Abstract
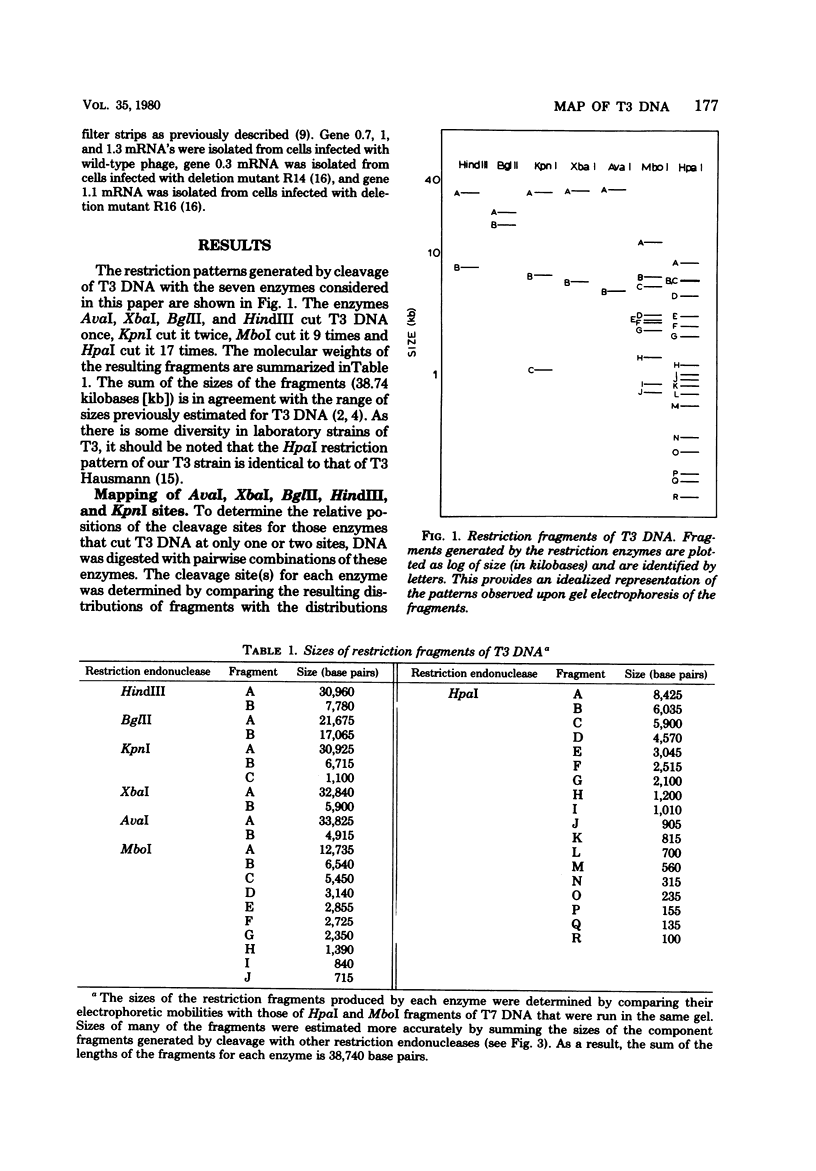
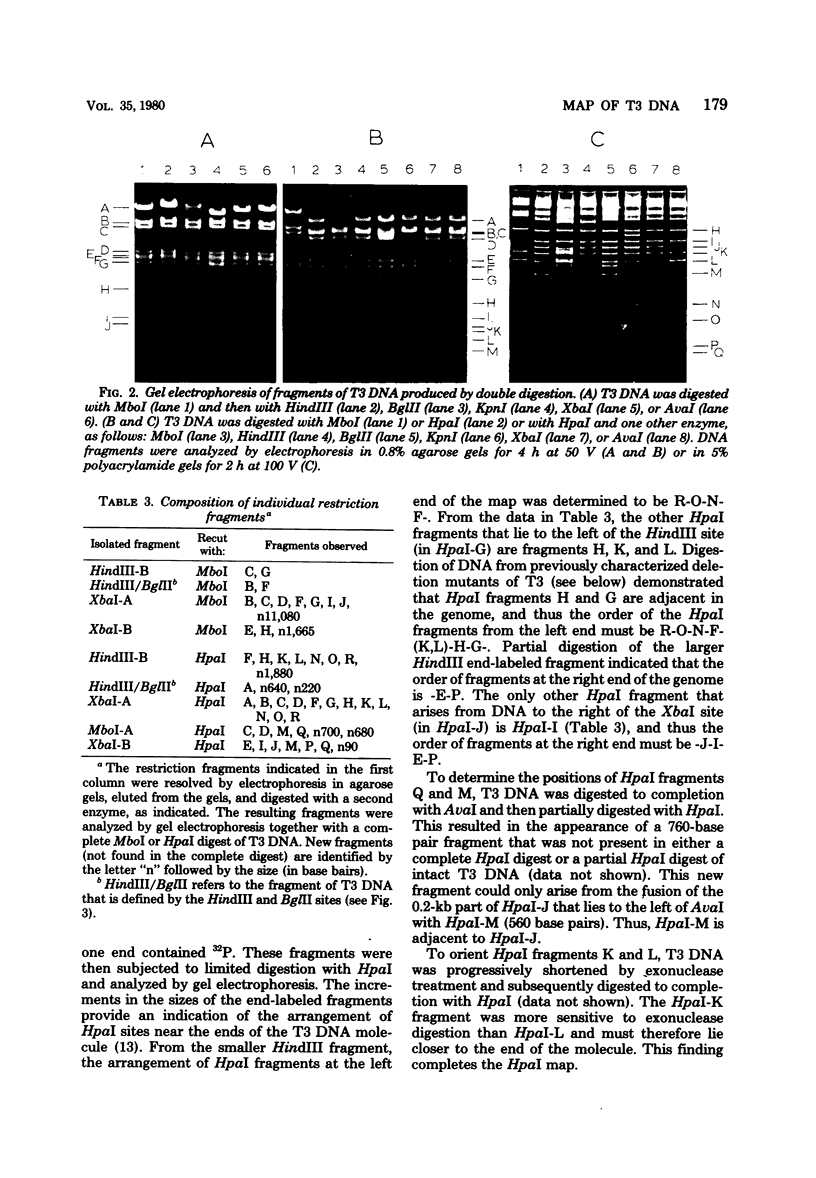
The DNA of bacteriophage T3 was characterized by cleavage with seven restriction endonucleases. AvaI, XbaI, BglII, and HindIII each cut T3 DNA at 1 site, KpnI cleaved it at 2 sites, MboI cleaved it at 9 sites, and HpaI cleaved it at 17 sites. The sizes of the fragments produced by digestion with these enzymes were determined by using restriction fragments of T7 DNA as molecular weight standards. As a result of this analysis, the size of T3 DNA was estimated to be 38.74 kilobases. The fragments were ordered with respect to each other and to the genetic map to produce a restriction map of T3 DNA. The location and occurrence of the restriction sites in T3 DNA are compared with those in the DNA of the closely related bacteriophage T7.
Full text
PDF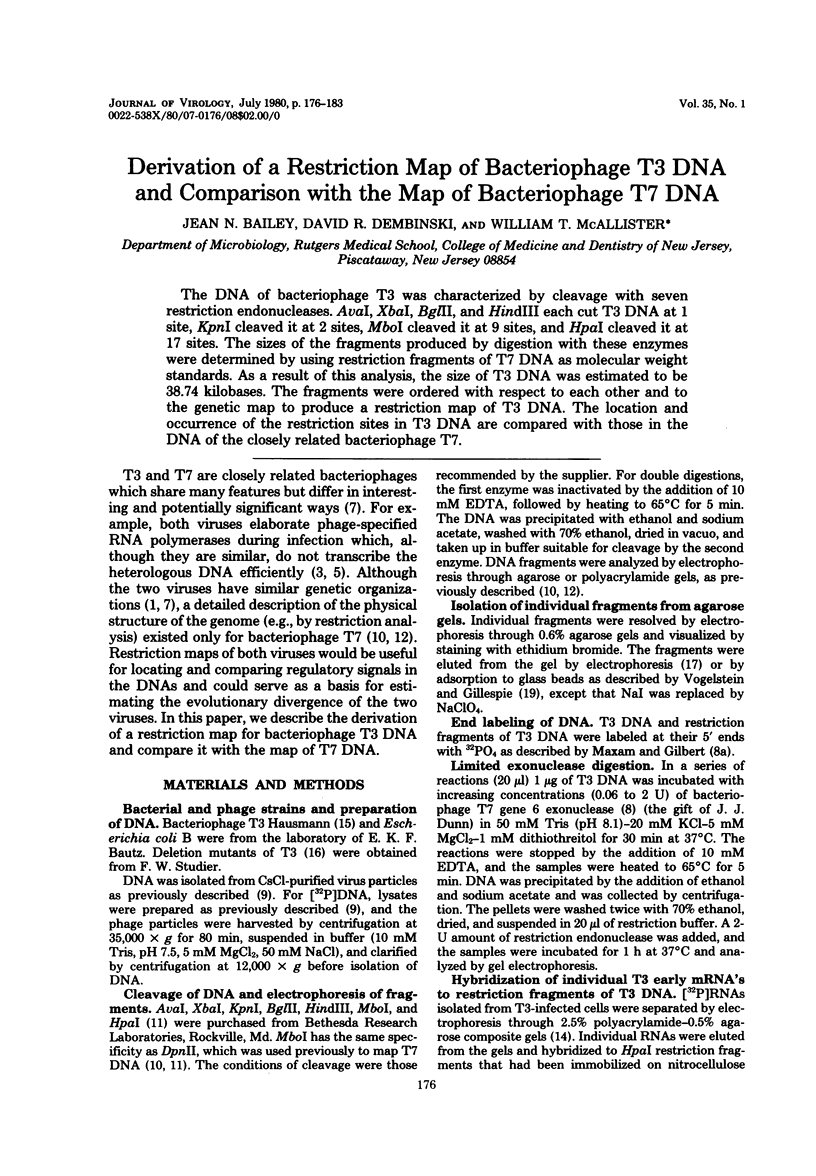




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BENDET I., SCHACHTER E., LAUFFER M. A. The size of T3 DNA. J Mol Biol. 1962 Jul;5:76–79. doi: 10.1016/s0022-2836(62)80062-x. [DOI] [PubMed] [Google Scholar]
- Beier H., Hausmann R. Genetic map of bacteriophage T3. J Virol. 1973 Aug;12(2):417–419. doi: 10.1128/jvi.12.2.417-419.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin M., McGrath J., Waskell L. New RNA polymerase from Escherichia coli infected with bacteriophage T7. Nature. 1970 Oct 17;228(5268):227–231. doi: 10.1038/228227a0. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Hyman R. W. A study in evolution: the DNA base sequence homology between coliphages T7 and T3. J Mol Biol. 1971 Dec 14;62(2):287–301. doi: 10.1016/0022-2836(71)90428-1. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Bautz F. A., Bautz E. K. Different template specificities of phage T3 and T7 RNA polymerases. Nat New Biol. 1971 Mar 17;230(11):94–96. doi: 10.1038/newbio230094a0. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. T7 early RNAs and Escherichia coli ribosomal RNAs are cut from large precursor RNAs in vivo by ribonuclease 3. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3296–3300. doi: 10.1073/pnas.70.12.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann R. Bacteriophage T7 genetics. Curr Top Microbiol Immunol. 1976;75:77–110. doi: 10.1007/978-3-642-66530-1_3. [DOI] [PubMed] [Google Scholar]
- Kerr C., Sadowski P. D. Gene 6 exonuclease of bacteriophage T7. I. Purification and properties of the enzyme. J Biol Chem. 1972 Jan 10;247(1):305–310. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McAllister W. T., Barrett C. L. Hybridization mapping of restriction fragments from the early region of bacteriophage T7 DNA. Virology. 1977 Oct 15;82(2):275–287. doi: 10.1016/0042-6822(77)90003-4. [DOI] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Gene. 1978 Nov;4(3):183–194. doi: 10.1016/0378-1119(78)90017-3. [DOI] [PubMed] [Google Scholar]
- Rosenberg A. H., Simon M. N., Studier F. W., Roberts R. J. Survey and mapping of restriction endonuclease cleavage sites in bacteriophage T7 DNA. J Mol Biol. 1979 Dec 25;135(4):907–915. doi: 10.1016/0022-2836(79)90519-9. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. Analysis of bacteriophage T7 early RNAs and proteins on slab gels. J Mol Biol. 1973 Sep 15;79(2):237–248. doi: 10.1016/0022-2836(73)90003-x. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Movva N. R. SAMase gene of bacteriophage T3 is responsible for overcoming host restriction. J Virol. 1976 Jul;19(1):136–145. doi: 10.1128/jvi.19.1.136-145.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. Relationships among different strains of T7 and among T7-related bacteriophages. Virology. 1979 May;95(1):70–84. doi: 10.1016/0042-6822(79)90402-1. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Simon M. N., Dunn J. J. Genetic and physical mapping in the early region of bacteriophage T7 DNA. J Mol Biol. 1979 Dec 25;135(4):917–937. doi: 10.1016/0022-2836(79)90520-5. [DOI] [PubMed] [Google Scholar]
- Upholt W. B. Estimation of DNA sequence divergence from comparison of restriction endonuclease digests. Nucleic Acids Res. 1977;4(5):1257–1265. doi: 10.1093/nar/4.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]