Abstract
PREP1 (PKNOX1) maps in the Down syndrome (DS) critical region of chromosome 21, is overexpressed in some DS tissues and might be involved in the DS phenotype. By using fibroblasts from DS patients and by overexpressing Prep1 in F9 teratocarcinoma and Prep1i/i MEF to single out the role of the protein, we report that excess Prep1 increases the sensitivity of cells to genotoxic stress and the extent of the apoptosis directly correlates with the level of Prep1. The apoptotic response of Prep1-overexpressing cells is mediated by the pro-apoptotic p53 protein that we show is a direct target of Prep1, as its depletion reverts the apoptotic phenotype. The induction of p53 overcomes the anti-apoptotic role of Bcl-XL, previously shown to be also a Prep1 target, the levels of which are increased in Prep1-overexpressing cells as well. Our results provide a rationale for the involvement of PREP1 in the apoptotic phenotype of DS tissues and indicate that differences in Prep1 level can have drastic effects.
INTRODUCTION
Prep1 belongs to the TALE class of homeodomain proteins and is essential for embryonic development. The human gene is referred to in the databases as PKNOX1; however, since this symbol is misleading it was suggested to name it PREP1 (1). In particular, Prep1 null mouse embryos die before gastrulation (Fernandez,L.C. and Blasi,F., manuscript in preparation), while 75% of the hypomorphic Prep1i/i embryos, that express 2–3% of Prep1 mRNA and up to 10% of the protein, show leaky embryonic lethality with defects in angiogenesis, hematopoiesis and eyes development. The other 25% live a normal-length life with major anomalies (2,3).
Prep1 is a transcription factor that, in combination with its major partners, Pbx proteins, regulates the overall size of the organism and individual organs as well as major developmental pathways. Indeed, hypomorphic Prep1i/i embryos and mice are much smaller and have also smaller organs, for example, pancreatic islets and fetal liver (2,4). Importantly, at least some of the phenotypes of Prep1i/i mice are also observed in the heterozygous state (4).
An important aspect of the Prep1i/i phenotype is the strong spontaneous apoptosis observed in E9.5 and E11.5 embryos, reproduced in E14.5 Prep1i/i mouse embryo fibroblasts (MEFs), which are also more sensitive to genotoxic stress (5).
The balance of pro- and antiapoptotic proteins at the mitochondrial outer membrane regulates its permeability, thus maintaining the organelle’s homeostasis and cotrolling apoptosis (6,7). The antiapoptotic Bcl-x gene is a transcriptional target of Prep1 (5) and its mRNA and gene product (Bcl-XL) are downregulated in Prep1i/i MEF and fetal liver cells. Indeed, Prep1i/i MEF have a compromised mitochondrial membrane potential, and show increased spontaneous and genotoxic stress-induced apoptosis, that are rescued by re-introduction of the Bcl-x gene (5).
Down syndrome (DS) is a human genetic disease due to trisomy of chromosome 21 that causes a distinctive phenotype with mental retardation, bone, blood and immune defects (8). The neurodegenerative and immune defects of DS patients correlate with an increased apoptosis rate (8–11), which was linked to increased expression of the proapoptotic tumor suppressor p53 (12,13).
PREP1 maps on chromosome 21 (21;q22.3) in humans and chromosome 17 in mice (14) and is overexpressed 1.5-fold in brain tissues of DS patients (15). The presence of PREP1 in the DS critical region of chromosome 21 (14) suggests that Prep1 might be involved in the phenotype of DS. However, the abnormally high apoptosis observed in embryos and MEF with reduced levels of Prep1 (Prep1i/i) (5) are in apparent discrepancy with the increased apoptosis of Prep1-overexpressing DS tissues (8,10–12).
We now report that the overexpression of Prep1 in Prep1i/i MEF and F9 teratocarcinoma cells (16), as well as human DS fibroblasts, also causes an increased sensitization to genotoxic stress in a p53-dependent manner, unlike hypomorphic Prep1i/i cells where apoptosis is mainly due to Bcl-XL depletion (5). Indeed, p53 is a direct transcriptional target of Prep1, is upregulated in Prep1-overexpressing cells and its downregulation in these cells prevents apoptosis. This shows that a correct balance of Prep1 is important in apoptotic homeostasis, as both its absence (5) and its overexpression induce apoptosis, although through diverse molecular targets: Bcl-x or p53.
MATERIALS AND METHODS
Cell culture
Human skin fibroblasts from spontaneous abortions (between 14 and 21 weeks of gestation) of normal and DS affected feti were provided by Dr. Franca Dagna Bricarelli (Galliera Genetic Hospital, Genoa) after karyotyping and maintained in RPMI medium supplemented with 10% of bovine fetal serum, 5 mM sodium pyruvate, 2 mM glutamine and streptomycin/penicillin at 37°C in a humidified incubator with 5% CO2. Primary Prep1i/i MEFs were obtained from 14.5-day embryos, after mating Prep1+/i heterozygous animals and genotyping and maintained as described elsewhere (5). Murine F9 teratocarcinoma (16) and Cos7 cells were maintained in Dulbecco's; modified Eagle's; medium (DMEM) supplemented with 10% of bovine fetal serum, 5 mM sodium pyruvate, 2 mM glutamine and streptomycin/penicillin at 37°C in a humidified incubator with 5% CO2.
Apoptotic treatments
3 × 105 MEF at passage 3 (5) and human skin fibroblasts, were plated in 6-cm dishes for fluorescence-activated cell sorting (FACS) analysis or at a density of 1 × 106 cells in 10-cm dishes for biochemical analysis. 6 × 105 F9 cells were plated in 6-cm dishes for FACS analysis or at a density of 2 × 106 cells in 10-cm dishes for biochemical analysis. After 24 h, each cell line was exposed to UV C (254 nm) at 1000 J/m2 (Vilber Lourmat, VL-115.C) or at 60 J/m2 (UV Stratalinker ™ 1800,Stratagene,) or treated with etoposide (Sigma) as indicated in the figures and previously described (5).
Flow cytometry
Apoptosis was measured with the Annexin V-FITC Apoptosis Detection KIT II (BD Pharmingen, San Diego, CA, USA) and analyzed by flow cytometry (FACSCAN, Becton–Dickinson).
Protein extraction and immunoblotting
Total cell extracts were prepared in RIPA buffer, clarified by centrifugation and quantitated as described in ref. 5. Protein extracts were fractionated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transfered to polyvinylidene difluoride (PVDF, Millipore). Membranes were incubated with primary antibodies for 1 h at room temperature and incubated with a peroxidase-conjugated secondary antibody for 1 h at room temperature. The ECL kit (Pierce, Rockford, IL, USA) was used to detect peroxidase activity following the manufacturer’s protocol. The resulting bands were quantitated by densitometric analysis (Personal Densitometer; Molecular Dynamics) as described (5). Antibodies: p53 (monoclonal antibody; JM-3036-100; MBL); Bcl-XL (monoclonal antibody; sc-8392; SantaCruz Biotechnology); caspase-3 (polyclonal antibodies; JM-3138-100; MBL); caspase-3 (polyclonal antibodies; Cell Signaling Techonology, Danvers, MA, USA); caspase-9 (monoclonal antibody; M054-3; MBL); β-actin (polyclonal antibodies; sc-1616; SantaCruz Biotechnology); tubulin-α (monoclonal antibody; N356; Amersham Lifescience); Prep1 (Meis 4.1; monoclonal antibody; 05-766; Upstate Biotechnology); and vinculin (V9131 monoclonal antibody; Sigma). Pbx1b (monoclonal antibody) was kindly provided by Dr. M. Cleary, Department of Pathology, Stanford University School of Medicine, Stanford, CA, USA.
Crude extracts used for the luciferase assay were obtained using ‘Reporter Lysis Buffer 1× Solution’ (Promega; see below). The collected supernatants were quantitated by the Bradford assay (Bio-Rad, Richmond, USA), fractionated on SDS-PAGE and transferred to PVDF (Millipore) as above.
RNA extraction, reverse transcription polymerase chain reaction and quantitative PCR
For RNA extraction the ‘RNeasy mini Kit’ (Qiagen) was used following the manufacturer’s instructions. Reverse transcription (RT) on total RNA was carried out with the ‘Superscript™ First-Strand Synthesis System for RT-PCR’ Kit (Invitrogen) using poly-A+ primers. For semiquantitative RT-polymerase chain reaction (PCR), 100 ng of cDNA were amplified with the following murine-specific primers:
Fwd-p53: 5′-CTTCCCAGCAGGGTGTCACGC-3′; Rev-p53: 5′-GTGCTGTGACTTCTTGTAGATGGC-3′; Fwd-Bcl-XL: 5′-CCACCACCTCCTCCCCGACC-3′; Rev-Bcl-XL 5′-GTACCGCAGTTCAAACTCATCGC-3′; Fwd-β-actin: 5′-GGCATCCTGACCCTGAAGT-3′; Rev-β-actin: 5′-CGGATGTCAACGTCACACTT-3′.
PCR was carried out on GeneAmp PCR System 2400 (Perkin Elmer), using a pre-PCR step of 2 min at 95°C followed by 28 cycles of 1 min at 95°C, 30 s at 63°C and 1 min at 72°C. PCR products were resolved on a 1.5% agarose 1× TAE gel and visualized by EtBr staining.
Triplicate RT-PCR reactions were carried out on an ABI/Prism 7900 HT Sequence Detector System (Applied Biosystems) at the IFOM-FIRC services (Milan) using 5 ng of the RT reaction; the following TaqMan Gene Expression Assays (Applied Biosystems) were used: murine BCL2L1 (NM_009743): Mm00437783_m1; murine β-actin (NM_007393.1): Mm00607939_s1; human TP53 (NM_000546): Hs00153349_m1; human PKNOX1(NM_197976.1): Hs00231814_m1; human β-actin (NM_001101.2): Hs99999903_m1.
Retroviral infection
Retroviral infection of MEF with pBabe-puro-Prep1 vectors was performed as described in ref. 5.
p-retro-Super vector containing a specific shRNA for murine p53 or a scrambled sequence were kindly provided by Dr. Silvia Soddu (Regina Elena Cancer Institute, Rome). Vectors were transfected in Phoenix packaging cells by the calcium phosphate method. Viral supernatants were used to perform four rounds of infections on Prep1 overexpressing F9 Teratocarcinoma cells after addition of 5 µg of Polybrene (Sigma).
p53-shRNA sequence: 5′-gatccccGTACATGTGTAATAGCTCCttcaagagaGGAGCTATTACACATGTACtttttggaaa-3′.
scrambled sequence: 5′-gatccccCTATAACGCTCGATATttcaagagaATATCGAGCGCCGTTATAGtttttggaaa-3′.
Hairpin-forming sequences are in capital letters.
Luciferase assay
Cos7 cells were transfected (in triplicate) with the plasmids described in Figure 5 using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. Plasmids: the reporter vector driven by the p53 regulatory region was provided by D. Reisman (17). The Prep1, Pbx1b and CMV β-Gal (Invitrogen) vectors were described previously (5,18). The mutated p53 promoter was prepared with the QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene) using the following primers: fwd: 5′-cctttctaaagtggtattttagctaatgaggg-3′; rev: 5′-ccctcattagctaaaataccactttagaaagg-3′.
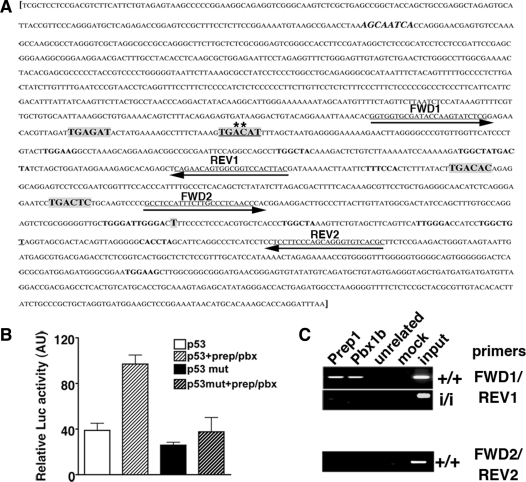
Figure 5.
p53 expression is directly controlled by Prep1-Pbx complex. (A) Murine p53 gene regulatory region [M26862 (17)]. Putative Prep1-Meis/Pbx-binding sites are in bold and in a larger font (low-affinity sites are in bold with the same font as the rest of the sequence). The canonical sequence is underlined and mutated bases are marked with an asterisk (A to G and C to T). The sequence in bold and italic is a canonical binding site on the reverse strand. Sequences underlined and with arrows are the primers used for the PCR amplification of genomic DNA from ChIP (see below). Primer set 1 (FWD1 and REV1) spans the region containing the Prep1/Pbx canonical binding site. Primer set 2 (FWD2 and REV2) spans the control region not containing any Prep1/Pbx binding site. The transcription start site (T) is shown. (B) Luciferase activity, in arbitrary units, assayed from extracts of transiently transfected Cos7 cells. The reporter (luciferase) vector containing either the WT (p53); (17)) or mutated (p53mut) regulatory region (–1327/+337 bp) of the murine trp53 gene [whole sequence shown in (A)] was co-transfected with Prep1 and/or Pbx1b expression vectors. The graph shows the β-galactosidase-normalized luciferase activity of total cell extracts. (C) ChIP was performed as described in ‘Materials and Methods’ section. Cross-linked chromatin from Prep1+/+ and Prep1i/i MEF was immunoprecipitated with antibodies against Prep1, Pbx1b and uPAR (as unrelated antibody). Amplification was performed using a specific primer set for the trp53 regulatory or control region (containing or not the Prep1/Pbx1-binding sites, respectively) and PCR products were fractionated on a 2% agarose 1 × TAE gel and visualized by EtBr staining. The picture shows a representative experiment out of three performed with the same outcome. mut: mutated.
Thirty hours after the transfection cells were washed with phosphate-buffered saline (PBS) and lysed with ‘Reporter Lysis Buffer 1× Solution’ (Promega) following the manufacturer’s protocol. The luciferase activity and the colorimetric β-gal reaction were measured as previously described in ref. 5.
Chromatin immunoprecipitation
Cross-linked chromatin was prepared from confluent MEF (at passage 4) as described (5,19). Briefly, 20-µg aliquots were incubated overnight with the appropriate antibodies (see below) or without antibodies (mock controls). The resulting material was processed as described (19). Four microliters of immunoprecipitated genomic DNA were amplified with primers spanning the Prep1/Pbx binding site (primer set 1): P53FWD1: 5′-GGTGGTGCGATACCAAGTATCTCG-3′; P53REV1: 5′-GTAAGTGGACCGCCACTGTTCTG-3′ or with primers amplifying the control region (not containing the Prep1/Pbx binding site; primer set 2): P53FWD2: 5′-GCCTCCATTTCTTGCCCTCAACC-3′; P53REV2: 5′-CGTGACACCCTGCTGGGAAGG-3′.
Amplification conditions were previously reported (19). Antibodies used for ChIP: anti-Pbx1b, provided by Dr. M. Cleary (Stanford University School of Medicine, CA, USA); anti-Prep1 polyclonal antibodies (18); anti-uPAR polyclonal antibodies, (20). Amplification products were fractionated on a 2% agarose gel in 1× TAE buffer and visualized by EtBr staining.
RESULTS
DS human fibroblasts express more Prep1 and are more sensitive to genotoxic stress
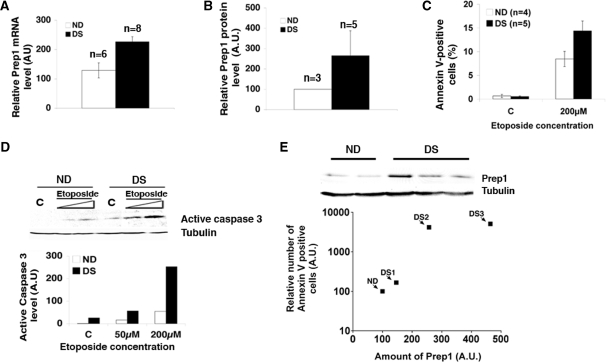
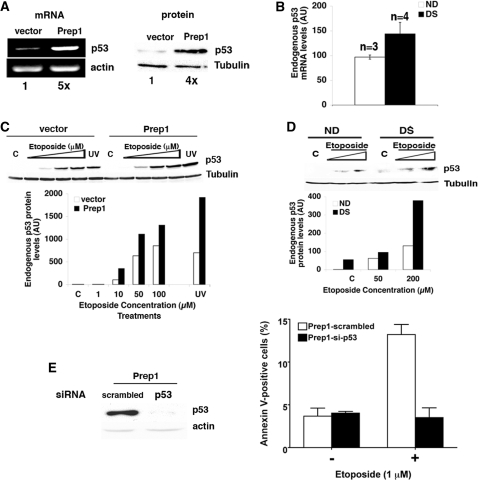
PREP1 maps on chromosome 21 (14): its level is increased in at least the brain of trisomy 21 DS patients (15). We find that in DS fibroblasts, Prep1 mRNA (Figure 1A) and protein (Figure 1B) are higher than in fibroblasts of normal donors (NDs), as expected. Several DS tissues are characterized by high levels of apoptotic cells (8,10–12); we, therefore, estimated the level of apoptosis of DS versus ND fibroblasts by measuring the relative amount of Annexin V-binding (flow-cytometry) cells after treatment with etoposide or UV-irradiation, both of which cause genotoxic stress. Figure 1C shows that untreated ND and DS fibroblasts have the same level of spontaneous apoptosis. However, upon a 24-h treatment with etoposide the number of apoptotic cells increased more substantially in DS than ND fibroblasts. The same results were obtained 24 h after UV irradiation (data not shown). Similar results were obtained with three different lines of DS fibroblasts. In agreement with this observation, the immunoblot of Figure 1D and its quantification show that after 24 h of etoposide treatment (or 24 h post-UV irradiation; data not shown), the level of endogenous active caspase 3 increases more sharply in DS than ND fibroblasts.
Figure 1.
Down syndrome fibroblasts show increased Prep1 levels and are more prone to genotoxic stress induced apoptosis. (A) Total mRNA from normal (ND) and Down syndrome (DS) skin fibroblasts was purified, retro-transcribed using a poly-A+ primer, quantitative real-time PCR analysis was performed using primers specific for human Prep1 (‘Material and Methods’ section) and the data normalized to β-actin mRNA values. (B) The average of Prep1 protein levels from three ND and five DS lines was calculated from the densitometric analysis of several immunoblots (data not shown). (C) ND (n = 4) and DS (n = 5) fibroblasts were treated (or not) with etoposide for 24 h. The number Annexin V-positive (i.e. apoptotic) cells was measured by FACS and values were expressed as percentage of total events. (D) Levels of active caspase 3 were detected by immunoblot with specific polyclonal antibodies using crude extracts from ND and DS fibroblasts treated (or not) with etoposide for 24 h at the concentrations indicated in the figure. The graph shows the results of the densitometric analysis, normalized to tubulin. C, untreated cells. (E) Cultures of three ND and three DS fibroblast lines were divided in two aliquots: one was treated with 200 µM etoposide for 24 h and the level of apoptosis measured by FACS using Annexin V staining and normalized to the level of apoptosis in the relative ND line. The other aliquot was used to determine the level of Prep1 in each line by immunoblot and densitometric analysis and normalized to the Prep1 level of the relative ND line. The relative Prep1 levels were plotted against the relative number of Annexin V positive cells for each DS line. A value of 100 was arbitrarily given to the Prep1 level and the number of Annexin V-positive cells in the three ND lines analyzed; therefore, a single ND value is reported in the graph.
We then exploited the variability in Prep1 content of the three lines of DS fibroblasts (Figure 1E, upper panel; see also the high standard deviation of Figure 1B) to correlate the levels of endogenous Prep1 with the susceptibility to genotoxic stress (Figure 1E, lower panel). Individual cultures of ND and DS fibroblasts were divided in two aliquots: one was used to establish the endogenous Prep1 level; the other was treated with etoposide (200 µM) for 24 h and the percentage of Annexin V-positive cells was measured by flow cytometry. As shown in Figure 1E, the relative amount of Annexin V-positive cells correlates with the endogenous level of Prep1, indicating that cells with a higher level of endogenous Prep1 are more susceptible to genotoxic stress.
Cells harboring an ectopic Prep1 expression vector recapitulate the apoptotic phenotype of DS fibroblasts
Many genes are present in the DS critical region of chromosome 21 and several of them may contribute to the DS phenotype (8). To single out the role of Prep1 in sensitizing cells to genotoxic stimuli, we tested the effect of Prep1 overexpression on the apoptotic behavior of murine teratocarcinoma (F9) cells (Figure 2A, left panel) and of MEFs from WT or Prep1i/i hypomorphic mutant mice (5,16). Figure 2A shows that the number of annexin V-positive cells was higher in Prep1-overexpressing than in control F9 cells both after etoposide treatment (Figure 2A, middle panel) and UV irradiation (Figure 2A, right panel). In agreement with these observations, the quantitation (Figure 2B) of an immunoblot analysis of endogenous caspase 9 in F9-overexpressing versus control cells after etoposide or UV irradiation (Supplementary Figure S1A) showed a more substantial increase in the former (Figure 2B). In addition, WT MEF overexpressing Prep1 displayed the same behavior (data not shown). Thus, Prep1 overexpression sensitizes cells to genotoxic stress-induced apoptosis.
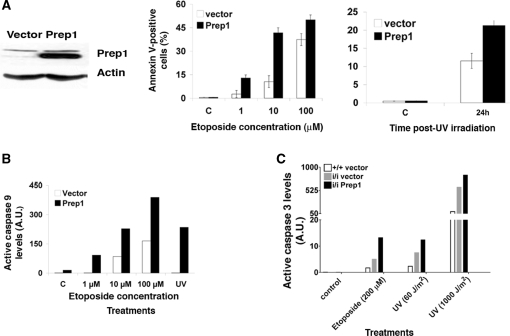
Figure 2.
Prep1 overexpression induces a strong apoptotic response in murine cells (A; left) Total extracts from murine F9 teratocarcinoma cells stably transfected with a Prep1-expressing or control vector (16) were resolved in an 8% SDS-PAGE and transferred to a PVDF membrane. The endogenous amount of Prep1 was checked by immunoblotting using a specific monoclonal antibody and normalized to β-actin content. (Middle) Cells were treated with etoposide as indicated or (Right) UV irradiated (UVC 254 nm; 60 J/m2). After 24 h of treatment or 24 h post-UV irradiation the number of AnnexinV-positive (apoptotic) cells was determined by FACS and plotted for overexpressing and control cells. Values are expressed as percentages of total events. C, untreated cells. (B) After 12 h of etoposide treatment (as indicated) or 12 h post-UV irradiation (UV C 254 nm; 60 J/m2), total cell extracts from Prep1-overexpressing or control F9 cells were resolved on 12% SDS-PAGE, transferred to a PVDF membrane and active caspases 9 levels were determined by immunoblotting and densitometric analysis, using tubulin for normalization. The densitometric analysis of the blot (Supplementary Figure S1A; one of three independent immunoblots is shown as representative experiment) is shown in the graph. (C) MEF from Prep1+/+ and Prep1i/i embryos infected with a retroviral vector (pBABE) either empty or containing the cDNA for human Prep1 were previously described [(5); Figure 6D]. Infected MEF were treated (or not) with etoposide (200 µM) for 24 h or irradiated with UV light (UV C 254 nm; 60 J/m2 or 1000 J/m2). Crude extracts were prepared after 24 h of drug treatment or 12 (1000 J/m2) or 24 h (60 J/m2) post-UV irradiation, fractionated in 12% SDS-PAGE, transferred to a PVDF membrane and the level of active caspase-3 determined by densitometric analysis (shown in the graph) of immunoblots (Supplementary Figure S1B). The experiment was repeated three times and the results from a representative experiment are shown.
We have shown that Prep1i/i MEF (expressing <10% of Prep1 protein) have a compromised mitochondrial membrane potential and increased spontaneous and induced apoptosis. Spontaneous apoptosis is rescued by Prep1 overexpression (5).We now, surprisingly, find that Prep1 overexpression increases the sensitivity to genotoxic stress. In fact, Prep1i/i MEF retrovirally infected to overexpress Prep1 show an increase of the endogenous levels of caspase 3 following genotoxic stress, as detected (Supplementary Figure S1B) and quantified (Figure 2C) by immunoblotting analysis. A similar activation of caspase 3 was also observed in Prep1-overexpressing WT MEF (data not shown). However, spontaneous (uninduced) apoptosis was not affected by Prep1 overexpression in F9 cells (Figure 2A middle panel).
We conclude that overexpression of Prep1 in F9 cells and Prep1i/i MEF increases their sensitivity and response to apoptotic (genotoxic) stimuli, similar to what observed in human skin fibroblasts from DS patients.
Prep1 overexpression, Bcl-x gene expression and apoptosis
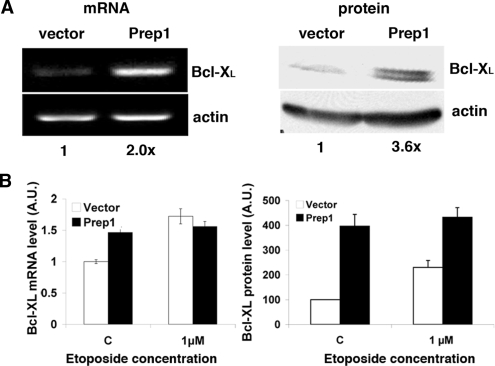
We have previously shown that the Bcl-x gene is a target of Prep1 and that its reduced expression in Prep1i/i MEF accounts for the increased spontaneous and genotoxic stress-induced apoptosis (5). We then analyzed the endogenous mRNA and protein levels of Bcl-XL. In Prep1-overexpressing F9 cells we find that both levels are increased (Figure 3A). The difference in the amount of protein and mRNA is probably due to the existence of post-transcriptional regulation, which has not been investigated. Moreover, they remain unaltered following genotoxic stress, whereas they increase in control cells (Figure 3B). These results are in line with the levels of spontaneous apoptosis being essentially identical in control versus overexpressing cells (Figures 1C and 2A). However, the increase in Bcl-XL is difficult to reconcile with the sensitization of Prep1-overexpressing cells to genotoxic-stress-induced apoptosis.
Figure 3.
Prep1 overexpression increases Bcl-XL expression (A; left) Total RNA was purified from untreated Prep1-overexpressing or control F9 cells, retrotranscribed using polyA+ primers and semiquantitative PCR analysis performed with specific primers for murine Bcl-XL and β-actin. The results of the densitometric analysis is shown under each lane. (Right) Crude extracts from the above cells were resolved by 12% SDS-PAGE and transferred to PVDF membrane. Endogenous Bcl-XL protein levels were analyzed by immunoblotting with specific monoclonal antibody and β-actin was used for normalization. Results of the densitometric analysis are shown under each lane. (B) Total mRNA and crude extracts were prepared and processed as above for qPCR (experiment performed in triplicate; left) or immunoblotting (right) from cell that were treated (or not) with the etoposide for 12 h. A value of 1 and of 100 was arbitrarily given to Bcl-XL mRNA (left) and protein (right) amount, respectively, in untreated cells infected with the control vector.
Apoptosis in Prep1 overexpressing mouse cells and human DS fibroblasts is p53-dependent
We therefore measured the endogenous mRNA and protein levels of the apoptosis-relevant protein p53 in cells overexpressing Prep1. We find that, in the absence of genotoxic stress, the level of endogenous p53 mRNA and protein are 5- and 4-fold, respectively, higher in Prep1-overexpressing than control F9 cells (Figure 4A) and in Prep1-overexpressing Prep1i/i MEF (data not shown). Similarly, q-PCR on polyA+ RNA from untreated ND versus DS fibroblasts showed a 50% higher content of p53 mRNA in the latter (Figure 4B), while also the protein levels appeared increased (Figure 4D; compare lane C in ND with lane C in DS). Upon etoposide treatment, higher levels of p53 accumulation (21,22) were detected in Prep1-overexpressing F9 cells and MEF versus their relative control (Figure 4C and data not shown, respectively) and also in DS versus ND fibroblasts (Figure 4D).
Figure 4.
Endogenous p53 expression levels increase in Prep1 overexpressing cells and its depletion rescues the apoptotic phenotype. (A; left) Total RNA was purified from untreated Prep1-overexpressing or control F9 cells, retrotranscribed using polyA+ primers and semiquantitative PCR analysis performed using specific primers for murine p53 and β-actin. Densitometric analysis results are shown under each lane. (Right) Crude extracts from the above cells were fractionated by 12% SDS-PAGE and transferred to PVDF membrane. Endogenous p53 levels were analyzed by immunoblotting with a specific monoclonal antibody. Tubulin was used for normalization. Densitometric analysis results are shown under each lane. (B) Total mRNA form ND (n = 3) and DS (n = 4) fibroblast lines was purified and processed as above. Quantitative real-time PCR was performed using primers specific for human p53 and the data are normalized to β-actin mRNA values. (C) Prep1-overexpressing or control F9 cells were treated with etoposide as shown or irradiated with UV C (254 nm; 60 J/m2). After 12 h of treatment or 12 h post-irradiation, crude extracts were prepared, resolved by 12% SDS-PAGE and transferred in PVDF membrane. The amount of p53 protein was determined by immunoblotting using a specific monoclonal antibody and the value normalized to tubulin. The graph reports the representative results of the above immunoblot (from one of three independent experiments), which was exposed for a short time to maintain p53 in the linear range for most of the lanes and in which p53 is not visible either in the C or 1-µM lanes. However, a longer exposure of the same blot shows the presence of p53 in the relevant C and 1-µM lanes (Supplementary Figure S1C). (D) Crude extracts were prepared from ND and DS fibroblasts treated (or not) with etoposide for 24 h, as indicated, fractionated and transferred as above. The levels of p53 were determined by densitometric analysis of the immunoblot, performed using specific monoclonal antibody and normalized to tubulin. C, untreated cells. The representative results of one of three independent experiments (using different ND and DS fibroblast lines) are shown. (E; left) Prep1 overexpressing F9 cells were infected with p-retro-super vector containing either a p53-specific or scrambled sequence shRNA and the level of endogenous p53 protein was detected from an aliquot of cells by immunoblotting using a monoclonal antibody and quantitated by densitometry and normalized to β-actin content. (Right) Another aliquot of knocked down or control Prep1-overexpressing F9 cells was treated with etoposide (1 µM) for 24 h and apoptotic cells were detected by flow cytometry using Annexin-V staining and their number plotted. The values were expressed as percentage of total events.
To confirm that genotoxic-stress-induced apoptosis in Prep1-overexpressing cells was indeed p53-dependent, we knocked-down p53 expression with a specific siRNA before subjecting them to treatment. Prep1-overexpressing F9 cells were infected either with a retroviral vector expressing a siRNA specifically targeted to p53 mRNA (see ‘Materials and Methods’ section) or a scrambled siRNA, as a control. The percentage of Annexin V-positive cells, in response to 1-µM etoposide, was then measured by flow cytometry. The left panel of Figure 4E shows the depletion of endogenous p53 protein obtained by the specific siRNA in Prep1 overexpressing F9 cells. When cells underwent genotoxic stress, p53-silenced cells displayed the same level of apoptosis as untreated cells, indicating that the Prep1-induced apoptosis is p53-dependent (Figure 4E, right). On the other hand scrambled siRNA-expressing cells showed an increase in the level of apoptosis, as expected (Figure 4E, right). We conclude, therefore, that Prep1 acts upstream of p53.
p53 is a transcriptional target of Prep1
We then investigated the possibility that Prep1 regulates p53 gene transcription. The sequence upstream of the murine p53 gene (17) contains two high affinity Prep1/Meis and Pbx canonical consensus sequences (TGACAT), as well as other low-affinity sites (Figure 5A) (23). We co-transfected COS cells with a luciferase reporter plasmid driven by the p53 regulatory region (p53-Luc, consisting of 1327 bp upstream and 337 bp downstream of the p53 transcription start site), along with expression vectors for Prep1 and Pbx1b, either alone or in combination. The p53-Luc reporter (17) had promoter activity of its own (Figure 5B) and was only marginally activated upon co-transfection with Prep1 or Pbx1b single expression vectors (data not shown). However, when the p53-Luc reporter was co-transfected with both Prep1 and Pbx1b expression vectors we observed a 2.5-fold increase in luciferase activity, indicating transcriptional activation of the reporter. Mutation of one canonical Prep1/Pbx binding sites in the regulatory region of the reporter plasmid reduced the activation by Prep1/Pbx1b heterodimer to almost basal levels, while causing a 30% reduction of the basal promoter activity. The results indicate that Prep1 and Pbx1 can indeed activate the p53 promoter.
We therefore asked whether a Prep1–Pbx1 complex was present on the regulatory region of p53 by using chromatin immunoprecipitation (ChIP). Since occupancy of chromatin regions corresponding to the regulatory elements of a given gene may be biased, in this kind of experiments, by the overexpression of specific proteins, we therefore decided to use MEF from WT and Prep1i/i hypomorphic mutant mice, using the latter cells as a negative control. Cross-linked, sonicated chromatin from the above cells was immunoprecipitated with antibodies against Prep1 and Pbx1b. DNA purified from the immunoprecipitates was analyzed by PCR amplifying both the regulatory region, containing the putative Prep1/Pbx binding sites, and the region immediately downstream the start site, that does not contain any canonical consensus-binding site (Figure 5A). PCR products were fractionated on agarose gels and detected by ethidium bromide staining. The results of Figure 5C show that chromatin immunoprecipitated with Prep1 and Pbx1b antibodies from WT (Figure 5C, upper +/+ panel), but not from Prep1i/i MEF (Figure 5C; i/i panel), is enriched in the p53 regulatory region, indicating the in vivo binding of the Prep1/Pbx heterodimer. The lack of enrichment in the immunoprecipitate from Prep1i/i MEF is a good control for the specificity of anti-Prep1 and anti-Pbx1 antibodies: in fact, Prep1i/i MEF also has a major reduction of all Pbx proteins (data not shown). No enrichment was detected in the downstream region (control), lacking Prep1/Pbx-binding sites (Figure 5C, bottom +/+ panel).
Overall, these results indicate that the p53 gene is a transcriptional target of the Prep1/Pbx complex.
DISCUSSION
Previously reported observations (5) and the results shown in this paper indicate that not only the absence of Prep1, but also its overexpression bias cells toward apoptosis. This appears incompatible, of course, unless the mechanisms are different in the two conditions. Indeed, we show that two different, although related, mechanisms are operative in Prep1-depleted versus overexpressing cells.
Here, we show that Prep1 overexpression causes an increase in genotoxic-induced, but not spontaneous apoptosis. We also illustrate that genotoxic-induced apoptosis is p53-dependent and that overexpression of Prep1 directly increases the level of p53. Finally, we demonstrate that a WT p53 reporter, but not a reporter mutated in the Prep1–Pbx1 binding site, is activated by the Prep1–Pbx1 heterodimer and that this complex binds the p53 promoter of WT, but not Prep1i/i MEF in vivo.
We have previously shown that in the absence of Prep1 Bcl-XL is decreased and that re-establishment of the Bcl-XL level rescues the spontaneous and induced apoptotic phenotype (5). In fact, we demonstrated that Bcl-x is a direct target of Prep1 since a wild-type Bcl-x reporter gene was activated by expression of a Prep1–Pbx1 heterodimer, while a reporter mutated in one Prep1–Pbx1 binding site was not. Moreover, we showed by ChIP that Prep1 and Pbx1 are bound to the Bcl-x promoter in wild-type MEF, but not in Prep1i/i MEF in vivo.
Therefore, the link between Prep1, Bcl-x expression and apoptosis is straightforward: a decrease in Prep1 levels leads to a decrease in Bcl-XL and altered mitochondrial homeostasis, priming cells to a greater sensitivity to genotoxic stress (5). On the contrary, when Prep1 is overexpressed, the p53 gene takes over. However, this appears dependent on the presence of genotoxic stress. In fact, untreated cells have increased levels of both Bcl-XL (as expected) and p53, but no Prep1-dependent difference in spontaneous apoptosis. The absence of differential apoptosis must signify that the two pro- and anti-apoptotic proteins must be quantitatively balanced. However, upon genotoxic stress of Prep1-overexpressing cells, p53 overcomes Bcl-XL and the balance is tilted in favor of the pro-apoptotic protein. This is most likely not only due to the effect of Prep1. Since Bcl-x gene expression is transcriptionally repressed by p53 (24,25), we would like to suggest that in overexpressing cells Prep1 activates both the Bcl-x and p53 genes. However, the stronger activation of p53 upon genotoxic stress leads to the repression of the Bcl-x gene, thus tilting the balance toward apoptosis.
These results shed some light on the role of Prep1 in regulating apoptosis. Although carried out in cell lines, the newly discovered mechanisms explain the apoptotic phenotype observed in zebra fish by morpholino regulation of prep1.1, and in Prep1i/i embryos. The fact that until now it has not been possible to produce Prep1-transgenic mice by oocyte injection is possibly due to a lethal effect (apoptosis) of Prep1 overexpression.
While increasing our knowledge on the molecular mechanisms underlying Prep1 phenotypes, the present results give valuable information on the DS phenotype. PREP1 in fact, maps on chromosome 21q23, i.e. in the DS critical region (14). In DS patients, Prep1 protein is overexpressed at least in the brain, where it over-activates the FABP7 gene (15). It is now well established that many tissues of DS patients present an excess apoptosis (8,10–12). We show here that fibroblasts from DS patients have a higher level of Prep1 and increased apoptosis when compared with ND fibroblasts. Moreover, the extent of apoptotic response to genotoxic stress directly correlates with the level of Prep1. These data suggest that Prep1 may be deeply involved in the apoptotic phenotype of DS tissues.
The rather strong differences between cells that do not drastically differ in Prep1 content recapitulate, however, similar effects observed in mouse embryos. Indeed, while Prep1–/– null embryos (i.e. 0% mRNA and protein) die before gastrulation at E7.5 (Fernandez and Blasi, submitted), Prep1i/i embryos (which express 2% mRNA and 4–10% protein) (2,4) die at E17.5 or survive (25%; see above). Finally, double heterozygous Prep1+/i–Prep1+/– embryos (which presumably express about 1% of the mRNA) die at an intermediate age (Rowan, Blasi and Maas, unpublished data). Therefore, minor differences in Prep1 level can have drastic effects.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Telethon Onlus (GGP06028 to FB) and Istituto Superiore di Sanità (ISS), Italy, Programma Malattie Rare and Ministero dell’Istruzione, dell’Università e della Ricerca (MIUR), Italy, Progetto Oncologia (to MPC). G.I. was supported by an A.I.R.C. (Associazine Italiana Ricerca sul Cancro) fellowship.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. F. Dagna Bricarelli, C. Baldo and the Galliera Genetic Bank – Network of Telethon Genetic Biobanks project GTB07001 for providing ND and DS fibroblasts; Silvia Soddu (Regina Elena Cancer Institute, Rome) for p-retro-super shp53 vector; David Reisman (Department of Biological Sciences University of South Carolina, U.S.A.) for the p53-luc reporter vector; Luis C. Fernandez (IFOM) for technical help. Francesca Bernassola (IFOM, Milan) and Andrea Brendolan (DIBIT, Milan) for critical reading of the manuscript.
REFERENCES
- 1.Mukherjee K, Bürglin TR. Comprehensive analysis of animal TALE homeobox genes: new conserved motifs and cases of accelerated evolution. J. Mol. Evol. 2007;65:137–153. doi: 10.1007/s00239-006-0023-0. [DOI] [PubMed] [Google Scholar]
- 2.Ferretti E, Villaescusa JC, Di Rosa P, Fernandez-Diaz LC, Longobardi E, Mazzieri R, Miccio A, Micali N, Selleri L, Ferrari G, et al. Hypomorphic mutation of the TALE gene Prep1 (pKnox1) causes a major reduction of Pbx and Meis proteins and a pleiotropic embryonic phenotype. Mol. Cell. Biol. 2006;26:5650–5662. doi: 10.1128/MCB.00313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Penkov D, Di Rosa P, Fernandez Diaz L, Basso V, Ferretti E, Grassi F, Mondino A, Blasi F. Involvement of Prep1 in the alphabeta T-cell receptor T-lymphocytic potential of hematopoietic precursors. Mol. Cell. Biol. 2005;25:10768–10781. doi: 10.1128/MCB.25.24.10768-10781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oriente F, Fernandez Diaz LC, Miele C, Iovino S, Mori S, Diaz VM, Troncone G, Cassese A, Formisano P, Blasi F, et al. Prep1 deficiency induces protection from diabetes and increased insulin sensitivity through a p160-mediated mechanism. Mol. Cell. Biol. 2008;28:5634–5645. doi: 10.1128/MCB.00117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Micali N, Ferrai C, Fernandez-Diaz LC, Blasi F, Crippa MP. Prep1 directly regulates the intrinsic apoptotic pathway by controlling Bcl-XL levels. Mol. Cell. Biol. 2009;29:1143–1151. doi: 10.1128/MCB.01273-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desagher S, Martinou JC. Mitochondria as the central control point of apoptosis. Trends Cell. Biol. 2000;10:369–377. doi: 10.1016/s0962-8924(00)01803-1. [DOI] [PubMed] [Google Scholar]
- 7.Borner C. The Bcl-2 protein family: sensors and checkpoints for life-or-death decisions. Mol. Immunol. 2003;39:615–647. doi: 10.1016/s0161-5890(02)00252-3. [DOI] [PubMed] [Google Scholar]
- 8.Antonarakis SE, Epstein CJ. The challenge of Down syndrome. Trends Mol. Med. 2006;12:473–479. doi: 10.1016/j.molmed.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Helguera P, Pelsman A, Pigino G, Wolvetang E, Head E, Busciglio J. ets-2 promotes the activation of a mitochondrial death pathway in Down's; syndrome neurons. J. Neurosci. 2005;25:2295–2303. doi: 10.1523/JNEUROSCI.5107-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zana M, Szecsenyi A, Czibula A, Bjelik A, Juhasz A, Rimanoczy A, Szabo K, Vetro A, Szucs P, Varkonyi A, et al. Age-dependent oxidative stress-induced DNA damage in Down's; lymphocytes. Biochem. Biophys. Res. Commun. 2006;345:726–733. doi: 10.1016/j.bbrc.2006.04.167. [DOI] [PubMed] [Google Scholar]
- 11.Roat E, Prada N, Ferraresi R, Giovenzana C, Nasi M, Troiano L, Pinti M, Nemes E, Lugli E, Biagioni O, et al. Mitochondrial alterations and tendency to apoptosis in peripheral blood cells from children with Down syndrome. FEBS Lett. 2007;581:521–525. doi: 10.1016/j.febslet.2006.12.058. [DOI] [PubMed] [Google Scholar]
- 12.Seidl R, Fang-Kircher S, Bidmon B, Cairns N, Lubec G. Apoptosis-associated proteins p53 and APO-1/Fas (CD95) in brains of adult patients with Down syndrome. Neurosci. Lett. 1999;260:9–12. doi: 10.1016/s0304-3940(98)00945-8. [DOI] [PubMed] [Google Scholar]
- 13.Sawa A, Oyama F, Cairns NJ, Amano N, Matsushita M. Aberrant expression of bcl-2 gene family in Down's; syndrome brains. Brain Res. Mol. Brain Res. 1997;48:53–59. doi: 10.1016/s0169-328x(97)00078-8. [DOI] [PubMed] [Google Scholar]
- 14.Berthelsen J, Viggiano L, Schulz H, Ferretti E, Consalez GG, Rocchi M, Blasi F. PKNOX1, a gene encoding PREP1, a new regulator of Pbx activity, maps on human chromosome 21q22.3 and murine chromosome 17B/C. Genomics. 1998;47:323–324. doi: 10.1006/geno.1997.5086. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez-Font MF, Bosch-Comas A, Gonzalez-Duarte R, Marfany G. Overexpression of FABP7 in Down syndrome fetal brains is associated with PKNOX1 gene-dosage imbalance. Nucleic Acids Res. 2003;31:2769–2777. doi: 10.1093/nar/gkg396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longobardi E, Blasi F. Overexpression of PREP-1 in F9 teratocarcinoma cells leads to a functionally relevant increase of PBX-2 by preventing its degradation. J. Biol. Chem. 2003;278:39235–39241. doi: 10.1074/jbc.M304704200. [DOI] [PubMed] [Google Scholar]
- 17.Reisman D, Eaton E, McMillin D, Doudican NA, Boggs K. Cloning and characterization of murine p53 upstream sequences reveals additional positive transcriptional regulatory elements. Gene. 2001;274:129–137. doi: 10.1016/s0378-1119(01)00623-0. [DOI] [PubMed] [Google Scholar]
- 18.Berthelsen J, Zappavigna V, Mavilio F, Blasi F. Prep1, a novel functional partner of Pbx proteins. EMBO J. 1998;17:1423–1433. doi: 10.1093/emboj/17.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrai C, Munari D, Luraghi P, Pecciarini L, Cangi MG, Doglioni C, Blasi F, Crippa MP. A transcription-dependent micrococcal nuclease-resistant fragment of the urokinase-type plasminogen activator promoter interacts with the enhancer. J. Biol. Chem. 2007;282:12537–12546. doi: 10.1074/jbc.M700867200. [DOI] [PubMed] [Google Scholar]
- 20.Resnati M, Pallavicini I, Daverio R, Sidenius N, Bonini P, Blasi F. Specific immunofluorimetric assay detecting the chemotactic epitope of the urokinase receptor (uPAR) J. Immunol. Methods. 2006;308:192–202. doi: 10.1016/j.jim.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Fridman JS, Lowe SW. Control of apoptosis by p53. Oncogene. 2003;22:9030–9040. doi: 10.1038/sj.onc.1207116. [DOI] [PubMed] [Google Scholar]
- 22.Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- 23.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugars KL, Budhram-Mahadeo V, Packham G, Latchman DS. A minimal Bcl-x promoter is activated by Brn-3a and repressed by p53. Nucleic Acids Res. 2001;29:4530–4540. doi: 10.1093/nar/29.22.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyashita T, Harigai M, Hanada M, Reed JC. Identification of a p53-dependent negative response element in the bcl-2 gene. Cancer Res. 1994;54:3131–3135. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.