Abstract
Adult bone marrow stromal cells (BMSCs) are capable of differentiating into cardiomyocyte-like cells in vitro and contribute to myocardial regeneration in vivo. Consequently, BMSCs may potentially play a vital role in cardiac repair and regeneration. However, this concept has been limited by inadequate and inconsistent differentiation of BMSCs into cardiomyocytes along with poor survival and integration of neo-cardiomyocytes after implantation into ischemic myocardium. In order to overcome these barriers and to explore adult stem cell based myocardial regeneration, we have developed an in vitro model of three-dimensional (3-D) cardiac muscle using rat ventricular embryonic cardiomyocytes (ECMs) and BMSCs. When ECMs and BMSCs were seeded sequentially onto a 3-D tubular scaffold engineered from topographically aligned type I collagen fibers and cultured in basal medium for 7, 14, 21, or 28 days, the maturation and co-differentiation into a cardiomyocyte lineage was observed. Phenotypic induction was characterized at morphological, immunological, biochemical and molecular levels. The observed expression of transcripts coding for cardiomyocyte phenotypic markers and the immunolocalization of cardiomyogenic lineage-associated proteins revealed typical expression patterns of neo-cardiomyogenesis. At the biochemical level differentiating cells exhibited appropriate metabolic activity and at the ultrastructural level myofibrillar and sarcomeric organization were indicative of an immature phenotype. Our 3-D co-culture system sustains the ECMs in vitro continuum of differentiation process and simultaneously induces the maturation and differentiation of BMSCs into cardiomyocyte-like cells. Thus, this novel 3-D co-culture system provides a useful in vitro model to investigate the functional role and interplay of developing ECMs and BMSCs during cardiomyogenic differentiation.
Keywords: Bone marrow stromal cells, Mesenchymal stem cells, Embryonic cardiac myocytes, Myocardial regeneration, Cardiac tissue engineering
Introduction
It is well established that postnatal bone marrow harbors a heterogeneous population of adult stem cells and precursor cells that includes hematopoietic stem cells (HSCs), bone marrow stromal cells or mesenchymal stem cells (BMSCs/MSCs), multipotent adult progenitor cells (MAPCs) and endothelial precursor cells (EPCs) [1]. These cells have a unique combination of surface antigenic markers and have the potential to generate different sets of differentiated progeny [2]. Adult stems cells, including BMSCs, exhibit a certain degree of developmental plasticity that enables them to differentiate across boundaries of lineage, tissue and germ layers [1]. This differentiation potential of BMSCs has prompted us to explore the ability of these cells to differentiate into myocardium.
Previous reports indicate that BMSCs can be directed to differentiate into multiple mesenchymal and non-mesenchymal cells in vitro including myocyte-like and cardiomyocyte-like cells, which may contract in vitro [3-7]. Preclinical and clinical studies have suggested that whole marrow isolates and/or cultured marrow stem cells may also contribute to cardiac repair in vivo and alleviate cardiac symptoms, although the mechanism for this remains obscure [8-11]. These cells have also been grafted onto ischemic heart tissue, but their extent of functional integration remains unresolved [12-13]. In addition, these cells may contribute to revascularization after myocardial injury [14]. Taken together these data suggest that BMSCs could play a vital role in cardiac regeneration, but this concept requires further validation.
Cumulative evidence demonstrates that exogenous stem cells when systemically infused, transplanted or directly infiltrated into cardiac lesions generate relatively few neo-cardiomyocytes, but some types of cells such as BMSCs and side population of HSCs contribute directly to the endothelial cell populations, especially near the infarcted zones [14-15]. A major issue that remains to be addressed is the extent to which introduced stem cells contribute directly to the formation of neo-cardiomyocytes versus their contribution to and/or stimulation of an enhanced local vascular response, which in turn may act as a supportive microenvironment for regeneration [16-17].
Unlike BMSCs, embryonic stem cells (ESCs) can be readily directed to differentiate into cardiomyocyte-like cells in vitro [18]. The ESC-derived three-dimensional (3-D) embryoid bodies (EBs) contained clusters of spontaneously beating cardiomyocytes. Subsequent reports have revealed that these ESC-derived cardiomyocytes undergo an orderly array of developmental gene expression that closely mimic that which occurs during in vivo cardiac embryogenesis [19]. Besides, these cells acquire the physiopharmacological characteristics of terminally differentiated cardiac myocytes [18-19]. The utility of ESCs in facilitating cardiac muscle regeneration is still in its incipient stages. A number of issues, including a propensity for some implanted ESCs to form benign or malignant teratomas in the heart, remain to be addressed.
Use of adult stem cells in the stimulation of mammalian cardiac muscle regeneration is in its infancy, and to date, it has been difficult to determine the efficacy of the procedures that have been employed. The outstanding question remains whether stem cells derived from the bone marrow or some other location within or outside of the heart can populate a region of myocardial damage and transform into tissue-specific cells and also exhibit functional synchronization [Carlson, 2007]. As a result, this necessitates the development of an appropriate in vitro 3-D model of cardiomyogenesis and prompts the development of a 3-D cardiac muscle construct for tissue engineering purposes, especially using the adult stem cell, BMSCs.
In this study, we investigated the cardiomyogenic differentiation potential of BMSCs in the milieu of embryonic cardiac myocytes (ECMs) when co-seeded onto a three-dimensional (3-D) tubular scaffold engineered from aligned type I collagen strands and cultured in basal medium. In this culture condition, cells underwent maturation and differentiation characteristic of myogenic lineage and acquired the cardiac phenotype.
Materials and Methods
Fabrication of tubular scaffold
The 3-D collagen type I tube served as a scaffold on which both rat BMSCs and ECMs co-differentiation cultures were carried out. The details of the production and properties of the collagen tubes have previously been described [20]. Briefly, a 25 mg/ml solution of bovine collagen type I was extruded with a device that contained two counter-rotating cones. The liquid collagen was fed between the two cones and forced through a circular annulus in the presence of an NH3-air (50-50 vol/vol) chamber. This process results in a hollow cylindrical tube of aligned collagen fibrils with an inner central lumen. The dimensions of tubes produced for this set of experiments had a length of 20 mm with a luminal diameter of 4 mm and an external diameter of 5 mm, leaving a wall thickness of 1 mm. The collagen tubes had a defined fiber angle of 18° relative to the central axis of the tube and had pores ranging from 1 to 10 μm. The tubes were sterilized using gamma radiation 1200 Gy followed by Stratalinker UV crosslinker 1800 (Stratagene) and were then placed in Mosconas's solution (in mM: 136.8 NaCl, 28.6 KCl, 11.9 NaHCO3, 9.4 glucose, 0.08 NaH2PO4, pH 7.4) (Sigma-Aldrich) containing 1 μl/ml gentamicin (Sigma-Aldrich) and incubated in 5% CO2 at 37°C until cellular seeding.
BMSCs isolation, expansion and maintenance
Procedures were performed in accordance with the guidelines for animal experimentation by the Institutional Animal Care and Use Committee (IACUC), School of Medicine, University of South Carolina. Rat BMSCs were isolated from the bone marrow of adult 300g Sprague Dawley®™ SD®™ rats (Harlan Sprague Dawley, Inc.) as described previously [21]. Briefly, after deep anesthesia, the femoral and tibial bones were removed aseptically and cleaned extensively to remove associated soft connective tissues. The marrow cavities of these bones were flushed with Dulbecco's Modified Eagle Medium (DMEM; Invitrogen) and combined. The obtained marrow plugs were triturated, and passed through needles of decreasing gauge (from 18 gauge to 22 gauge) to break up clumps and cellular aggregates. The resulting single-cell suspensions were centrifuged at 200g for 5 minutes. Nucleated cells were counted using a Neubauer chamber.
Cells were plated at a density of 5 × 106 − 2 × 107 cells per T75 cm2 flasks in basal media composed of DMEM supplemented with 10% fetal bovine serum (FBS, lot-selected; Hyclone), gentamicin (50 μg/ml) and amphotericin B (250 ng/ml) and incubated in a humidified atmosphere of 5% CO2 at 37°C for 7 days. The medium was replaced, and ch anged three times per week until the cultures became ∼70% confluent (between 12 and 14 days). Cells were trypsinized using 0.05% trypsin-0.1% EDTA and re-plated at a density of 1 × 106 cells per T75 cm2 flasks. After three passages, attached marrow stromal cells were devoid of any non-adhering population of cells.
Phenotypic characterization of BMSCs by flow cytometry and confocal microscopy
Qualitative evaluation for various cell surface markers was performed on cells grown in the Lab-tek™ chamber slide system™ (Nunc) using a Zeiss LSM 510 Meta confocal scanning laser microscope (Carl Zeiss, Inc.) and quantitative analysis of the same set of markers was performed by single-color flow cytometry using a Coulter® EPICS® XL™ Flow Cytometer (Beckman Coulter, Inc.) as described previously [Valarmathi et al., 2009]. Briefly, the passage 3 maintained BMSCs were trypsinized, pelleted at 200g for 5 minutes and washed twice with Mosconas's solution, pH 7.4. Cells were re-suspended in staining buffer (1.5% bovine serum albumin in PBS, pH 7.4) (BSA, Sigma-Aldrich) and incubated for 30 minutes at 4°C with appropriate dilutions of FITC-conjugated mouse anti-rat CD11b, CD31, CD44, CD45, CD90 and OX43 monoclonal antibodies for direct immunostaining (Table 1). Similarly, the cells were incubated with FITC-unconjugated mouse anti-rat CD34, CD73 and CD106 monoclonal antibodies followed by incubation with appropriate dilutions of FITC-conjugated anti-mouse secondary antibodies for indirect immunostaining (Table 1). FITC labeled mouse anti-rat-IgGs antibodies served as the isotype controls. The stained cells were washed twice with Moscona's solution and the data acquired immediately or alternately the cells were fixed in ice-cold 0.5% paraformaldehyde (Sigma-Aldrich) and stored in the dark at 4°C until acquired in flow cytometry. The obtained data were analyzed using Expo32 ADC software (Beckman Coulter, Inc.).
Table 1. Primary antibodies used in this study.
| Primary Antibodies | Dilutions | Source | Cell Target |
|---|---|---|---|
| BMSCs characterization markers | |||
| CD11b | 1:50 | BD Pharmingen | Leukocytes |
| CD31 | 1:10 | Abcam | Endothelial |
| CD34 | 1:50 | Santa Cruz Biotechnology | Endothelial |
| CD44 | 1:10 | Gene Tex, Inc | Leukocytes |
| CD45 | 1:50 | BD Pharmingen | Hematopoietic |
| CD73 | 1:50 | BD Pharmingen | BMSCs |
| CD90 | 1:50 | BD Pharmingen | BMSCs |
| CD106 | 1:50 | BD Pharmingen | Endothelial |
| OX43 | 1:10 | Gene Tex, Inc | Endothelial |
| Cardiac myocyte differentiation markers | |||
| Sarcomere myosin (MF20) | 1:100 | DSHB | Cardiomyocyte |
| Cardiac myosin heavy chain (α/β MHC) | 1:100 | Abcam | Cardiomyocyte |
| Cardiac troponin I (cTnI) | 1:100 | Santa Cruz Biotechnology | Cardiomyocyte |
| Cardiac α-actinin | 1:100 | Santa Cruz Biotechnology | Cardiomyocyte |
| Connexin 43 (Cx43) | 1:100 | Santa Cruz Biotechnology | Cardiomyocyte |
| N Cadherin | 1:100 | Santa Cruz Biotechnology | Cardiomyocyte |
| Desmin | 1:100 | Abcam | Cardiomyocyte |
| GATA4 | 1:100 | Santa Cruz Biotechnology | Cardiomyocyte |
| ANP | 1:100 | Santa Cruz Biotechnology | Cardiomyocyte |
| BNP | 1:100 | Santa Cruz Biotechnology | Cardiomyocyte |
Purification and enrichment of BMSCs by magnetic cell sorting
The adherent populations of BMSCs were further purified by indirect magnetic cell labeling method using an autoMACS™ Pro Separator (Miltenyi Biotech). The cells were subjected to CD90 positive selection by incubating the cells with FITC-labeled anti-CD90 antibodies (BD Pharmingen), followed by incubation with anti-FITC magnetic microbeads (Miltenyi Biotech), and passed through the magnetic columns as per the manufacturer's instructions. The resultant enriched CD90+/CD34-/CD45- fractions were expanded by subcultivation and subjected to flow cytometric analysis.
Labeling of BMSCs with green fluorescent protein (GFP) for lineage tracing
Lentiviral vectors and lentivirus production
Lentiviral construct generation and lentivirus preparation were performed as described previously [22]. In brief, lentiviral expression plasmid pWPI-GFP, along with packaging plasmid pCMVΔR8.91 and envelope plasmid pMD.D(VSV-G) were co-transferred with a mass ratio of 3:2:1 into a 30-40% confluent 293T cells in serum-free medium using ProFection® Mammalian transfection system (Promega) following the manufacture's instructions, and incubated in a humidified atmosphere of 5% CO2 at 37°C for 16 hours. After 16 hours the serum-free medium was replaced with fresh DMEM medium supplemented with 10% FBS to collect viral particles for the next 24 hours. The collected viral supernatants were filtered through 0.22 μm, pooled and subjected to ultracentrifugation at 26,000 rpm using a SW28 rotor for 2 hours. The supernatant was removed and the deposited viral particles were resuspended in 100 μl of PBS, pH 7.4 and stored at -70°C until further use. The viral titre was determined by standard HeLa titre procedure using GFP as a marker.
Lentiviral transduction
For lentiviral transduction, BMSCs (0.2 × 106 cells/well) and the desired number of viral particles (MOI = 5) were resuspended in 1 ml of serum-free medium (serum-free DMEM) and were seeded onto six-well culture plates precoated with retronectin (20 μg/ml/well) and incubated overnight in a humidified atmosphere of 5% CO2 at 37°C. After overnight transduction, the serum-free medium was replaced with complete medium (DMEM with 10% FBS) and cells were further incubated for 48 to 72 hours before the transduced GFP gene expression was analyzed. The GFP marked BMSCs were expanded further by subculturing. GFP expression was observed using confocal microscopy and the transduction efficiency was estimated.
Embryonic ventricular cardiac myocytes (ECMs) isolation
Isolation of embryonic ventricular primary cells was performed as described in detail previously [23-24]. Briefly, under deep euthanasia, E15 timed pregnant Sprague Dawley®™ SD®™ rats (Harlan Sprague Dawley, Inc.) were subjected to cervical dislocation and the embryos were removed from the adherent placental membranes. The hearts were dissected out from these E15 embryos and pooled in ice-cold Moscona's solution. Atria were removed from the ventricles under a dissecting microscope (Olympus SZ60) and discarded. Isolated ventricles were rinsed with ice-cold Krebs-Ringer bicarbonate-I (KRB-I) with 1 mg/ml BSA fractionV, 2 mg/ml glucose, 100 U/ml penicillin, and 75 U/ml streptomycin, pH 7.4. The ventricles were minced thoroughly and incubated with a KRB-II-collagenase type II (Worthington) solution (120 U/ml) in a 37°C shaking water bath. Tissues were dissociated by repeated (at 8 minute intervals) triturations. The process was repeated until all tissues were completely dissociated. Then, the cells were pelleted at 800g for 8 minutes, the supernatant was discarded, and the obtained pellet was resuspended in 5 ml of KRB-II solution (20 mg/ml BSA fractionV, 2 mg/ml glucose, 100 U/ml penicillin, and 75 U/ml streptomycin, pH 7.4) and passed through a 20 μm nylon mesh. The cells were once again pelleted and resuspended in complete medium composed of DMEM (Invitrogen) supplemented with 10% FBS (Atlas Biologicals), gentamicin (50 μg/ml) and amphotericin B (250 ng/ml).
Creation of ECMs tubular myotubes
Isolated ventricular primary cells were quantified using a hemocytometer and plated at a density of 1 × 106 cells/20 mm of collagen-gel tubular scaffold. Cells were seeded into the lumen and on the surface, incubated in a humidified atmosphere of 5% CO2 at 37°C for 7 days, and observed for spontaneous rhythmic beating and/or contractility. These ECM-myotubes (ECMs only) were cultured in basal medium (DMEM with 10% FBS) for further 7, 14, 21 or 28 days.
BMSCs and ECMs myotube co-culture
The expanded and purified population of CD90+ BMSCs (GFP unlabeled or GFP labeled) were seeded into the lumen and on the surface of the generated tubular ECM-myotubes at a density of 0.2 × 106 cells/20 mm tube and cultured in basal medium consisting of DMEM supplemented with 10% FBS and gentamicin (50 μg/ml) for 7, 14, 21 or 28 days. The cultures (ECMs/BMSCs-GFP unlabeled or ECMs/BMSCs-GFP labeled or ECMs only) were terminated at these regular intervals and the collected samples were subjected to RT-qPCR, immunofluorescence, morphometric, ultrastructural, and biochemical analyses.
Real-time polymerase chain reaction
Total cellular RNA extraction from three independent collagen tube cultures (ECMs/BMSCs-GFP unlabeled or ECMs only) maintained in basal medium were performed using TRIzol® Plus RNA purification kit (Invitrogen) according to manufacturer's instructions. The quality and quantity of the obtained RNA was analyzed on the Agilent 2100 Bioanalyzer using the Agilent RNA 6000 nano kit (Agilent Technologies, Inc.) according to manufacturer's instructions. The reverse transcriptase (RT) reaction was performed using 500 ng of total RNA in a final reaction volume of 20 μl using an iScript™ cDNA synthesis kit (Bio-rad Laboratories) according to manufacturer's recommendations. Gene-specific primers for cardiac myosin heavy chain alpha (myosin, heavy chain6, cardiac muscle, alpha - Myh6/α-MHC), cardiac myosin heavy chain beta (myosin, heavy chain 7, cardiac muscle, beta - Myh7/β-MHC), cardiac α-actin (actin, alpha, cardiac muscle 1 - Actc1), cardiac troponin I (troponin I type 3, cardiac - Tnni3, cTnI), GATA binding protein 4 (Gata4), atrial natriuretic peptide (natriuretic peptide precursor A – Nppa/ANP), brain natriuretic peptide (natriuretic peptide precursor B – Nppb/BNP) and connexin 43 (gap junction protein, alpha 1 – Gja1/Cx43) and; the endogenous normalizer reference gene, acidic ribosomal phosphoprotein P0 (Arbp) were designed using web based software Primer3 [25], synthesized commercially (Integrated DNA Technologies, Inc.), and evaluated for an annealing temperature of 58°C as shown in Table 2.
Table 2. RT-qPCR primer sequences used in this study.
| Genes | Forward Primer | Reverse Primer | Product Length (bp) |
Annealing Temperature (°C) |
GenBank Accession No |
|---|---|---|---|---|---|
| Tnni3 | 5′-ACGTGGAAGCAAAAGTCACC-3′ | 5′-CCTCCTTCTTCACCTGCTTG-3′ | 201 | 58 | NM_017144.1 |
| Myh6 | 5′–TGATGACTCCGAGGAGCTTT–3′ | 5′–TGACACAGACCCTTGAGCAG–3′ | 234 | 58 | NM_017239.2 |
| Myh7 | 5′-CCTCGCAATATCAAGGGAAA-3′ | 5′-TACAGGTGCATCAGCTCCAG-3′ | 198 | 58 | NM_017240.1 |
| Actc1 | 5′–CACGGCATTATCACCAACTG–3′ | 5′–AACAATGCCTGTGGTTCTCC–3′ | 240 | 58 | NM_019183.1 |
| Gja1 | 5′-TCCTTGGTGTCTCTCGCTTT-3′ | 5′-GAGCAGCCATTGAAGTAGGC-3′ | 167 | 58 | NM_012567.2 |
| Gata4 | 5′-TCAAACCAGAAAACGGAAGC-3′ | 5′-CTGCTGTGCCCATAGTGAGA-3′ | 192 | 58 | NM_144730.1 |
| Nppa | 5′-ATACAGTGCGGTGTCCAACA-3′ | 5′-CGAGAGCACCTCCATCTCTC-3′ | 209 | 58 | NM_012612.2 |
| Nppb | 5′-GGAAATGGCTCAGAGACAGC-3′ | 5′-CGATCCGGTCTATCTTCTGC-3′ | 164 | 58 | NM_031545.1 |
| Arbp | 5′–CGACCTGGAAGTCCAACTAC–3′ | 5′–ATCTGCTGCATCTGCTTG–3′ | 109 | 58 | NM_022402.1 |
Real-time PCR conditions were optimized as described previously [26]. All RT-qPCRs were performed with SYBR Green I chemistry in a MyiQ™ single-color real-time PCR detection system (Bio-rad Laboratories). For qPCRs, iQ5 SYBR Green I supermix, 3 pmol/μl of each forward and reverse primers, and 5 μl cDNA template were used in a final reaction volume of 50 μl. PCR cycling parameters included an initial denaturation of 8 minutes and 45 seconds at 95°C followed by 45 cycles of 30 seconds at 95°C, 30 seconds at 58°C, and 30 seconds at 72°C. Data collection was enabled at 72°C in each cycle. CT (threshold cycle) values were calculated using the MyiQ optical system software, version 2.0. The calibrator control included BMSCs day 0 sample and the target gene expression was normalized by a non-regulated reference gene expression, Arpb. The mathematical model previously described in detail [27], was used to determine the expression ratio of genes.
Immunofluorescence staining and confocal microscopy
Collagen tube cultures (ECMs/BMSCs-GFP unlabeled or ECMs/BMSCs-GFP labeled or ECMs only) were collected at day 7, 14, 21 or 28 and processed according to previously described protocols [21]. After incubation, the tubes were rinsed twice in Mosconos's solution, pH 7.4 and fixed in 2% paraformaldehyde at 4°C for 12-16 hours. Each of the samples was then permeabilized in PBS, pH 7.4 containing 0.1% Triton X-100 and 100 mM glycine for 30 to 40 minutes at room temperature and blocked in 1.5% BSA, 100 mM glycine in PBS for 1 hour at room temperature. The primary antibodies used are shown in Table 1. Primary antibodies were used at 1:100 dilutions in blocking buffer for 12-16 hours at 4°C. Secondary antibodies (Alexa fluor® 488, 546, 633 obtained from Molecular Probes, Invitrogen) were used at 1:200 dilutions in blocking buffer for 2 hours at room temperature in the dark. Alexa fluor® 488 phalloidin (1:100 in PBS) was used to stain filamentous actin. Nuclei were stained with DAPI (4, 6-diamidino-2-phenylindole, 100 ng/ml; Sigma-Aldrich). Images of the stained cells in the collagen tubular constructs were visualized using a confocal microscope (Zeiss LSM 510 Meta CSLM). Negative controls for staining included only secondary antibodies.
Nuclear morphometric analysis
For nuclear morphometric analysis, the captured images of the immunostained collagen-gel tube cultures (BMSCs only or ECMs only or ECMs/BMSCs) were separated into their red, green and blue channels respectively to isolate the DAPI (blue) stained nuclei. These images of the blue stained nuclei were converted into 16-bit monochrome images. Changes in the nuclear morphology of the BMSCs alone, ECMs alone and the ECMs/BMSCs co-cultured cells were analyzed by using the integrated morphometry subroutine of MetaMorph 6.1. Each complete DAPI stained nucleus in the 16-bit monochrome image field was isolated as a region of interest (ROI) and descriptors of nuclear morphology measured.
Descriptors of nuclear morphology (A, P, EFF and EOV)
The descriptors of area (A), perimeter (P), the elliptical form factor (EFF) and the equivalent oblate volume (EOV) were measured by the imaging software for each thresholded nucleus in the image. The perimeter of the nucleus was measured from the mid-point of each pixel comprising the thresholded border or margin. The EFF was defined as the ratio of the breadth (caliper width of the nucleus perpendicular to the longest chord, B) to the length (longest chord through the major axis of the nucleus, L), thus, EFF = L/B. The EOV of the nucleus was an estimate of the volume of an oblate spheroid with its minor axis matching the minor axis of the nucleus.
Values of A, P, EFF and EOV were calculated directly from the integrated morphometry subroutine of Meta-Morph image analysis software. Using the software's optical calipers, the measurements were refined by setting specific boundary conditions for A, P, EFF and EOV for acceptance of the fluorescent signal from the labeled DAPI labeled nuclei and to minimize or eliminate the contributions of any non-specific and background auto-fluorescence. Background fluorescence was minimal or non-detectable in these sample images. Incomplete nuclei on the edges of the image field were excluded from analysis.
Atrial and brain natriuretic peptide (Nppa/Nppb) quantification assays
Production of both Nppa (ANP)/Nppb (BNP) hormonal peptides released into the cell culture medium was measured by enzyme linked immunosorbent assay (ELISA) using a plate reader (Bio-rad, Bench Mark Plus, Microplate Spectrophotometer). The cell culture supernatants of tube cultures (ECMs/BMSCs or ECMs only) that were maintained at various time points (7 or 14 days) as replicates were collected and stored immediately at -70°C. Upon thawing, to measure the levels of both ANP and BNP, the cell culture media was centrifuged at 2,000g for 10 minutes to remove any debris. Aliquots of 50 μl supernatant (1:4 dilution) were analyzed for ANP using AssayMax Rat ANP ELISA Kit (ERA 7010-1, Assaypro), and for BNP using AssayMax Rat BNP-45 (rBNP-45) ELISA Kit (ERB 1202-1, Assaypro) according to manufacturer's instructions, respectively. The absorbance was read at 450 nm immediately after the addition of reagents. The concentration of rat ANP and BNP was determined using the standard solutions prepared in parallel, and expressed in (ng/mL).
Transmission electron microscopic (TEM) analysis of tubular constructs
To visualize the ultrastructural characteristics of the co-differentiating cells in the tubular construct, day 21 samples were processed for transmission electron microscopy (TEM). In brief, the tubular scaffolds that were seeded with cells (BMSCs only or ECMs only or BMSCs/ECMs) and maintained in basal culture conditions were dissected longitudinally and rinsed twice in PBS, pH 7.4. The tubes were fixed in 2.5% glutaraldehyde (Sigma-Aldrich) in PBS overnight at 4°C, rinsed in PBS, and treated with 1% tannic acid and 2% glutaraldehyde in PBS for one hour. The samples were then post fixed in 1% OsO4 and 1.5% potassium ferricyanide in PBS for 1 hour at room temperature, rinsed in water, and dehydrated through a graded ethanol series up to 100%. Then, the samples were impregnated with increasing concentrations of PolyBed 812 up to pure resin, using acetonitrile as an intermediate solvent, embedded and cured at 60°C for two days. The embedded samples were cut on an ultramicrotome (Leica Ultracut R) and 100 nm sections were collected on Cu grids. The sections were then stained with 2% uranyl acetate for 40 minutes at 37°C, and Hanaichi lead stain for 6 minutes at room temperature and examined using a JEOL200CX TEM (Tokyo, Japan) operated at 120 kV. Images were captured using Axiovision, Software Engine Version 5.42.479 by AMT (Advanced Microscopy Techniques, Danvers, MA).
Statistical analysis
Data were represented as mean ± standard error of the mean (mean ± SEM). The results of biochemical assays were grouped by day for the various culture conditions whereas the nuclear morphometric analyses data were categorized according to various culture conditions and experimental data were analyzed by ANOVA using SigmaStat 3.5 software. For RT-qPCR data, the differences in expression (cardiomyogenic markers) between control (day 0) and treated samples (day 7, 14, 21 or 28) were assessed in-group means for statistical significance by applying ‘Pair Wise Fixed Reallocation Randomization Test’ using Relative Expression Software Tool (REST©) [28]. In all cases, values of p < 0.05 were considered statistically significant.
Results
Phenotypic characterization of input BMSCs
Analysis of cell surface markers by flow cytometry of BMSCs revealed that the fluorescent intensity and distribution of the cells stained for CD11b, CD31, CD34, CD44, CD45 and CD106 were not significantly different from the intensity and distribution of cells stained with isotype controls (Figure 1 A-E, H). In addition, these cells were negative for the rat endothelial cell surface marker OX43 (Figure 1, I), an antigen expressed on all vascular endothelial cells of rat, indicating that these cultures were devoid of any hematopoietic stem and/or progenitor cells as well as differentiated bone-marrow-derived endothelial cells. In contrast, BMSCs exhibited a high expression of CD73 and CD90 surface antigens (Figure 1, F-G), which are consistent characteristics of undifferentiated BMSCs. Phenotypic characterization using the same set of markers on BMSCs by confocal microscopy also revealed that the cells were negative for CD11b, CD31, CD34, CD44, CD45, CD106, OX43 and, strongly positive for CD73 and CD90 (data not shown). The expression profiles of these surface molecules were consistent with previous reports [7, 21, 29].
Figure 1.
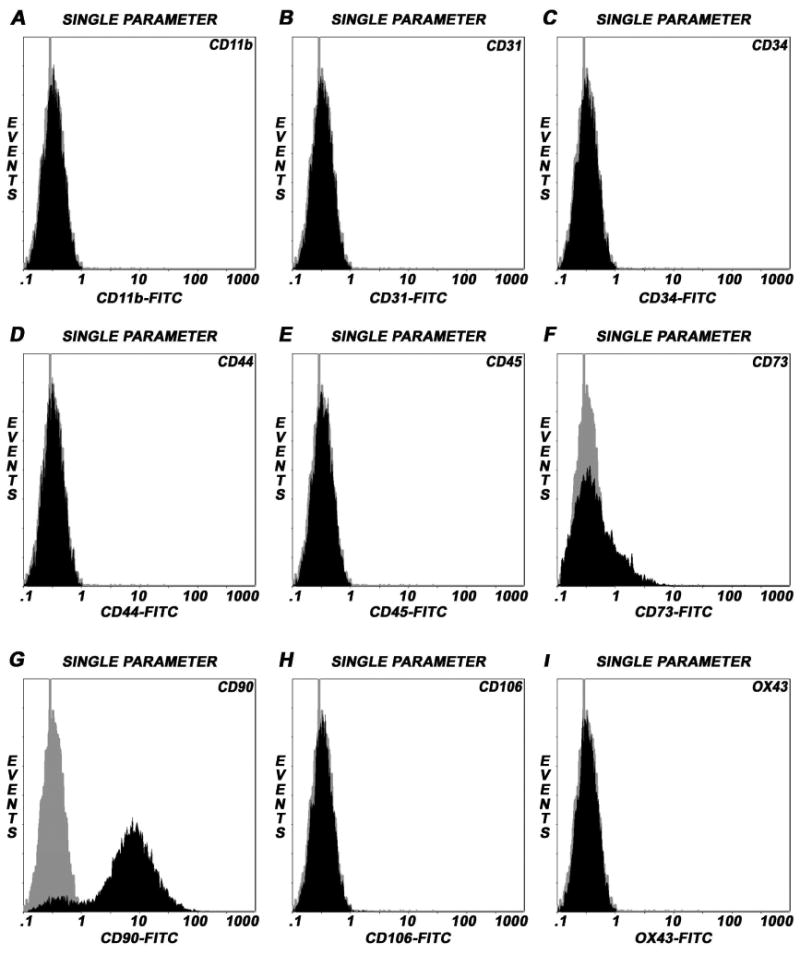
Immunophenotyping of undifferentiated rat BMSCs by flow cytometry. Single parameter histograms showing the relative fluorescence intensity of staining (abscissa) and the number of cells analyzed, events (ordinate). Isotype controls were included in each experiment to identify the level of background fluorescence. The intensity and distribution of cells stained for hematopoietic and endothelial markers; CD11b, CD31, CD34, CD44, CD45, CD106 and OX43 (black, shaded peaks) were not significantly different from those of isotype control (grey, shaded peaks) (Panels A-E, H-I), indicating that these cultures were devoid of any potential hematopoietic and/or endothelial cells of bone marrow origin. The fluorescent intensity was greater (shifted to right) when BMSCs were stained with CD73 and CD90 (black) compared to isotype control (grey) (Panels F, G). The predominant population of BMSCs consistently expressed CD90 surface molecule, a property of rat bone marrow-derived mesenchymal/stromal stem cells.
mRNA analysis of cardiomyogenic markers
To determine the differential gene expression profiles between BMSCs/ECMs co-culture and ECMs only culture, real-time quantitative RT-PCR (RT-qPCR) analyses of cardiogenic differentiation markers were carried out at the defined time points (7, 14, 21 and 28 days) on BMCSs/ECMs co-cultured in tubular scaffolds in basal medium. Additionally, RT-qPCR on ECMs cultured in the tubular scaffolds in the same medium condition was performed at the same time points.
ECMs cultured in tubular scaffold in basal medium constitutively expressed transcripts coding for key cardiogenic lineage specific markers (Figure 2A-B). α-MHC remained upregulated up to 28 days with a marked expression seen at day 14, whereas β-MHC showed a sustained upregulation up to 28 days. α-Actc1 showed consistent upregulation up to day 28 days with a alternating peak expressions noticeable around day 14 and day 28. cTnI showed a sustained upregulation up to day 28, with a marked expression around day 14 (Figure 2A). In addition, in ECMs tube cultures, the transcript levels of Gata4 showed a sustained upregulation up to 28 days with a marked expression seen at day 21. The natriuretic peptide ANP showed upregulation at day 7 and thereafter demonstrated a remarkable consistent upregulation over 21 days. BNP showed consistent upregulation over 28 days. Cx43 showed consistent downregulation over 21 days (Figure 2B).
Figure 2.
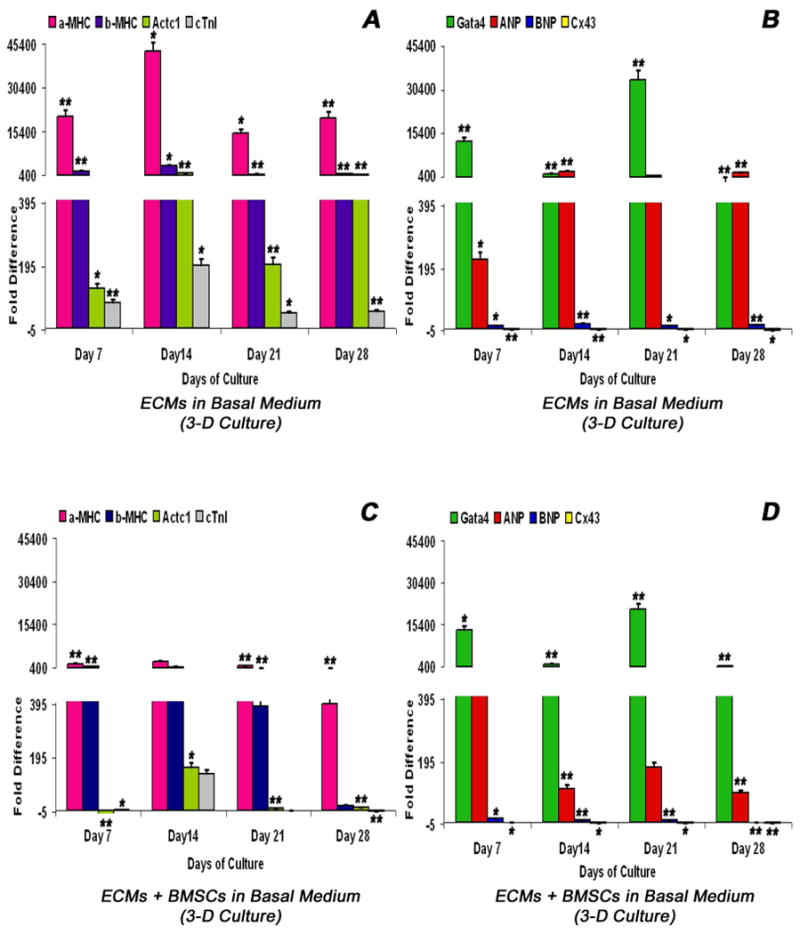
Real-time reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) analysis of various key cardiogenic markers, α-MHC (myosin, heavy chain6, cardiac muscle, alpha - Myh6), β-MHC (myosin, heavy chain 7, cardiac muscle, beta - Myh7), α-actin cardiac (actin, alpha, cardiac muscle 1 -Actc1), cTnI (troponin I type 3, cardiac - Tnni3), Gata4 (GATA binding protein 4 - Gata4), ANP (natriuretic peptide precursor A – Nppa), BNP (natriuretic peptide precursor B – Nppb) and Cx43 (gap junction protein, alpha 1 – Gja1) as a function of time (abscissa). ECMs cultured in collagen-gel tubular scaffolds (3-D culture) in basal medium (A, B). ECMs and BMSCs co-cultured in collagen-gel tubular scaffolds (3-D culture) in basal medium (C, D). The calibrator control included BMSCs day 0 sample and; the target gene expression was normalized by a non-regulated reference gene expression, Arbp. The expression ratio (ordinate) was calculated using the REST-XL version 2 software. The values are means ± standard errors for three independent cultures (n=3), *p < 0.05; **p < 0.001.
In contrast, in BMSC/ECM tube co-cultures the expression pattern of key cardiogenic marker transcripts showed moderate differences in their expression pattern, especially the structural and contractile filamental genes. α-MHC showed consistent sustained levels of upregulation throughout 28 days, whereas the β-MHC showed similar sustained high levels of expression until day 21 and subsequently returned to basal level. α-Actc1 showed an initial dowregulation at day 7 followed by sharp upregulation at day 14 and thereafter immediately returned to basal level. Similarly, the cTnI showed progressive upregulation till day 14 and thereafter showed gradual downregulation over the remaining observed period of time (Figure 2C). Besides, in BMSCs/ECMs tube co-cultures, the transcript levels of Gata4 showed a sustained upregulation up to 28 days with a marked expression seen at day 21. ANP showed a marked upregulation at day 7 followed by a variable level of upregulation up to day 28. BNP showed consistent upregulation up to day 21 and thereafter returned to basal level. Cx43 revealed consistent downregulation beyond day 7 (Figure 2D).
Expression of cardiomyogenic markers in tubular scaffold
To validate the findings of mRNA expression patterns of ECMs and BMSCs/ECMs for important cardiac myocyte phenotypic markers in these collagen-gel tube cultures and to determine whether these messages were translated into proteins, immunocytochemical staining of cardiac specific markers was undertaken. This was accomplished by staining collagen-gel tube cultures using antibodies directed against cardiac specific transcription factors, structural and contractile filaments, hormones and junctional complex proteins. Immunostaining and confocal laser scanning microscopic analysis established that these myotubes were composed of cells that were positive for a variety of cardiomyocyte specific markers, cardiac α/β-myosin heavy chain (α/β-MHC), sarcomeric myosin heavy chain (sMHC), cardiac troponin I (cTnI), cardiac α actinin, desmin, N-cadherin, connexin 43 (Cx43), GATA-binding protein 4 (Gata4), atrial natriuretic peptide (ANP) and brain natriuretic peptide (BNP). Representative staining patterns obtained for these cardiac-related proteins for ECMs only tube cultures are shown in Figure 3 (A-L) and in Figure 4(A-L); similarly, for BMSCs/ECMs tube co-cultures are shown in Figure 5(A-L) and in Figure 6(A-L), respectively.
Figure 3.
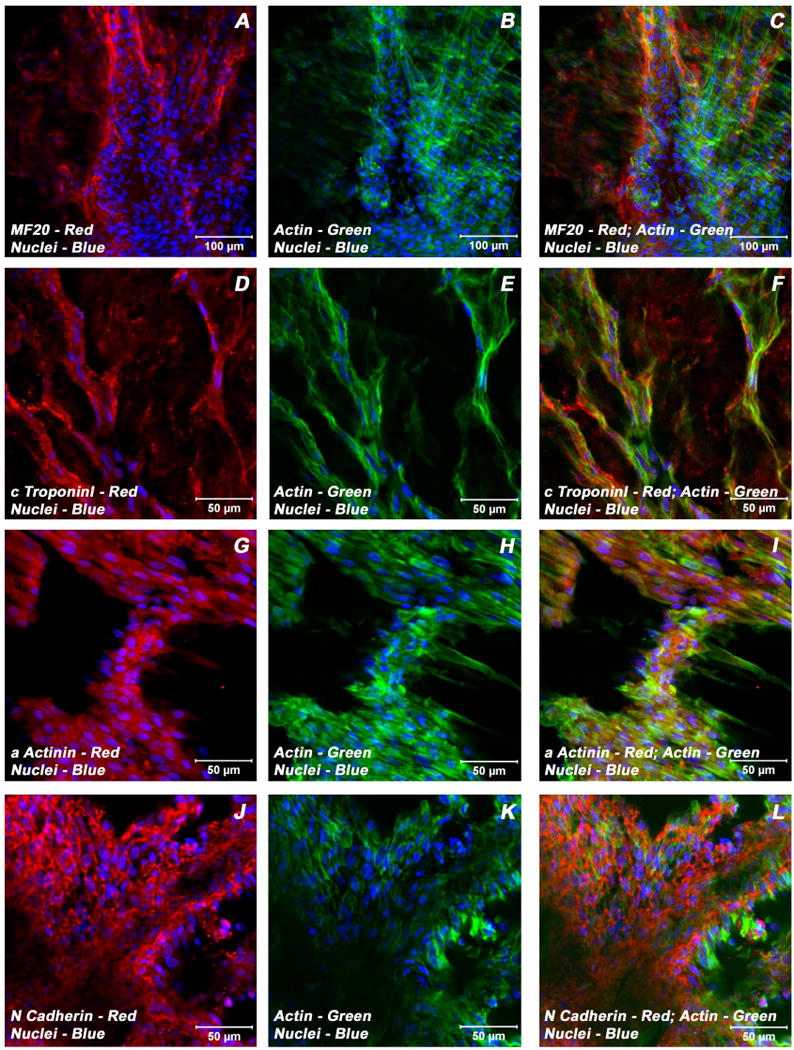
Expression pattern of various cardiomyogenic markers in ECMs containing myotube constructs by confocal microscopy. Localization of key cardiac myocyte phenotypic markers of day 21 embryonic cardiac myocyte (ECM) tube cultures in basal medium demonstrated the expression of structural and contractile proteins, sMHC (A, C), cTnI (D, F), α-actinin (G, I), microfilament, actin (B-C, E-F, H-I, K-L) and the adherent junction protein, N cadherin (J, L). Dual immunostaining of ECMs tube cultures in basal medium revealed areas of elongated and/or round to polyhedral type of cells arranged in multilayers and organized into cord-like or trabecular type of cellular arrangement (A-L). Nuclei of these cells were large and either oval or elongated and fusiform in appearance and were centrally positioned. Cells were also stained for nuclei (blue, DAPI) and fibrillar actin (green, Alexa 488 phalloidin). Merged images (A-L). (A-C, scale bar 100 μm; D-L, scale bar 50 μm).
Figure 4.
Expression pattern of various cardiogenic markers in ECMs containing myotube constructs by confocal microscopy. Localization of key cardiac myocyte phenotypic markers of day 21 embryonic cardiac myocyte (ECM) tube cultures in basal medium demonstrated the expression of transcription factor, Gata4 (A, C), peptide hormone, ANP (D, F), structural intermediate filament, desmin (G, I), gap junction protein, Cx43 (J, L) and the microfilament, actin (B-C, E-F, H-I, K-L). Dual immunostainings of ECMs in the tubular construct showed areas of cells on the exterior surface that were aligned and overlapping in an orderly manner (A-C, G-I), whereas the cells on the luminal surface were elongated and/or round to polygonal in nature and were arranged in multilayers (D-F). Nuclei of these cells were large and either oval or elongated and fusiform in appearance and were centrally positioned. Cells were also stained for nuclei (blue, DAPI) and fibrillar actin (green, Alexa 488 phalloidin). Merged images (A-L). (A-C, G-L scale bar 50 μm; D-F scale bar 100 μm).
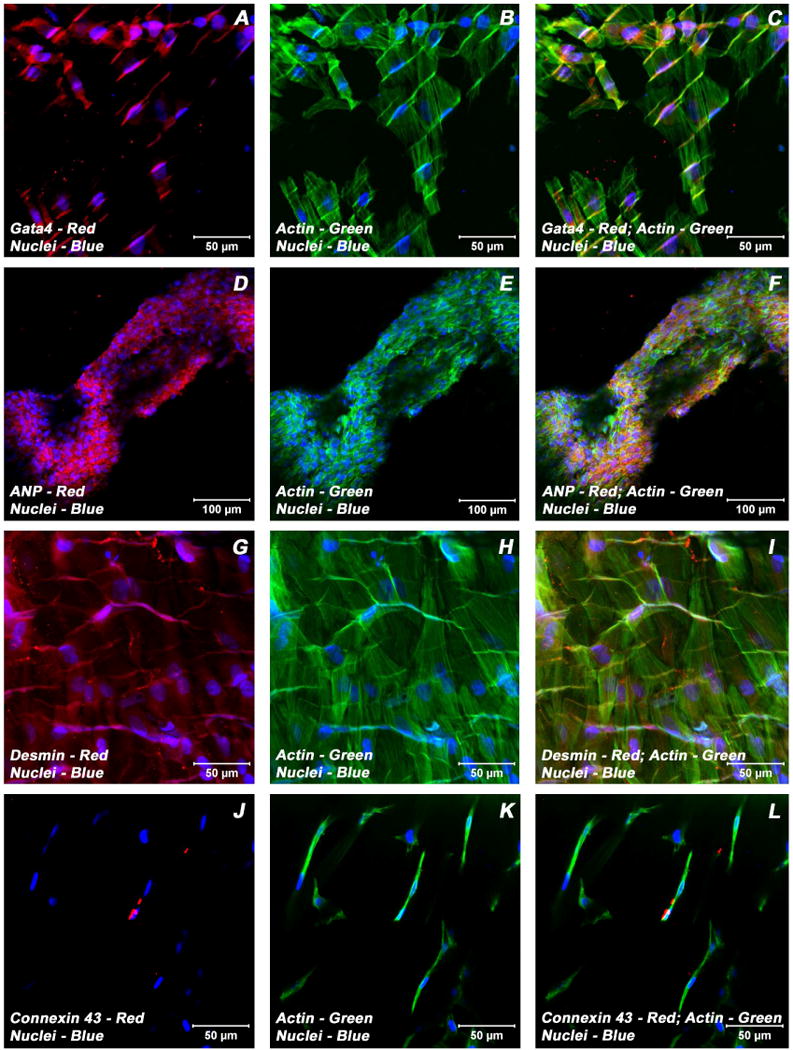
Figure 5.
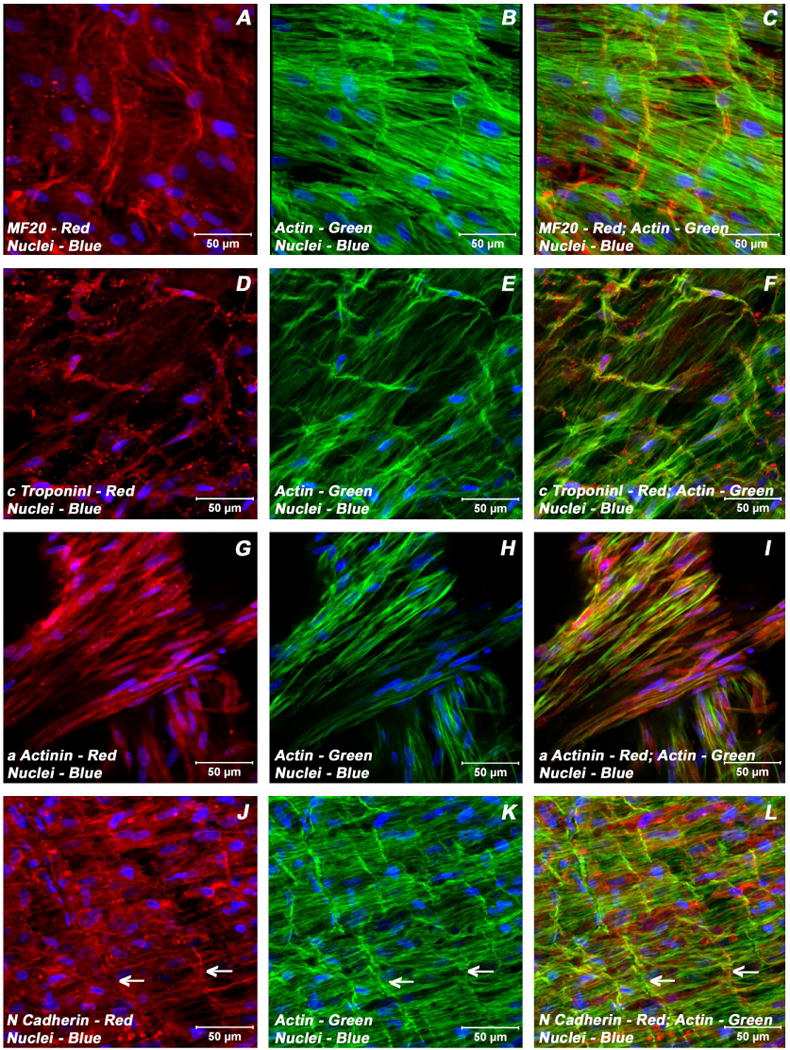
Expression pattern of various cardiogenic markers in BMSCs/ECMs containing myotube constructs by confocal microscopy. Localization of key cardiac myocyte phenotypic markers of day 21 embryonic cardiac myocytes (ECMs) and bone marrow stromal cells (BMSCs) tube co-cultures in basal medium demonstrated the expression of structural and contractile proteins, sMHC (A, C), cTnI (D, F), α-actinin (G, I), microfilament, actin (B-C, E-F, H-I, K-L) and the adherent junction protein, N cadherin (J, L). The myofibers or trabeculae of cells were arranged in a pseudosyncytial pattern (A-C, E-F, J-L). Specialized cell junctions such as intercalated disk were noticed along with characteristic branching and cross bridging of these trabeulated cells (white arrows, J-L). Nuclei of these cells were large and either oval or elongated and fusiform in appearance and were centrally located. Cells were also stained for nuclei (blue, DAPI) and fibrillar actin (green, Alexa 488 phalloidin). Images (A-C) show a projection representing 9 sections collected at 2 μm intervals (16 μm). Merged images (A-L). (A-L scale bar 50 μm).
Figure 6.
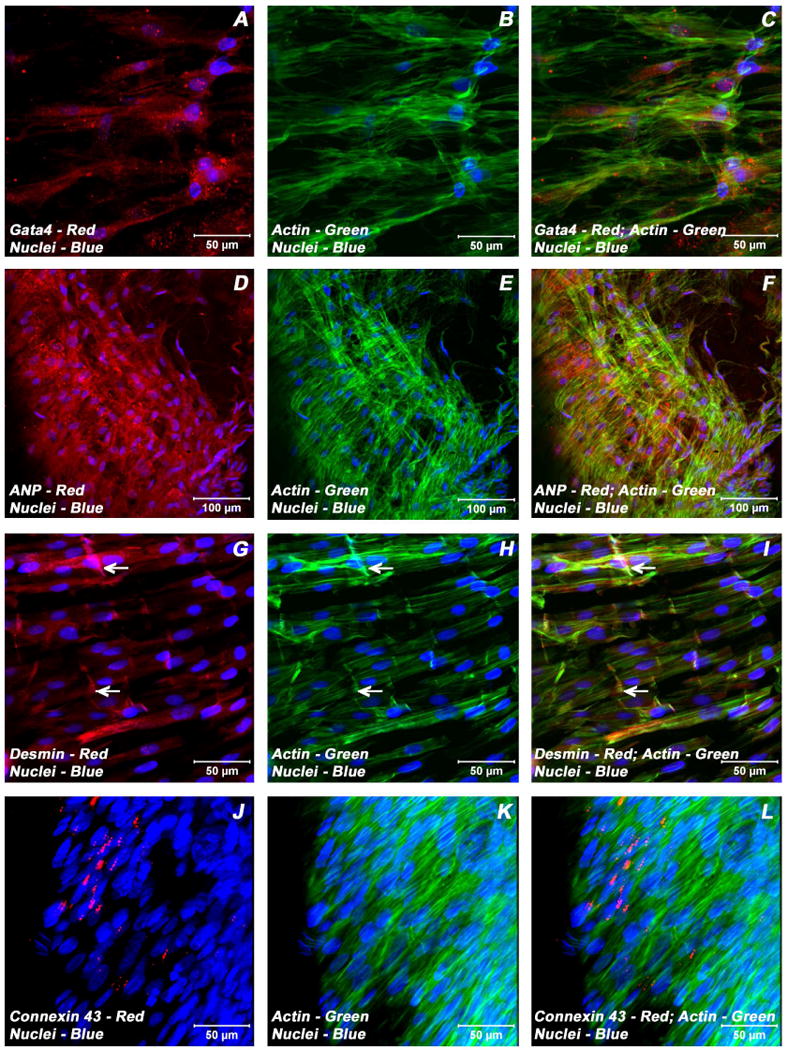
Expression pattern of various cardiogenic markers in BMSCs/ECMs containing myotube constructs by confocal microscopy. Localization of key cardiac myocyte phenotypic markers of day 21 embryonic cardiac myocytes (ECMs) and bone marrow stromal cells (BMSCs) tube co-cultures in basal medium demonstrated the expression of transcription factor, Gata4 (A, C), peptide hormone, ANP (D, F), structural intermediate filament, desmin (G, I), gap junction protein, Cx43 (J, L) and the microfilament, actin (B-C, E-F, H-I, K-L). The co-differentiating cells appeared as parallely arranged trabeculae and displayed the characteristic branching and cross bridges to give a pseudosyncytial arrangement (G-L). These trabeculae displayed specialized cell junctions, the intercalated disks (white arrows, G-I). Nuclei of these cells were large and either oval or elongated and fusiform in appearance and were centrally located. Cells were also stained for nuclei (blue, DAPI) and fibrillar actin (green, Alexa 488 phalloidin). Images (J-L) show a projection representing 9 sections collected at 3 μm intervals (24 μm). Merged images (A-L). (A-C, G-L scale bar 50 μm; D-F scale bar 100 μm).
Examination of day 21 ECMs tube cultures revealed the presence of myocytes mainly on the luminal and outer surfaces of the tubular construct. On the external surface, ECMs were aligned and overlapping in an orderly manner whereas the cells on the luminal surface exhibited cord-like or trabecular type of cellular arrangement that resemble those of in vivo embryonic trabecular myocytes. The aligned myocytes showed the presence of developing sarcomeric units, which were positive for actin, myosin and desmin (Figure 3A-C; Figure 4 G-I), and indicative of progressive maturation and differentiation towards in vivo neonatal-like ventricular myocytes. On the contrary, in BMSCs/ECMs tube co-cultures, the differentiating cells appeared to be elongated and organized into interlacing bundles depicting varying degrees of alignments throughout the construct. These multilayers of cells were composed of differentiating and maturing cardiac myocytes with evidence of evolving Z-disks and the cardiac specific sarcomeric organization which were positive for N-cadherin and desmin (Figure 5J-L, Figure 6G-I) and the cardiac related biosynthetic activities, ANP (Figure 6D-F). In the BMSCs/ECMs tube co-culture, the myofibers or trabeculae were mainly parallel but displayed the characteristic branching and cross bridges to give a pseudosyncytial arrangement. Nuclei of the cells were large, oval and were centrally placed (Figure 6G-I). Intercalated disks were identified in the trabeculae as prominently stained lines crossing fibers either transversely or in a staggered, zigzag manner (Figure 5J-L). They were sites of specialized cell junctions (Figure 6G-L).
Characterization of GFP-labeled BMSCs in myotubes
In order to localize and characterize the phenotypic nature of differentiating BMSCs amidst the developing ECMs in the milieu of collagen-gel tubes, BMSCs were transfected with GFP-expressing lentivirus. Day 21 co-culture myotube constructs harboring these GFP-BMSCs/ECMs were subjected to immunostaining and confocal microscopic analysis using the same battery of cardiomyogenic phenotypic markers as aforementioned. Similar to what was observed within unlabelled-BMSCs/ECMs co-culture myotubes, the GFP-marked BMSCs were positive for a variety of markers that are associated with differentiating cardiac myocytes such as cardiac α/β-MHC, sarcomeric MHC, cardiac cTnI, desmin, N-cadherin, Cx43 (Figure 7A-I). In the co-culture tubes the resident ECMs and untransformed GFP-BMSCs were distinguished from the transdifferentiated GFP-BMSCs by their lack of detectable co-localization signals. Representative staining pattern and typical co-localization obtained for these cardiac-related proteins within GFP-BMSCs/ECMs myotube co-cultures are shown in Figure 7(A-I).
Figure 7.
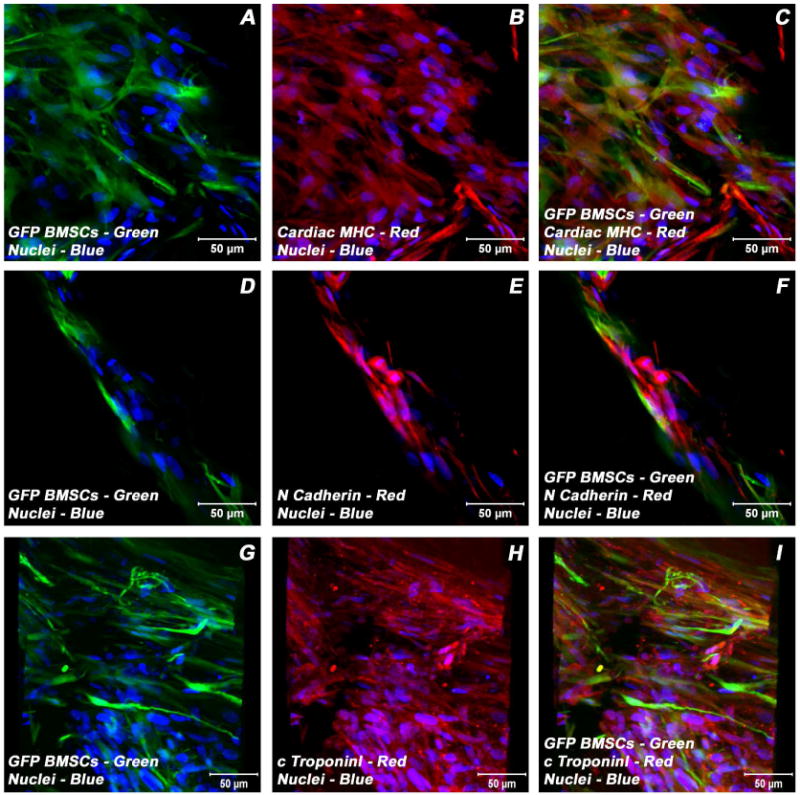
Expression pattern of various cardiogenic markers in GFP-BMSCs/ECMs containing myotube constructs by confocal microscopy. Immunostaining of key cardiac myocyte phenotypic markers of day 21 embryonic cardiac myocytes (ECMs) and green fluorescent protein labeled bone marrow stromal cells (GFP-BMSCs) tube co-cultures in basal medium demonstrated the expression of contractile proteins, α/β-MHC (A-C), cTnI (G-I), the adherent junction protein, N cadherin (D-F) and GFP (A, C, D, F, G, I). The transdifferentiated GFP-BMSCs were delineated by their co-localization signals (yellow). Nuclei of these cells were large and either oval or elongated and fusiform in appearance and were centrally placed. Cells were also stained for nuclei (blue, DAPI) and GFP-BMSCs (green, GFP). Images (G-I) show a projection representing 16 sections collected at 10 μm intervals (150 μm). Merged images (A-I). (A-I scale bar 50 μm).
Quantitation of atrial and brain natriuretic peptides (Nppa/Nppb)
To further confirm the functional competence of the co-differentiating BMSCs/ECMs cells at the biochemical level, we analyzed the level of Nppa (ANP) and Nppb (BNP) hormones secreted into the culture media by these cells. Analysis of ECM tube culture supernatants showed a significant decrease in the level of Nppa and Nppb proteins from day 7 to day 14 (Figure 8A-B), whereas the BMSCs/ECMs co-culture supernatants showed a significant increase in the level of Nppa protein and showed a slight reduction in the level of Nppb protein from day 7 to day 14 (Figure 8A-B). Thus, indicating that the co-differentiating cells were metabolically active.
Figure 8.
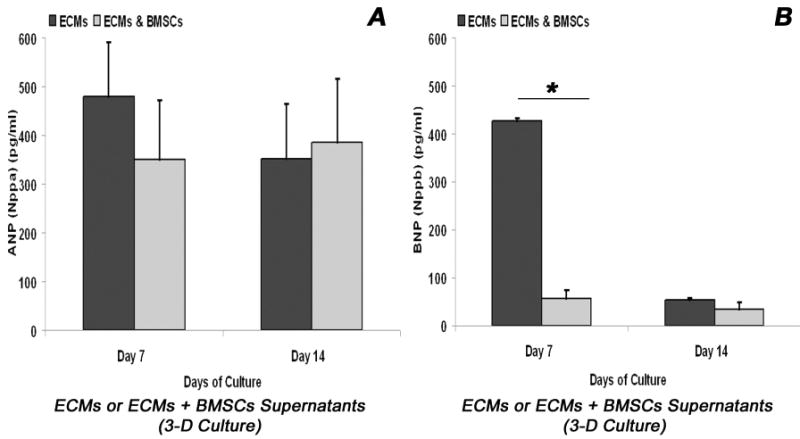
Biochemical analysis of cardiomyogenic differentiation of BMSCs/ECMs co-culture in 3-D collagen-gel tubular scaffold. Quantitative analysis of ANP and BNP hormones in myotube culture supernatants over a period of 14 days measured by enzyme linked immunosorbent assay, ELISA. (A) Quantification of ANP (Nppa protein) levels was compared between ECMs only cultures and BMSCs/ECMs co-cultures. *p < 0.05. The data represent the mean ± SEM of triplicate cultures (n = 3). (B) Quantification of BNP (Nppb protein) levels was compared between ECMs only cultures and BMSCs/ECMs co-cultures. *p < 0.05. The data represent the mean ± SEM of triplicate cultures (n = 3).
Nuclear morphometric analysis
Morphometric analysis of the nuclei of stem cells (BMSCs) alone, cardiac myocytes (ECMs) alone, and the stem cell-cardiac myocyte (BMSCs/ECMs) co-cultures at day 21 of culture revealed significant differences in nuclear A, P, and EOV. The value of A, P, and EOV for the nuclei of the BMSCs alone were greater than the ECMs alone or the hybrid cells in the BMSCs/ECMs co-cultures (Figure 9A-C). However, the nuclear EFF of the BMSCs was significantly smaller than either the ECMs or the hybrid cells in the BMSCs/ECMs co-cultures (Figure 9D). These observations indicate that the morphology of the nuclei of the stem cells were characterized by being significantly larger and more rounded, in comparison to the smaller, more elongated and fusiform nuclei characteristic of the cardiac myocytes. The nuclei in the hybrid cells in the BMSCs/ECMs co-cultures expressed an elongated, fusiform morphology in which the parameters of A, P, EFF and EOV were not significantly different in from the values characteristic of the ECMs, but were significantly different from the values of the parameters describing the nuclei of the stem cells, BMSCs (Figure 9A-D).
Figure 9.
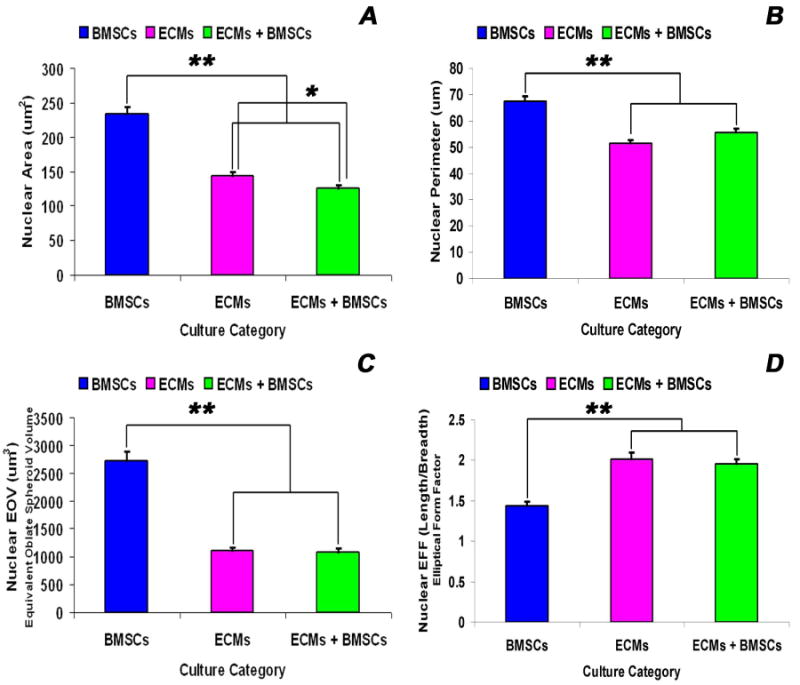
Nuclear morphometric analysis of BMSCs, ECMs and ECMs/BMSCs cultured in the tubular scaffold under basal conditions. The descriptors of nuclear morphology, viz., area - A (A), perimeter - P (B), the equivalent oblate volume – EOV (C) and the elliptical form factor – EFF (D) were measured by the imaging software for each thresholded and image isolated nucleus. Values of A, P, EFF and EOV were calculated directly from the integrated morphometry subroutine of Meta-Morph image analysis software. The nuclear parameters (A, P, EOV and EFF) of BMSCs nuclei were significantly greater than the nuclear parameters of ECMs and ECMs/BMSCs nuclei (p < 0.001). There were no differences between the nuclear parameters (P, EOV and EFF) for the ECMs and ECMs/BMSCs nuclei (p = 0.101, p = 0.426, p = 0.657), except the nuclear area (ECMs > ECMs/BMSCs, p = 0.008). The data represent the means ± SEM. *p < 0.05; **p < 0.001.
TEM analysis of myotube construct
TEM analysis of the day 21 BMSCs only tube constructs showed the ultrastructural characteristics of immature cells. The predominant population of cells showed nuclei with opened out chromatin, ribosomes and polyribosomes as well as pinocytotic vesicles indicative of active, undifferentiated cells. In addition, it revealed the presence of junctional complexes in the form of gap junctions and fasciae adherens (Figure 10A). On the contrary, the TEM analysis of the day 21 ECMs only myotube constructs demonstrated the ultrastructural characteristics of typical developing embryonic cardiac myocytes (Figure 10B). The majority of cells revealed immature nuclei with euchromatin and nucleolus. The cytoplasm of these cells showed the presence of random distribution of myofilaments as well as organized early sarcomeric units containing the Z-bodies and Z-disks (Figure 10B). Most of these developing sarcomeric units were located around the periphery of the nucleus. These cells also possessed numerous cytoplasmic mitochondria and fasciae adherens (Figure 10B).
Figure 10.
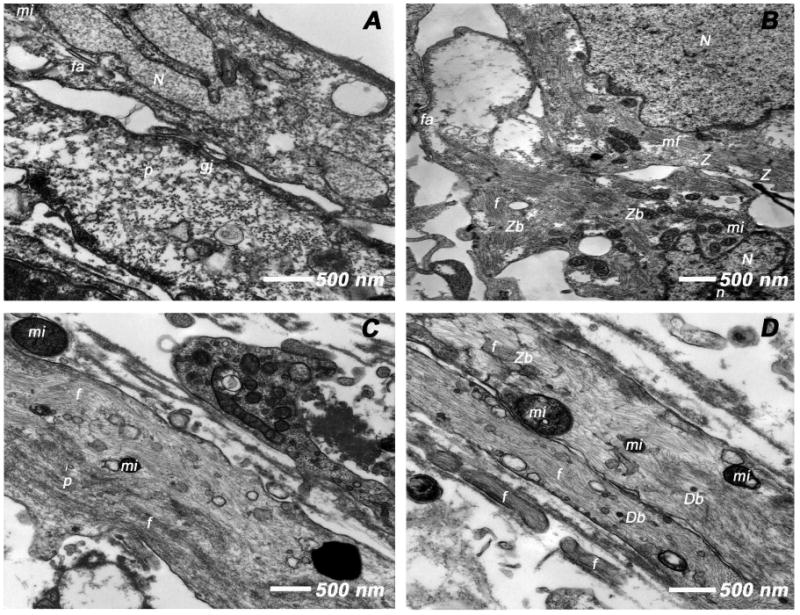
Transmission electron microscopic (TEM) analysis of day 21 tubular constructs. (A) BMSCs cultured in tubular constructs in basal medium revealed euchromatic nuclei (N), cytoplasmic pinocytotic vesicles (p) and junctional complexes (gj, fa), the ultrastructural characteristics of active, immature cells. (B) ECMs under the same culture conditions showed active euchromatic nuclei (N) with nucleolus (n). The cytoplasm revealed myofibrils (mf) myofilaments (f), early sarcomeric units, Z-bodies (Zb) and Z-disks (Z) and numerous mitochondria (mi). (C-D) BMSCs/ECMs co-culture under similar culture conditions demonstrated cytoplasmic myofilaments (f), Z-bodies (Zb), pinocytotic vesicles (p), mitochondria (mi) and numerous dense bodies (Db).
Similar to what was observed in day 21 ECMs tube cultures, the TEM analysis of the day 21 BMSCs/ECMs co-culture myotube constructs revealed the typical appearance of differentiating cells acquiring the ultrastructural characteristics of developing cardiac myocytes. These cells were elongated, multilayered and exhibited orderly registry. These aligned cells revealed areas of myofilaments that were randomly dispersed throughout the cytoplasm, indicative of early myofibrillogenesis (Figure 10C). At some areas these myofibrils were arranged in parallel bundles and oriented towards the long axis of the cells (Figure 10D). The presence of rudimentary Z-bodies that develops into a Z-disk (Z) was seen amidst the myofibrils indicative of developing sarcomeric units. In addition, the cells showed numerous cytoplasmic mitochondria and pinocytotic vesicles characteristics of active cardiac myocytes (Figure 10C-D).
Discussion
The 3-D tubular scaffold engineered from micro-topographically aligned collagen type I strands has been utilized previously as a model of maturation of ventricular myocytes and other cardiovascular structures [Evans et al., 2003; Yost et al., 2004; Valarmathi et al., 2009]. Here, we report a novel co-culture of two different types of cells, BMSCs (postnatal) and ECMs (embryonic) on a 3-D tubular construct in basal medium that recapitulates many aspects of their synergistic and synchronized maturation and co-differentiation, a phenomenon that seems to be a pivotal event found during the early phase and subsequent cascade of in vivo myocardial regeneration.
Previous studies demonstrated that populations of characterized or uncharacterized BMSCs are capable of differentiating into cardiac myocytes when cultured on two-dimensional (2-D) planar surfaces, especially under the influence of either the chemical factors and/or cellular interactions; however, these reports are not without controversy due to the lack of quantitative data [5-6, 31]. All these studies excluded the influence of physical factors that are crucial to the maturation and differentiation of functioning cardiac myocytes [32-33]. One of the other confounding factors introduced in most of these in vitro and ex vivo studies was that unpurified heterogeneous populations of BMSCs were utilized [5-6, 30-33, 35]. Here we show that under appropriate in vitro combination of physical, chemical and cellular environmental cues (local environmental cues) BMSCs can be induced to undergo a cardiomyogenic differentiation pathway. To accomplish this, we developed a 3-D co-culture system in which a pure population of CD90+ rat BMSCs were co-seeded with rat ECMs and cultured on a highly aligned porous and biocompatible collagen-fiber tubular scaffold for differentiation and regeneration purposes.
Here, we examined two different types of cultures, the ECMs only culture and the BMSCs/ECMs co-culture and their maturation and differentiation in basal medium alone (without any cardiogenic supplements). We also compared and specifically assessed the consequence of BMSCs presence on the expression of cardiogenic markers in ECMs and vice versa. Both these two types of cultures supported the ECMs in vitro continuum of cardiomyogenic differentiation and, simultaneously promoted the maturation and differentiation of BMSCs into cells characteristic of embryonic cardiac myocytes. To determine the in vitro differentiation of rat ECMs and rat BMSC-derived cardiac myocyte-like cells a plethora of cardiomyogenic markers associated with murine cardiomyogenesis in vivo were used. The cells within the tube constructs were shown to express cardiac specific genes, proteins, ion channels, receptors, and signal transduction machinery in a pattern that closely recapitulates the developmental pattern of in vivo murine neo-cardiomyogenesis. In addition, spontaneous contraction of the differentiated cultures was evident in these myotube constructs.
To characterize the expression pattern of key cardiogenic gene transcripts in the 3-D tube constructs, we examined the expression of various cardiac associated markers: α/βMHC, sMHC, cTnI, α actinin, desmin, N-cadherin, Cx43, Gata4, ANP and BNP. Constitutive expression of these markers was detected at low to very low levels in undifferentiated input BMSCs. RT-qPCR results showed that cardiomyogenic differentiation cultures of ECMs and BMSCs/ECMs in basal culture conditions for 28 days resulted in increased expression of transcripts coding for various filament proteins associated with cardiac structural maturation and cytoskeletal development, and contraction such as α-MHC, β-MHC, α-actinin and troponin I. The peak expression of all these contractile related proteins appeared around day 14 in both ECMs only and BMSCs/ECMs co-cultures indicating that the differentiating cells began to acquire a distinctive cardiac type phenotype characteristic of differentiating and maturing cardiomyocytes. The temporal down regulation of all these four structural/contractile elemental proteins in BMSCs/ECMs tube co-cultures when compared with ECMs only tube cultures during this observed period may represent continual development and remodeling of the developing ECMs in correspondence with BMSCs. This highlights the fact that quantitation of cadiomyogenic differential gene expression pattern is mandatory to understand the synergistic interaction of embryonic or neonatal cardiomyocytes with BMSCs in co-differentiation condition as well as directed differentiation of BMSCs towards a cardiomyogenic lineage.
As demonstrated by immunostaining for various cardiogenic markers, day 21 tube cultures in basal medium showed that ECMs as well as BMSCs were able to adhere and undergo maturation and differentiation into cardiac myocytes. The ECMs and ECMs/BMSCs co-cultured cells expressed cardiac α/β-MHC, cardiac cTnI, cardiac α-actinin and desmin. In addition, immunostaining using the putative MF20 antibody revealed that these cells were positive for sMHC, another contractile protein typically expressed by cardiac myocytes. These cells also expressed the junctional complex proteins; N-cadherin, a adherent junction protein and, Cx43, a gap-junction protein, suggesting the presence of eletromechanical coupling among ECMs as well as between ECMs and BMSCs in co-culture. This is consistent with the observation that the cells in the beating tubes contract synchronously. In addition to the structural and contractile proteins, BMSC-derived cardiomyocytes specifically expressed the cardiac transcription factor, Gata4. Immunostaining localized the expression of Gata4 in the nuclei of sMHC (MF20) positive cells. Thus, we were able to corroborate the mRNA expression profile with protein expression pattern.
Apart from the known advantages of our tubular scaffold, that is, the orientation of the aligned collage-fibers can be varied and the dimensions are scalable, these collagen-gel tubes are optically transparent. This allowed direct observation of the fate of BMSCs amidst the developing ECMs. These myotubes were seeded with GFP-labeled BMSCs, which enhanced their visualization and real-time observation of their locations and morphological transformations in correspondence to ECMs. Immunostaining revealed that the predominant population of GFP-marked BMSCs expressed various key cardiogenic markers that are associated with cardiac specific genes, proteins, ion channels, receptors, and signal transduction machinery (α/β-MHC, sMHC, N-Cadherin, Desmin, Cx43, Gata4) in a pattern that closely resembles that observed for ECMs. The expression of these cardiac specific genes by majority of BMSCs suggest that these cells have the ability to transform into cardiac phenotype under appropriate inductive cues provided by the ECMs. The detection of certain untransformed GFP-marked BMSCs in these constructs may be suggestive of their inherent differential cardiomyogenic potential.
Having established the cytodifferentiation of GFP-BMSCs towards cardiac phenotype, the nuclear transformation of these cells was validated. In order to assess and delineate the nuclear morphological changes of transdifferentiating BMSCs in the milieu of ECMs cells and oriented collagen-gel fibers, morphometric analysis was carried out on day 21 tube cultures. The obtained results suggest that the nuclei of the stem cells in contact with cardiac myocytes undergo a transformation assuming the physical morphology characteristic of the cardiac myocytes. There were no very large or giant nuclei present in the hybrid cells in the stem cell-cardiac myocyte co-culture (BMSCs/ECMs), suggesting that there was no obvious fusion of stem cell and cardiac myocyte nuclei to produce large, tetraploid or polyploid nuclei.
The peptide hormone ANP is expressed in developing atrial and ventricular cardiac myocytes, but down regulated in adult ventricular myocytes, was detected in these differentiating cells at mRNA and protein levels by immunohistochemistry, RT-qPCR and ELISA. Similarly, these differentiating cardiac myocytes expressed yet another peptide hormone, BNP. The expression of these proteins by ECMs and/or BMSC-derived cardiomyocytes indicates that they exhibit appropriate metabolic activity. Although these cells in these tubular construct expressed appropriate structural and contractile proteins at mRNA and protein levels, ultrastructural analysis of myofibrillar and sarcomeric organization indicate an immature phenotype. Further work is ongoing to investigate the physiopharmacological characteristics such as electromechanical coupling of these cells (BMSCs/ECMs) and, whether mechanotransduction in the form of applied mechanical forces such as circumferential strain and fluid shear stress can render these nascent cardiac myocytes seen in our cardiac muscle construct to undergo remodeling and acquire more mature cardiac phenotype, as our 3-D tubular myotubes construct is more amenable to these types of experiments.
Finally, our morphological, morphometric, molecular, immunological and biochemical data reveal the intrinsic cardiomyogenic differentiation potential of BMSCs under appropriate 3-D microenvironmental conditions. The favorable physicochemical microenvironment provides by the aligned collagen-gel fibers and the growth factors routinely present in the basal medium renders these co-cultured cells, BMSCs and ECMs to reprogramme in synchrony towards cardiomyogenic lineage and commitment. Thus, creating the notion that in the presence of BMSCs, ECMs are capable of undergoing partial dedifferentiation, a step necessary for the further successive but progressive synchronized redifferentiation of both BMSCs and ECMs towards cardiac myocytes. Therefore, we propose that the phenomenon of partial dedifferentiation followed by redifferentiation of in vitro cardiomyocytes in the vicinity of adult marrow-derived stem cells may be a primary event that occurs during the adult marrow stem cell based myocardial regeneration in vivo.
Conclusions
Our data indicate that the scaffold provides a highly conducive microenvironment that allows BMSCs and ECMs to co-differentiate into cardiomyocytes in basal medium. Moreover, this clearly demonstrates that adult BMSCs under appropriate in vitro combination of physical, chemical and cellular environmental cues could be induced to undergo cardiomyogenic differentiation pathway and recapitulates many aspects of in vivo neo-myogenesis in a three-dimensional context. Thus, this co-culture system provides an in vitro model and a prospect to elucidate various molecular mechanisms underpinning the integration and orderly maturation and differentiation of BMSCs into neo-cardiomyocytes during cardiac regeneration.
Acknowledgments
The authors thank Dr. Robert L. Price for his critical reading of the manuscript, Dr. Daping Fan for his generous contribution of GFP lentivirus and Dr. Udai P. Singh for his flow cytometry technical assistance and; Ms. Cheryl Cook and Ms. Lorain Junor for their excellent technical support, cell culture and myotube preparations respectively.
Footnotes
Disclosures: All authors have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Korbling M, Estrov Z. Adult stem cells for tissue repair: A new therapeutic concept? N Engl J Med. 2003;349:570–582. doi: 10.1056/NEJMra022361. [DOI] [PubMed] [Google Scholar]
- 2.Bunting KD, Hawley RG. Integrative molecular and developmental biology of adult stem cells. Biol Cell. 2003;95:563–578. doi: 10.1016/j.biolcel.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Wakitani S, Saito T, Caplan AI. Myogenic cells derived from rat bone marrow mesenchymal stem cells exposed to 5-azacytidine. Muscle Nerve. 1995;18:1417–26. doi: 10.1002/mus.880181212. [DOI] [PubMed] [Google Scholar]
- 4.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 5.Makino S, Fukuda K, Miyoshi S, Konishi f, Kodama H, Pan J, et al. Cardiomyocytes can be generated from marrow stromal cells in vitro. J Clin Invest. 1999;103:697–705. doi: 10.1172/JCI5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuda K. Development of regenerative cardiomyocytes from mesenchymal stem cells for cardiovascular tissue engineering. Artif Organs. 2001;25:187–193. doi: 10.1046/j.1525-1594.2001.025003187.x. [DOI] [PubMed] [Google Scholar]
- 7.Valarmathi MT, Davis JM, Yost MJ, Goodwin RL, Potts JD. A three-dimensional model of vasculogenesis. Biomaterials. 2009;30:1098–112. doi: 10.1016/j.biomaterials.2008.10.044. [DOI] [PubMed] [Google Scholar]
- 8.Toma C, Pittenger MF, Cahill KS, Cahill KS, Byrne BJ, Kessler PD. Human mesenchymal stem cells differentiate to a cardiomyocyte phenotype in the adult murine heart. Circulation. 2002;105:93–98. doi: 10.1161/hc0102.101442. [DOI] [PubMed] [Google Scholar]
- 9.Mangi AA, Noiseux N, Kong D, He H, Rezvani M, Ingwall JS, et al. Mesenchymal stem cells modified with Akt prevent remodeling and restore performance of infarcted hearts. Nat Med. 2003;9:1195–1201. doi: 10.1038/nm912. [DOI] [PubMed] [Google Scholar]
- 10.Chen SL, Fang WW, Ye F, Liu YH, Qian J, Shan SJ, et al. Effect on left ventricular function of intracoronary transplantation of autologous bone marrow mesenchymal stem cell in patients with acute myocardial infarction. Am H Cardiol. 2004;94:92–5. doi: 10.1016/j.amjcard.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 11.Couzin J, Vogel G. Cell therapy. Renovating the heart. Science. 2004;304:192–4. doi: 10.1126/science.304.5668.192. [DOI] [PubMed] [Google Scholar]
- 12.Balsam LB, Wagers AJ, Christensen JL, Kofidis T, Weissman IL, Robbins RC. Haematopoietic stem cells adopt mature haematopoietic fates in ischemic myocardium. Nature. 2004;428:668–73. doi: 10.1038/nature02460. [DOI] [PubMed] [Google Scholar]
- 13.Murry CE, Soonpaa MH, Reinecke H, Nakajima H, Nakajima HO, Rubart M, et al. Haematopoietic stem cells do not transdifferentiate into cardiac myocytes in myocardial infarcts. Nature. 2004;428:664–8. doi: 10.1038/nature02446. [DOI] [PubMed] [Google Scholar]
- 14.Jackson KA, Majka SM, Wang H, Pocius J, Hartley CJ, Majesky MW, et al. Regeneration of ischemic cardiac muscle and vascular endothelium by adult stem cells. J Clin Invest. 2001;107:1395–402. doi: 10.1172/JCI12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laflamme MA, Murry CE. Regenerating the heart. Nat Biotechnol. 2005;23:845–856. doi: 10.1038/nbt1117. [DOI] [PubMed] [Google Scholar]
- 16.Orlic D. The strength of plasticity: stem cells for cardiac repair. Int J Cardiol. 2004 1:16–9. doi: 10.1016/s0167-5273(04)90005-8. [DOI] [PubMed] [Google Scholar]
- 17.Carlson BM. Principles of regenerative biology. Amsterdam: Elsevier; 2007. Stimulation of regeneration; pp. 279–304. [Google Scholar]
- 18.Bohler KR, Czyz J, Tweedie D, Yang HT, Anisimov SV, Wobus AM. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ Res. 2002;91:189–201. doi: 10.1161/01.res.0000027865.61704.32. [DOI] [PubMed] [Google Scholar]
- 19.Sauer H, Wartenberg M, Sachinidis A, Hescheler J. Development of the cardiovascular system in embryoid bodies derived from embryonic stem cells. In: Sell S, editor. Stem cell handbook. Totowa, NJ: Humana Press; 2004. pp. 229–238. [Google Scholar]
- 20.Yost MJ, Baicu CF, Stonerock CE, Goodwin RL, Price RL, Davis M, et al. A novel tubular scaffold for cardiovascular tissue engineering. Tissue Eng. 2004;10:273–84. doi: 10.1089/107632704322791916. [DOI] [PubMed] [Google Scholar]
- 21.Valarmathi MT, Yost MJ, Goodwin RL, Potts JD. A three-dimensional tubular scaffold that modulates the osteogenic and vasculogenic differentiation of rat bone marrow stromal cells. Tissue Eng. 2008;14:491–504. doi: 10.1089/tea.2007.0235. [DOI] [PubMed] [Google Scholar]
- 22.Fan D, Qiu S, Overton CD, Yancey PG, Swift LL, Jerome WG, et al. Impaired secretion of apolipoprotein E2 from macrophages. J Biol Chem. 2007;282:13746–53. doi: 10.1074/jbc.M611754200. [DOI] [PubMed] [Google Scholar]
- 23.Evans HJ, Sweet JK, Price RL, Yost MJ, Goodwin RL. Novel 3D culture system for study of cardiac myocyte development. Am J Physiol Heart Circ Physiol. 2003;285:H570–H578. doi: 10.1152/ajpheart.01027.2002. [DOI] [PubMed] [Google Scholar]
- 24.Simpson DG, Terracio L, Terracio M, Price RL. Modulation of cardiac myocyte phenotype in vitro by the composition and orientation of the extracellular matrix. J Cell Physiol. 1994;161:89–105. doi: 10.1002/jcp.1041610112. [DOI] [PubMed] [Google Scholar]
- 25.Rosen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, MIsener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Totowa, NJ: Human Press; 2000. pp. 365–86. [DOI] [PubMed] [Google Scholar]
- 26.Valarmathi MT, Yost MJ, Goodwin RL, Potts JD. The influence of proepicardial cells on the osteogenic potential of marrow stromal cells in a three-dimensional tubular scaffold. Biomaterials. 2008;29:2203–16. doi: 10.1016/j.biomaterials.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 27.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reyes M, Dudek A, Jahagirdar B, Koodie L, Marker PH, Verfaillie CM. Origin of endothelial progenitors in human postnatal bone marrow. J Clin Invest. 2002;109:337–46. doi: 10.1172/JCI14327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang JS, Shum-Tim D, Galipeau J, Chedrawy E, Eliopoulos N, Chiu RC. Marrow stromal cells for cellular cardiomyoplasty: feasibility and potential clinical advantages. J Thorac Cardiovasc Surg. 2000;120:999–1005. doi: 10.1067/mtc.2000.110250. [DOI] [PubMed] [Google Scholar]
- 31.Martin-Rendon E, Sweeney D, Lu F, Girdlestone J, Navarrete C, Watt SM. 5-Azacytidine-treated human mesenchymal stem/progenitor cells derived from umbilical cord, cord blood and bone marrow do not generate cardiomyocytes in vitro at high frequencies. Vox Sang. 2008;95:137–48. doi: 10.1111/j.1423-0410.2008.01076.x. [DOI] [PubMed] [Google Scholar]
- 32.Rose RA, Jiang H, Wang X, Helke S, Tsoporis JN, Gong N, et al. Bone marrow-derived mesenchymal stromal cells express cardiac-specific markers, retain the stromal phenotype, and do not become functional cardiomyocytes in vitro. Stem Cells. 2008;26:2884–92. doi: 10.1634/stemcells.2008-0329. [DOI] [PubMed] [Google Scholar]
- 33.Pijnappels DA, Schalij MJ, Ramkisoensing AA, van Tuyn J, de Vries AA, van der Laarse A, et al. Forced alignment of mesenchymal stem cells undergoing cardiomyogenic differentiation affects functional integration with cardiomyocyte cultures. Circ Res. 2008;103:167–76. doi: 10.1161/CIRCRESAHA.108.176131. [DOI] [PubMed] [Google Scholar]
- 34.Zammaretti P, Jaconi M. Cardiac tissue engineering: regeneration of the wounded heart. Curr Opin Biotechnol. 2004;15:430–4. doi: 10.1016/j.copbio.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Wang JS, Shum-Tim D, Chedrawy E, Chiu RC. The coronary delivery of marrow stromal cells for myocardial regeneration: pathophysiologic and therapeutic implications. J Thorac Cardiovasc Surg. 2001;122:699–705. doi: 10.1067/mtc.2001.116317. [DOI] [PubMed] [Google Scholar]


