metabolomics to begin the process of annotating the contribution of the intestinal microbiome to whole body metabolism in mice.
- 22.Mattila I, Seppanen-Laakso T, Suortti T, Oresic M. Application of lipidomics and metabolomics to the study of adipose tissue. Methods Mol Biol. 2008;456:123–130. doi: 10.1007/978-1-59745-245-8_9. [DOI] [PubMed] [Google Scholar]
- 23. Handschin C, Spiegelman BM. The role of exercise and PGC1alpha in inflammation and chronic disease. Nature. 2008;454:463–469. doi: 10.1038/nature07206. The remarkable story of the mechanistic link between exercise and its diverse benefits to human health, identifying the transcriptional co-activator PGC-1a as both a regulator of mitochondrial biogenesis and the stimulator of a wide range of protective gene clusters.
- 24. Narkar VA, Downes M, Yu RT, et al. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134:405–415. doi: 10.1016/j.cell.2008.06.051. This paper reveals what many have thought to be a fond but wild dream, that it was possible to understand biochemistry so well, that we could replace hard work (exercise) with easy chemistry - or in this case pharmacology.
- 25. Choi CS, Befroy DE, Codella R, et al. Paradoxical effects of increased expression of PGC-1alpha on muscle mitochondrial function and insulin-stimulated muscle glucose metabolism. Proc Natl Acad Sci U S A. 2008;105:19926–19931. doi: 10.1073/pnas.0810339105. Study beginning the process of taking the detailed cell biology of transcriptional co-activation of mitochondrial biogenesis to the processes of whole body metabolism and glucose regulation. Take home: biology still has surprises in store!
- 26.Koves TR, Ussher JR, Noland RC, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7:45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 27.Yan B, A J, Wang G, et al. Metabolomic investigation into variation of endogenous metabolites in professional athletes subject to strength-endurance training. J Appl Physiol. 2009;106:531–538. doi: 10.1152/japplphysiol.90816.2008. [DOI] [PubMed] [Google Scholar]
- 28.Chorell E, Moritz T, Branth S, et al. Predictive Metabolomics Evaluation of Nutrition-Modulated Metabolic Stress Responses in Human Blood Serum During the Early Recovery Phase of Strenuous Physical Exercise. J Proteome Res. 2009 doi: 10.1021/pr900081q. [DOI] [PubMed] [Google Scholar]
- 29.Champagne CM, Bray GA, Kurtz AA, et al. Energy intake and energy expenditure: a controlled study comparing dietitians and non-dietitians. J Am Diet Assoc. 2002;102:1428–1432. doi: 10.1016/s0002-8223(02)90316-0. [DOI] [PubMed] [Google Scholar]
- 30. Cao H, Gerhold K, Mayers JR, et al. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell. 2008;134:933–944. doi: 10.1016/j.cell.2008.07.048. One of the first studies to use metabolomics as a key to revealing new insights into mechanisms of metabolic regulation. The identification of metabolic products as part of the language of inter-organ cross talk may be logical in retrospect, but it is nonetheless an elegant application of systems biology.
- 31.McDevitt RM, Bott SJ, Harding M, et al. De novo lipogenesis during controlled overfeeding with sucrose or glucose in lean and obese women. Am J Clin Nutr. 2001;74:737–746. doi: 10.1093/ajcn/74.6.737. [DOI] [PubMed] [Google Scholar]
- 32. Parks EJ, Skokan LE, Timlin MT, Dingfelder CS. Dietary sugars stimulate fatty acid synthesis in adults. J Nutr. 2008;138:1039–1046. doi: 10.1093/jn/138.6.1039. Once again proving the value of actually measuring the truth of biology, this study used very careful flux determinations to quantify lipogenesis after varying proportions of fructose in a fixed carbohydrate bolus 'meal'. Results beg the question: what does fructose do to peripheral lipid metabolism?
- 33.Puri P, Baillie RA, Wiest MM, et al. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology. 2007;46:1081–1090. doi: 10.1002/hep.21763. [DOI] [PubMed] [Google Scholar]
- 34.Zivkovic AM, German JB, Esfandiari F, Halsted CH. Quantitative lipid metabolomic changes in alcoholic micropigs with fatty liver disease. Alcohol Clin Exp Res. 2009;33:751–758. doi: 10.1111/j.1530-0277.2008.00892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]