Abstract
The Hu RNA-binding protein family consists of four members: HuR/A, HuB, HuC and HuD. HuR expression is widespread. The other three neuron-specific Hu proteins play an important role in neuronal differentiation through modulating multiple processes of RNA metabolism. In the splicing events examined previously, Hu proteins promote skipping of the alternative exons. Here, we report the first example where Hu proteins promote inclusion of an alternative exon, exon 6 of the HuD pre-mRNA. Sequence alignment analysis indicates the presence of several conserved AU-rich sequences both upstream and downstream to this alternatively spliced exon. We generated a human HuD exon 6 mini-gene reporter construct that includes these conserved sequences. Hu protein over-expression led to significantly increased exon 6 inclusion from this reporter and endogenous HuD. Studies using truncated and mutant HuD exon 6 reporters demonstrate that two AU-rich sequences located downstream of exon 6 are important. RNAi knockdown of Hu proteins decreased exon 6 inclusion. An in vitro splicing assay indicates that Hu proteins promote HuD exon 6 inclusion directly at the level of splicing. Our studies demonstrate that Hu proteins can function as splicing enhancers and expand the functional role of Hu proteins as splicing regulators.
INTRODUCTION
Alternative splicing is a process in which multiple messenger RNAs (mRNAs) are generated from one pre-messenger RNA (pre-mRNA) molecule and, as a result, multiple proteins are produced with potentially diverse functions. Similar to transcription, alternative pre-mRNA splicing provides an important mechanism of gene expression regulation. An analysis of high-throughput transcriptome sequencing indicates that 92–94% of human genes undergo alternative splicing (1,2). The most extensive alternative splicing occurs in brain tissues (3). Alternative splicing plays a key role in supporting the complex functions of the nervous system. However, our understanding of the regulatory mechanisms that control brain-specific alternative splicing remains very limited. A small number of brain-specific RNA-binding proteins have been identified that regulate alternative splicing (4).
Hu proteins have recently been identified as RNA processing regulators (5). Research carried out by our laboratory and others demonstrates that Hu proteins bind to intronic AU-rich elements to regulate alternative RNA processing. Three Hu-protein-regulated alternative splicing events have been characterized including the alternative splicing of neurofibromatosis type 1 (NF1) (6), apoptosis receptor Fas (7) and Ikaros (8). In all three examples, Hu proteins function as splicing repressors. Using the human calcitonin/calcitonin gene-related peptide (CGRP) system, we showed that Hu proteins could also suppress polyadenylation (9).
Hu proteins were originally cloned as the autoimmune antigens in patients with paraneoplastic encephalomyelitis, a neurodegenerative disorder (10). The Hu protein family consists of four members, HuR/A, HuB/Hel-N1, HuC and HuD (11). HuA (HuR) is widely expressed, while HuB, HuC and HuD are exclusively expressed in neuronal tissue. Every mammalian neuron is known to express at least one of the latter three Hu proteins (10,11). The neuron-specific Hu proteins have been shown to play important roles in neuronal differentiation (12–14) and function (15). The widely expressed family member, HuR, plays roles in muscle differentiation, adipogenesis, suppression of the inflammatory response and modulation of gene expression in response to chronic ethanol exposure and amino acid starvation (16–23).
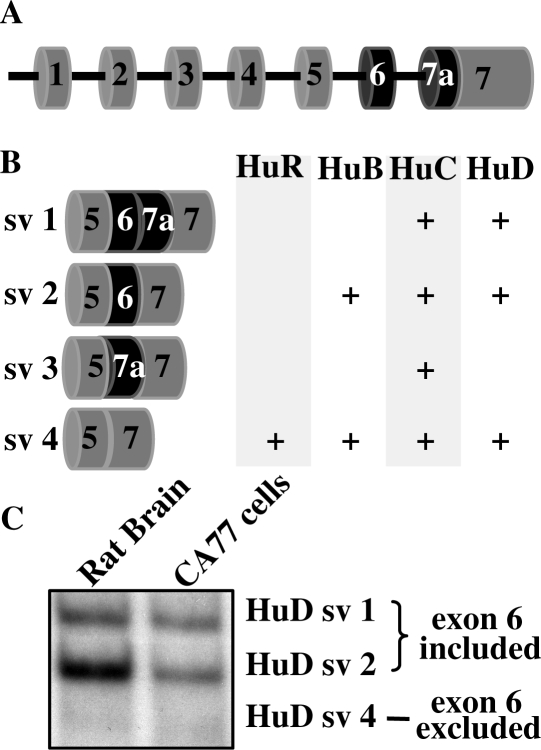
The biological functions of Hu proteins are carried out through their ability to bind to specific target mRNAs and affect their expression. Each Hu protein consists of three RNA-recognition motifs (RRMs) and a hinge domain between RRM2 and the C-terminal RRM3 (11). Hu proteins, through their first two RRMs, recognize and bind to AU-rich RNA sequences with an empirical preference for U-rich sequences (24). By binding to RNA, Hu proteins are involved in a wide range of post-transcriptional regulation of gene expression both in the nucleus and cytoplasm (5). The RRM domains are highly homologous between different Hu proteins (11). In contrast, the hinge domain, which is encoded by the region of pre-mRNA that undergoes alternative splicing, retains the highest variability. As shown in Figure 1A and B, within the hinge region, which is encoded by exons 5, 6 and part of 7 of the Hu pre-mRNA, only one isoform is generated for HuR, while multiple isoforms are generated for HuB, HuC and HuD as a result of the alternative splicing (11). The function of the hinge domain as well as the differential function of the Hu protein splice variants is poorly understood although persistence of each isoform through evolution supports functional differences (11).
Figure 1.
Hu splice variants (sv) and alternative splicing of HuD in rat brain and CA77 cells. (A) Schematic diagram of the Hu exon-intron structure. The RRM1 domain is located in exon 2 and part of exon 3. The RRM2 domain is located in part of exon 3, exon 4 and part of exon 5. The hinge domain is located in part of exon 5, exon 6 and part of exon 7. RRM3 is located on exon 7. (B) Splice variants of Hu proteins. Schematic diagram showing different splice variants of Hu protein family members. (C) RT-PCR analysis of HuD alternative splicing using RNA isolated from rat brain and CA77 cells. The HuD splice variants are indicated on the right.
To further investigate the function of Hu proteins as splicing regulators, we carried out a search for additional Hu-binding targets. Our search revealed several blocks of AU-rich sequences located in the introns surrounding the alternatively spliced HuD exon 6 and suggested that the HuD pre-mRNA itself is a potential target of Hu-mediated splicing regulation. We hypothesized that these AU-rich sequences may be important for the regulated HuD exon 6 splicing event. We generated a human HuD exon 6 mini-gene reporter construct that includes these AU-rich sequences. Hu protein over-expression in cells with low Hu protein expression led to significantly increased but varying levels of exon 6 inclusion from these reporters. Studies using truncated and mutated HuD exon 6 reporters demonstrated that two AU-rich sequences located downstream of exon 6 are required for the Hu-mediated regulation of exon 6 inclusion. Furthermore, we carried out an RNAi knockdown experiment in a cell line where Hu proteins are abundantly expressed and found that exon 6 inclusion decreased with reduced level of endogenously expressed Hu proteins. An RNA gel-shift analysis demonstrated that an RNA containing the wild type but not mutated AU-rich sequence can bind Hu proteins. Finally, an in vitro splicing assay provided the definitive evidence that Hu proteins promote HuD exon 6 inclusion directly at the level of splicing.
MATERIALS AND METHODS
Bioinformatic analysis of the genomic sequence of HuD exon 6
The method we used to search for potential binding sites of Hu proteins was previously described (6). Exon 6 of HuD was one of the potential targets. The alignment of the DNA sequences surrounding exon 6 (500 nt upstream and downstream) of human, mouse and rat was performed with MegAlign of DNASTAR (DNASTAR Inc., Madison, WI, USA) using the clustal W method. Only the highly conserved sequences flanking exon 6 are shown in Figure 2 (200 nt upstream and 233 nt downstream).
Figure 2.
Genomic sequences surrounding exon 6 of HuD are conserved across species. The HuD genomic sequence from Homo sapiens (AL592182), Mus musculus (AL627425) and Rattus norvegicus (NW_047718) were aligned and compared. Conserved sequences are shaded in black. The bold lines under the sequence represent the conserved AU-rich sequences. The nucleotides of exon 6 are indicated in capital letters.
Plasmids
The human HuD exon 6 reporter constructs used in transfection experiments consist of HuD exon 6 with part of the flanking introns inserted into the first intron of the human metallothionein 2A (HMT) gene. To generate the HMT-HuD exon 6 (E6) reporter, the human HuD sequence was polymerase chain reaction (PCR)-amplified from HeLa cell genomic DNA using primers HuD1 5′ and HuD1 3′. The PCR products were digested with BglII and BamHI and cloned into the RSV-HMT reporter linearized with BglII (Figure 3A). Primers HuD1 and HuD2-7 were used to generate the truncated HuD exon 6 reporter constructs (E6-T1-6) shown in Figure 4A. Primers HuD8 and HuD6 were used to generate the truncated HuD exon 6 reporter constructs E6-T7 shown in Figure 4D. The reporters containing mutated AU-rich sequences (Figure 5A) were generated by a PCR-mediated mutagenesis procedure using primers HuD9-11. The constructs used to generate RNA splicing substrates for experiments, shown in Figure 8A, were generated by PCR-mediated cloning using E6-T5 or Mut3 reporter as template and HuD12 5′ and HuD12 3′ as primers. The PCR products were restriction digested with HindIII and XbaI and cloned into the pGEM-3Zf(–) vector (Promega) digested with HindIII and XbaI. All of the plasmids were sequenced and the DNA primer sequences are shown in Table 1.
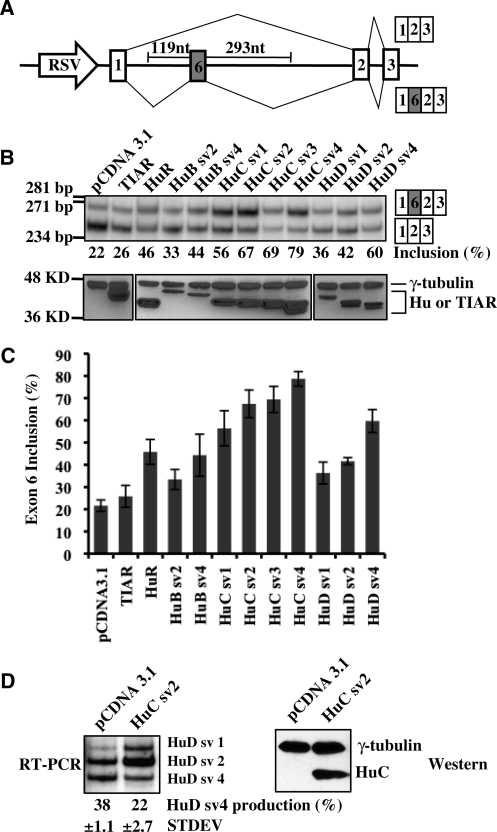
Figure 3.
Hu proteins promote HuD exon 6 inclusion. (A) A schematic diagram of the HuD splicing reporter construct E6. The two potential splicing products are indicated. (B) E6 reporter was co-transfected into HeLa cells with pCDNA3.1 vector, TIAR, or different Hu protein isoforms. Splicing of the reporter was analyzed by RT-PCR on total RNA isolated from the transfected cells (top). The percentage of exon 6 inclusion is indicated below the RT-PCR gel. Expression of co-transfected proteins was analyzed by western blot assay (bottom). γ-Tubulin was used as a loading control. The molecular markers for DNA or protein size are indicated on the left of the gels. (C) Graphic representation of RT-PCR results for exon 6 inclusion of the E6 reporter shown in (B), with error bars indicating standard deviations. (D) F9 cells were transfected with pCDNA3.1 vector or HuC protein. Splicing of the endogenous HuD was analyzed by RT-PCR on total RNA isolated from the transfected cells (top). The percentage of HuD sv4 production is indicated below the RT-PCR gel. Expression of the over-expressed protein was analyzed by western blot (bottom). γ-tubulin was used as a loading control.
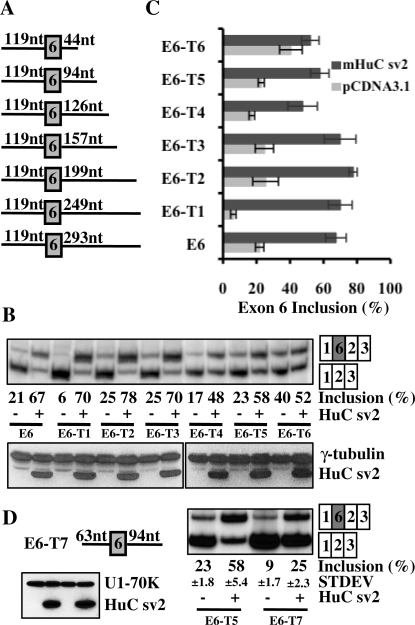
Figure 4.
Minimal sequences required for Hu-mediated inclusion of exon 6. (A) Schematic diagrams depicting the six truncated E6 reporters that contain varying lengths of sequence downstream of exon 6. (B) The pcDNA3.1 or HuC sv2 expression vector was co-transfected with the truncated reporters into HeLa cells. Splicing was analyzed using RT-PCR shown in the top panel. Expression of HuC sv2 was analyzed by western blot assay shown in the bottom panel. (C) Graphic representation of the RT-PCR results shown in (B). (D) Schematic diagram of the truncated E6 reporter E6-T7. Splicing of this reporter in HeLa cells in the absence or presence of HuC sv2 over-expression was analyzed using RT-PCR shown in the right panel. Expression of HuC sv2 was analyzed by western blot assay shown in the bottom panel.
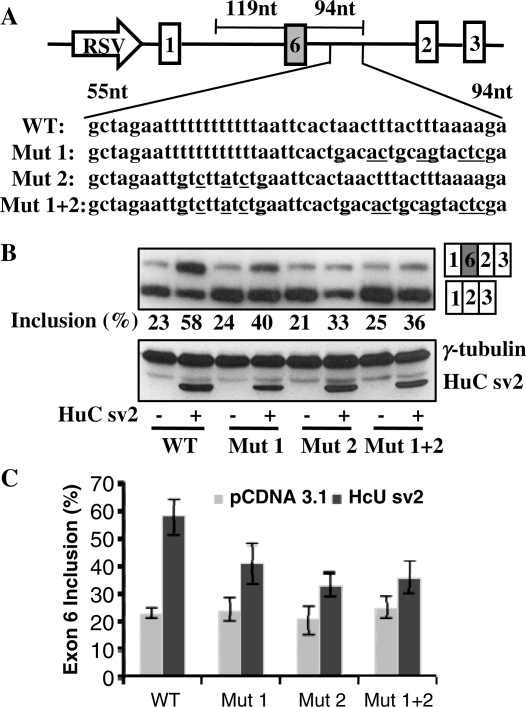
Figure 5.
Identification of two AU-rich elements important for Hu-mediated exon 6 inclusion. (A) Schematic diagram of the wild type (WT) and mutant E6-T5 reporters. The sequences in the AT-rich region in the wild type and mutant reporters are shown. (B) Wild type or mutant reporters were co-transfected with pcDNA3.1 or HuC sv2 expression vector. Splicing was analyzed by RT-PCR. Expression of HuC sv2 shown in the western blot analysis. (C) Graphic representation of the RT-PCR results shown in (B).
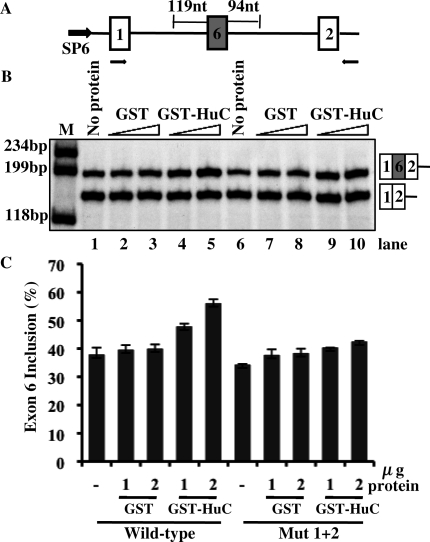
Figure 8.
HuC promotes the inclusion of HuD exon 6 in an in vitro splicing assay. (A) A schematic diagram of the construct used to generate the RNA splicing substrate for the in vitro splicing assay. The black arrows below the diagram indicate the position of primers used for RT-PCR analysis. (B) Lanes 1–5 indicate the splicing activity of the wild-type transcript in the presence of buffer alone, increasing amounts (1 and 2 µg) of GST alone, or increasing amounts GST-HuC sv4. Lanes 6–10 indicate the splicing activity of the Mut 1 + 2 transcript under the same conditions. (C) Graphic representation of the RT-PCR results shown in (B).
Table 1.
List of oligonucleotides used in this study
| Oligonucleotide | Sequence | Restriction site |
|---|---|---|
| HuD1 5′ | ACAGATCTAGTGGGGAAAGTTGTT | BglII |
| HuD1 3′ | ACGGATCCGGAGCCATTCAAAGGACCAGA | BamHI |
| HuD2 | ACGGATCCGCACCTGAACCATAGGAG | BamHI |
| HuD3 | ACGGATCCCACACAAATACATTTTGTG | BamHI |
| HuD4 | ACGGATCCCTGAGGTCCGAAATTAAAATTAAG | BamHI |
| HuD5 | ACGGATCCCATGCTGGTGGAAGAAAAAGG | BamHI |
| HuD6 | ACGGATCCTCTTTTAAAGTAAAGTTAGTG | BamHI |
| HuD7 | ACGGATCCCTGTGCCAACATGGACATCTG | BamHI |
| HuD8 | ACAGATCTGTTCCATTGAGCGATCTTGCCATC | BglII |
| HuD9 | ACGGATCCTCGAGTACTGCAGTGTCAGTGAATTAAAAAAAAAAAATTCTAG | BamHI |
| HuD10 | ACGGATCCTCTTTTAAAGTAAAGTTAGTGAATTCAGATAAGACAATTCTAGCC | BamHI |
| HuD11 | ACGGATCCTCGAGTACTGCAGTGTCAGTGAATTCAGATAAGACAATCTAGCC | BamHI |
| HuD12 5′ | ACAAGCTTCATGGATCCCAACTG | HindIII |
| HuD12 3′ | AGTCTAGACTGGAGGACAGGGAAGGGTAGAG | XbaI |
| DS8 | TTGACCATTCACCACATTGGTGTGC | |
| HMT3 | ATCTGGGAGCGGGGCTGT | |
| HuD endo-splicing-5′ | TTGCCAACAACCCCAGCCAGAAGTCCAG | |
| HuD endo-splicing-3′ | GTTATAGACGAAGATGCACCAGCCTG | |
| HuB endo-5′ | TGAGCTCTTGTCCTCAGTCCA | |
| HuB endo-3′ | GTACCTCTTGTCCATATTCAA | |
| HuC endo-5′ | AGCAGGCAGACCCATACACCT | |
| HuC endo-3′ | GGCCTGAGTAGGGCACCATTG | |
| HuD endo-5′ | TTAGTGGCCTTCCCAAGACCATG | |
| HuD endo-3′ | TCTGTAGCACCGCTGGGCTTCTG | |
| β-actin 5′ | TGGGCGACGAGGCCCAGAGCA | |
| β-actin 3′ | GTCAGGTCCCGGCCAGCCAGG |
The mammalian expression plasmids and GST fusion protein production plasmids for HuB and HuC protein isoforms were generated as described previously (9). The HuD mammalian expression plasmid pcHuD was a gift from Dr. Nora Perrone-Bizzozero at University of New Mexico, School of Medicine. Expression plasmids of HuD splicing variants, sv1 and sv4, were generated by PCR-mediated cloning using pcHuD as template.
The RNAi target sequences for HuB, HuC and HuD in CA77 cells were chosen using the siRNA designing tool from the siDESIGN® Center (www.dharmacon.com). The target sequences for HuB and HuC, both located on their exon 2, are GTCCTGTAAGCTTGTAAGA (HuB-1), AGCAAGACCAACCTAATAG (HuB-2) and TGAATCCTGCAAGTTGGTT (HuC-1), AGGACGAGTTCAAGAGTCT (HuC-2). The target sequence for HuD, located on its exon 4, is CACGCATCCTGGTTGATCA. The shRNA expression plasmids containing these target sequences were generated by oligonucleotide-mediated ligation into the vector pU6P [a gift from Dr. Guangbin Luo at Case Western Reserve University (25)]. The target sequence for eGFP GCCACCTACGGCAAGCTG was cloned into the vector pU6P as a control.
Cell culture and cell transfection
The HeLa and F9 cells were maintained according to the instructions from American Type Culture Collection (ATCC). CA77, a cell line derived from rat medullary thyroid carcinoma (a gift from Drs. Alison Hall at Case Western Reserve University, Cleveland, OH, and Andrew Russo at University of Iowa, Iowa City, IA, USA), were cultured in DMEM/F-12 media (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and 1% Pen/Strep (Invitrogen) (9). Transfection of HeLa cells were performed as described previously (6,26). In co-transfection experiments, 1 µg of the HuD exon 6 reporter plasmid and 0.5 µg of either Hu or TIAR expression plasmid were used. The over-expression of HuC in F9 cells was carried out using the Nucleofector Kit V with the Nucleofector II device (Lonza, formerly Amaxa). The transfection was performed with the standard protocol using 1.0 µg of the HuC sv2 expression vector and program D-023. The cells were grown for 48 h after transfection before harvest. The RNAi-mediated knockdown of Hu proteins in CA77 cells was carried out using the Nucleofector Kit V with the Nucleofector II device (Lonza). The transfection was performed with the standard protocol using 0.5 µg of the Hu expression plasmid or 1.5 µg of the HuD exon 6 truncated reporter 5 (E6-T5) with 4 µg of the various Hu shRNA plasmids and program T-030. The cells were grown for 72 h after transfection before protein or RNA harvest.
RNA and protein analysis
Procedures for total RNA and protein isolation and semi-quantitative reverse transcription (RT)-PCR analysis were described previously (26). Primers DS8 and HMT3 located on HMT exon 1 and exon 3 were used to analyze the HuD E6 reporter RNA. Low PCR cycle numbers were used to analyze reporter RNA isolated from HeLa (18–20 cycles) and CA77 (20–23 cycles) cells, respectively. Endogenous HuD alternative splicing in F9 cells that were transfected with HuC was analyzed using 24 PCR cycles with primers HuD endo-splicing-5′ and HuD endo-splicing-3′ as listed in Table 1. Endogenous Hu expression in CA77 cells treated with shRNAs was analyzed using 27–29 PCR cycles with primers HuB endo-5′ and HuB endo-3′, HuC endo-5′ and HuC endo-3′, or HuD endo-5′ and HuD endo-3′, as listed in Table 1. Quantification of HuD exon 6 inclusion was determined by phosphorimager analysis using a Typhoon Trio (GE Healthcare). The results shown are representative of at least three independent transfections for each experiment. The effect of Hu proteins on splicing of the reporter pre-mRNA was calculated as a percentage of HuD exon 6 inclusion [exon 6 inclusion/(exon 6 inclusion + exon 6 exclusion)]. The effect of HuC protein on splicing of the endogenous HuD pre-mRNA was calculated as a percentage of HuD sv4 production [(HuD sv4)/(HuD sv1 + sv2 + sv4)]. Western blot analysis using proteins isolated from the transfected cells was carried out with anti-Xpress (Invitrogen), anti-Myc (Invitrogen) or anti-HA antibodies (Covance). Anti-γ-tubulin (Sigma) and Anti-U1-70K antibody was used as a loading control in western blot analysis.
RNA gel mobility shift assay
A gel mobility shift assay was performed using recombinant GST or GST-HuC sv4 prepared from bacteria and wild type or mutant RNA oligonucleotides obtained from Thermo Fisher Scientific Inc. (Figure 7). The oligonucleotides were 32P-labeled at the 5′-end using T4 polynucleotide kinase (Invitrogen). The gel mobility shift assay was carried out in a volume of 25 µl containing 20 mM creatine phosphate, 2 mM adenosine triphosphate (ATP), 1 µg/µl bovine serum albumin (BSA), 2 µg/µl heparin and 80 fmol 32P-labeled RNA oligonucleotides in the absence or presence of 2, 10 and 50 ng of GST or GST-HuC sv4 protein. Reaction mixtures were incubated at 30°C for 30 min, then the mixture was separated on an 8% non-denaturing polyacrylamide gel in 1× TG buffer (0.05 M Tris and 0.05 M glycine).
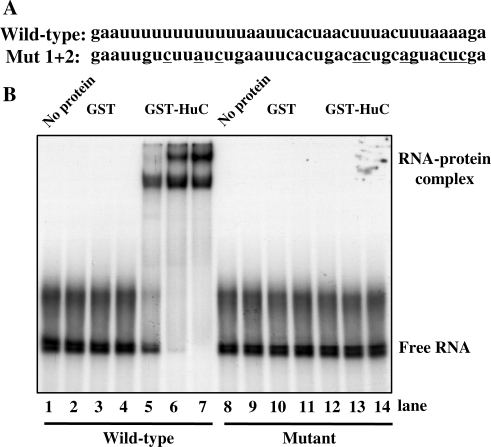
Figure 7.
Hu proteins bind to the AU-rich elements downstream of HuD exon 6. (A) The sequences of the wild type and mutant RNA oligonucleotides are shown. (B) RNA gel mobility shift analysis. The wild type (lanes 1–7) or mutant (lanes 8–14) RNA was incubated with no protein (lanes 1 and 8), increasing amounts (2, 10 and 50 ng) of GST protein alone (lanes 2–4 and 9–11) or GST-HuC sv4 (lanes 5–7 and 12–14).
In vitro splicing assay
HeLa cell nuclear extracts were prepared using S3 suspension culture and standard procedure (26). The plasmid used as template for in vitro transcription was digested with XbaI and RNA transcription was carried out as described previously (26). The full-length RNA substrates were excised from a urea polyacrylamide gel electrophoresis (PAGE) gel under ultraviolet (UV) light and recovered by eluting with buffer X [0.5M NH4Ac, 0.5% sodium dodecyl sulfate (SDS) and 5 mM ethylenediaminetetraacetic acid (EDTA)] followed by ethanol precipitation.
The in vitro splicing assay was carried out in a volume of 25 µl containing 44% (vol/vol) HeLa cell nuclear extract, 20 mM creatine phosphate, 2 mM ATP, 0. mM MgCl2, 1.5% polyethylene glycol, 0.15 mM dithiothreitol and 50 fmol RNA substrates in the absence or presence of 1 or 2 µg of GST or GST-HuC sv4 protein. Reaction mixtures were incubated at 30°C for 60 min, then the RNA was recovered from the reactions by phenol/chloroform extraction and ethanol precipitation. The resulting RNA was analyzed by semi-quantitive RT-PCR (16 cycles) as reported in a previous study (27) using the primers HuD12 5′ and HuD12 3′ (Table 1). Quantification of HuD exon 6 inclusion was determined using a Typhoon Trio (GE Healthcare). The results shown are representative of three independent splicing assays. The splicing products were cloned into a TA cloning vector pGEM-T (Promega) and confirmed by DNA sequencing.
RESULTS
Conserved AU-rich sequences are located near exon 6 in the HuD pre-mRNA
Previous studies have demonstrated that Hu proteins can function as splicing repressors to block exon inclusion by binding to AU-rich sequences (6). A computational analysis to search for additional Hu-binding targets identified more than 20 potential targets (6). The alternatively spliced HuD exon 6 is one of the potential targets. Analysis of the genomic sequences surrounding exon 6 in the HuD gene in human, mouse and rat revealed several conserved AU-rich sequences (Figure 2). The most conserved sequences are located within 140 nt upstream and 271 nt downstream of exon 6. In this conserved region, two and four blocks of AU-rich sequences are located upstream and downstream of exon 6, respectively, and a small block of AU-rich sequence is located in exon 6 (Figure 2). We hypothesized that these AU-rich sequences were targets of Hu proteins and therefore were important for the regulated HuD exon 6 splicing event.
Hu proteins promote HuD exon 6 inclusion
Recently, Hu proteins have been demonstrated to regulate alternative splicing by binding to intronic AU-rich elements (6–9,28). In all cases, interestingly, Hu proteins block inclusion of affected alternative exons (6–9,28). In the case of exon 6 of HuD, we postulated that Hu proteins may promote inclusion of this exon because in differentiated neurons, where Hu proteins are abundantly expressed, this exon is predominantly included (Figure 1C) (11). To determine if Hu proteins affect inclusion of this exon, we co-transfected HeLa cells, where none of the neuronal Hu proteins are expressed (6), with a mini-gene reporter (E6) containing HuD exon 6 with its flanking intronic sequences and individual members of the Hu protein family. The reporter contains exon 6, 119 nt of upstream, and 293 nt of downstream intronic sequences that include the conserved AU-rich sequences. The HuD sequence was inserted into the first intron of the human MT2A gene. Transcription of the reporter is directed by a Rous sarcoma virus (RSV) promoter (Figure 3A).
The mammalian Hu protein family consists of four members, HuA/HuR, HuB (HelN1), HuC and HuD. Except for HuR, which has only one protein isoform, each of the Hu proteins has different isoforms generated through alternative splicing in the coding region (Figure 1) (11). The major alternative splicing events that generate different isoforms of Hu proteins occur at the hinge region between the second and third RRMs. The naturally occurring alternative splicing events lead to production of the different Hu protein isoforms. Thus, an interesting question is whether these different Hu family members and their different isoforms have redundant functions in regulating splicing. In order to address this question, we either obtained or cloned all of the different isoforms of all of the Hu proteins (HuR, two mouse HuB splicing variants, four mouse HuC splicing variants and three human HuD splicing variants) and tested them in HeLa cells by transfecting each individually together with the E6 reporter.
An RT-PCR assay was carried out to analyze the splicing pattern of exon 6 (Figure 3B, top) and western blot analysis was used to show the protein expression level of transfected Hu cDNA plasmid (Figure 3B, bottom). It is clear from the results that every Hu protein member is capable of promoting inclusion of exon 6. However, different Hu members exhibit different potency in this splicing regulation. Overall, HuC protein isoforms are most potent as splicing promoters, followed by HuD, HuR and HuB (Figure 3B). Note that some of the Hu proteins, such as the HuB protein isoforms, are over-expressed at lower levels than others, which may explain the weaker splicing regulatory activity for these proteins. However, HuR is expressed at a similar level as HuC protein isoforms, yet is not as potent as HuC in promoting exon 6 inclusion. As a negative control, TIAR has no effect on the inclusion of HuD exon 6 (Figure 3B and C). To test the effect of over-expression of Hu proteins on alternative splicing of the endogenous HuD pre-mRNA, we transfected F9 cells, a mouse teratocarcinoma cell line in which HuD is endogenously expressed. In F9 cells, a significant amount of HuD sv4 isoform (38% of total HuD mRNA), which results from exon 6 exclusion, is produced (Figure 3D). HuC sv2 is used in this and the following experiments because of its consistent expression level and strong ability to promote exon 6 inclusion. The production of the HuD sv4 isoform is decreased to 22% upon HuC over-expression (Figure 3D). These experiments demonstrate that Hu proteins are capable of promoting HuD exon 6 inclusion from either the endogenous or the reporter HuD pre-mRNA.
Minimal sequences required for Hu-mediated inclusion of exon 6
Several blocks of AU-rich sequences are located surrounding HuD exon 6 (Figure 2). To test the importance of these AU-rich sequences on Hu-mediated exon 6 inclusion, we generated six truncated reporters that contain 119 nt upstream of exon 6 and varying lengths of sequences downstream of exon 6 (Figure 4A). These reporters were co-transfected into HeLa cells with the HuC sv2 expression plasmid. As shown in Figure 4B and C, HuC sv2 promotes exon 6 inclusion from the truncated reporters E6-T1, E6-T2, E6-T3, E6-T4 and E6-T5 at similar levels as from the parental reporter E6. However, when only 44 nt of the downstream sequence was included in the reporter E6-T6, the reporter did not respond to HuC sv2 (Figure 4B and C). To investigate the role of upstream AU-rich of exon 6, we generated a truncated reporter E6-T7 that contains 63 nt upstream of exon 6 and 94 nt downstream of exon 6 (Figure 4D). The only difference between this reporter and E6-T5 is that the AU-rich sequences upstream of exon 6 are deleted in E6-T7. Interestingly, although the baseline inclusion of exon 6 is decreased from the 23% with E6-T5 to 9% with E6-T7, addition of HuC sv2 resulted in a similar fold of increase of exon 6 inclusion from E6-T7 as E6-T5 (2.8-fold versus 2.5-fold) (Figure 4D). These results suggest that the upstream AU-rich sequences plays a minor, if any, role in Hu-medicated exon 6 inclusion and that the sequence between 44 and 94 nt downstream of exon 6 contains important elements required for Hu-mediated exon 6 inclusion.
Identification of two AU-rich elements important for Hu-mediated exon 6 inclusion
Two blocks of AU-rich sequences are located between 44 and 94 nt downstream of exon 6 (Figure 5A). Importantly, these AU-rich sequences are highly conserved in human, mouse and rat (Figure 2). To determine if these sequences are necessary for Hu-mediated inclusion of exon 6, we carried out mutational analysis. We mutated the two AU-rich sequences either individually or in combination using E6-T5 as a parental reporter, which contains the shortest sequence, 94 nt downstream of exon 6, which still responds to over-expression of Hu protein (Figure 5A). We generated three mutant reporters, mutant reporter 1 (Mut 1) with point mutations in the downstream AU-rich sequence, mutant reporter 2 (Mut 2) with mutations in the upstream AU-rich sequence and mutant reporter 1 + 2 (Mut 1 + 2) with mutations in both AU-rich sequences. The reporters were co-transfected into HeLa cells with the HuC sv2 expression plasmid. Compared to the wild-type E6-T5 reporter pre-mRNA that is processed to include exon 6 at 58% upon HuC sv2 over-expression, the Mut 1, Mut 2 and Mut 1 + 2 reporters showed significantly reduced exon 6 inclusion upon HuC sv2 over-expression (40%, 33% and 36%, respectively) (Figure 5C). These results indicate that the AU-rich elements located between nucleotides 44 and 94 downstream of exon 6 are important for Hu-mediated HuD exon 6 inclusion in HeLa cells.
Decreased exon 6 inclusion in CA77 cells with reduced level of Hu proteins
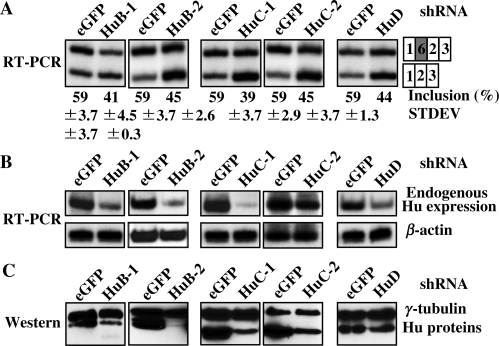
To determine if Hu proteins are required for inclusion of exon 6 in cells that endogenously express Hu proteins, we carried out an RNAi knockdown experiment in CA77 cells. In these cells, exon 6 of HuD is predominantly included (Figure 1C) and all of the Hu protein family members are expressed (9). We generated shRNA constructs that target HuB, HuC or HuD coding sequence. In CA77 cells, the pre-mRNA of the E6-T5 reporter was spliced to predominantly include exon 6 (59%, Figure 6A) when co-transfected with eGFP shRNA plasmid. However, when the reporter was co-transfected with the shRNA plasmid targeting HuB, HuC or HuD, exon 6 inclusion was consistently reduced to 41% (HuB-1), 45% (HuB-2), 39% (HuC-1), 45% (HuC-2) and 44% (HuD), respectively (Figure 6A). Due to the lack of Hu member-specific antibodies, we used an RT-PCR assay to analyze the mRNA expression level of endogenous Hu proteins in CA77 cells treated with the shRNAs (Figure 6B). Different Hu shRNAs exhibit different potency in reduction of Hu expression. The shRNA for HuC-1 appeared to be the most potent, followed by HuB-2 and HuB-1 (Figure 6B). To further test the Hu knockdown efficiency, we also co-transfected the epitope-tagged Hu expression vector with the various shRNA constructs in CA77 cells. As shown in Figure 6C, the level of the transfected Hu proteins was significantly reduced after co-transfection with shRNA expression vector, which is indicative of efficient knockdown. These experiments demonstrate that reduction of endogenous Hu proteins can decrease inclusion of HuD exon 6.
Figure 6.
Decreased inclusion of HuD exon 6 in CA77 cells with reduced level of Hu proteins. E6-T5 reporter was co-transfected with shRNA plasmid of eGFP, HuB-1, HuB-2, HuC-1, HuC-2 or HuD into CA77 cells. (A) Splicing of the reporter was analyzed by RT-PCR. (B) RT-PCR analysis of endogenous HuB, HuC and HuD expression in CA77 cells transfected with shRNA plasmids. The β-actin in the shRNA-treated cells used as a control. (C) To test the knockdown efficiency of the shRNA plasmids, they were co-transfected with Hu expression plasmid into CA77 cells. Expression of the over-expressed Hu with eGFP or Hu shRNA plasmids are shown in the western blot analysis.
HuC binds the AU-rich elements in the downstream intron sequence of HuD exon 6
Hu proteins have been shown to have strong affinity for AU-rich sequences (9,24). To determine if the AU-rich sequences downstream of the HuD exon 6 are targets for Hu proteins, we carried out RNA gel mobility shift assays using a wild type or mutant RNA oligonucleotide and a recombinant GST–HuC fusion protein. As shown in Figure 7, we detected two slow-moving complexes that formed on the wild-type RNA, which is indicative of binding of HuC protein to the RNA. Importantly, the mutant RNA in which the two AU-rich blocks were disrupted did not support formation of these complexes (Figure 7). Formation of two RNA–protein complexes at higher concentrations of HuC protein suggests that more than one HuC protein may associate with the RNA simultaneously.
HuC promotes the inclusion of HuD exon 6 in an in vitro splicing assay
The experiments discussed above demonstrate that Hu proteins promote HuD exon 6 inclusion in HeLa and CA77 cells and that two AU-rich elements are important for this regulation. To provide definitive evidence that Hu proteins promote HuD exon 6 inclusion directly at the level of splicing, we carried out in vitro splicing analysis using HeLa cell nuclear extract. The splicing substrate used in this experiment is shown in Figure 8A and was generated from the reporter used in the cell transfection assay. Both a wild-type substrate and a mutant one in which the AU-rich elements were disrupted were generated. We first carried out the classical in vitro splicing analysis using uniformly 32P-labeled RNA containing the HuD exon 6, 94 nt of downstream intronic sequence and exon 2 of the HMT gene. However, the splicing efficiency of this substrate was exceedingly low and no splicing products were observed (data not shown). This result is not surprising as alternative exons usually associate with suboptimal splicing signals. In the case of HuD exon 6, the polypyrimidine tract of its 3′ splice site has very short U-runs and the +5 position of its 5′ splice site is a uridine instead of the canonical guanidine (Figure 2). Thus, in order to examine in vitro splicing activity of this particular splicing event, we used RT-PCR to detect the splicing products, as pioneered in previous studies (27). As shown in Figure 8B and C, increasing amounts of GST-HuC, but not of GST protein, increased production of spliced RNA that contains exon 6 with the wild-type substrate (lanes 1–5). Importantly, little change was observed with the mutant substrate (lanes 6–10). These results demonstrate that Hu proteins directly affect splicing to promote inclusion of HuD exon 6.
DISCUSSION
A novel enhancer function of Hu proteins in splicing regulation
Hu proteins are known to bind specifically to AU-rich sequences. Previous studies in our laboratory and others demonstrate that Hu proteins function as repressors, negatively regulating splicing and polyadenylation (6,7,9,28). Here, we report the first example where Hu proteins function as splicing enhancer proteins, promoting inclusion of HuD exon 6. The ability of a splicing regulator to function as both an enhancer and a repressor is consistent with an important theme in splicing regulators. Well-studied examples that follow this theme include Nova-1 (29–32), SR proteins (33,34), CELF proteins (35) and Fox-1/Fox-2 (36–39).
In this study, we showed that Hu proteins promote inclusion of exon 6 of the HuD pre-mRNA by binding to evolutionarily conserved AU-rich intronic sequences. Since members of the Hu family proteins show similar exon–intron structure, it is possible that exon 6 in the pre-mRNA of the other three Hu members undergoes similar regulation. Of the four Hu protein transcripts, inclusion of exon 6 has never been observed with HuR/HuA pre-mRNA. Inclusion of exon 6 of the HuB pre-mRNA appears to undergo neuron-specific regulation. This exon is preferentially included in neurons and shows cell-type specific splicing pattern in cell lines (40,41). Exon 6 of the HuC pre-mRNA appears to be alternatively included, but no cell-specificity has been reported (11).
When we compared intronic sequences surrounding exon 6 in the HuB/C/D genes among human, mouse and rat, we found a good correlation between the presence of conserved AU-rich sequences and regulated inclusion of exon 6. For example, several conserved sequences are located around exon 6 in the HuB and HuD pre-mRNA, while no significant AU-rich blocks were found in the HuC sequence (data not shown). Based on this observation, we speculate that Hu proteins may also regulate inclusion of exon 6 of the HuB pre-mRNA in a similar way to that of the HuD pre-mRNA. However, the function of Hu proteins on alternative splicing of HuB exon 6 remains to be investigated experimentally.
Biological function of alternative inclusion of HuD exon 6
Hu proteins are involved in diverse biological processes. The neuronal Hu proteins, HuB, HuC and HuD, play an important role in neuronal development. Over-expression of HuD accelerates neurite outgrowth in E19 rat cortical neurons, PC12 cells, and retinoic acid-induced embryonic stem cells (42–44).
During mouse development, inclusion of exon 6 appears to be regulated. The HuB sv4 isoform, which does not include exon 6, is expressed early in mouse embryonic development at Day E10 and its expression is attenuated by Day E19. Expression of HuB sv2, the longer isoform that includes exon 6, is dramatically increased in differentiating embryonic neurons and is moderately expressed in adult neurons (13).
The splicing pattern involving HuD exon 6 is changed when it is ectopically expressed in small-cell lung cancers. In these cancer cells, exon 6 inclusion is significantly decreased (45). The HuB splicing pattern was also reported to change in human brain tumor medulloblastoma cells in which expression HuB sv2 is downregulated and HuB sv4 is upregulated (41). The biological significance of these changes is not clear.
Auto-regulation is very common in splicing regulators. In most cases, such regulation serves to tightly control the expression of these splicing regulators. For example, Nova-1 has been shown to repress its exon 4 inclusion by binding to exon 4 (46). HnRNP A1 binds to RNA elements surrounding exon 7b that leads to increased skipping of this exon (47). Exon 11 of the PTB pre-mRNA is repressed by PTB in an auto-regulating feedback loop that leads to degradation (48). In these examples, nonsense-mediated decay is usually triggered by the alternative splicing, which leads to reduced expression of these splicing regulators. In the case of HuD, since exon 6 contains 39 nt, change of the splicing pattern does not cause a reading-frame shift. In order to understand the biological function of the regulated splicing of HuD, it is important to determine the differential function of these isoforms. A previous study suggests that exon 6 encodes for part of the nuclear export signal (NES) in the hinge region. Therefore, isoforms that contain exon 6 may have different localization than isoforms that do not (44).
At the molecular level, all of the Hu protein isoforms are capable of promoting HuD exon 6 inclusion. However, differences do exist in the ability of each protein isoform to regulate alternative splicing. Within the same Hu protein (e.g. HuD), the shortest isoform (sv4) that lacked exon 6 promoted the highest level of exon 6 inclusion (Figure 3). Note that these differences were observed using splicing reporters. Thus, it remains to be determined if Hu protein isoforms affect splicing of the endogenously expressed pre-mRNAs differentially.
Potential mechanisms of Hu-mediated HuD exon 6 inclusion
How do Hu proteins promote inclusion of HuD exon 6 by binding to two AU-rich sequences downstream of the exon? We envision at least two potential mechanisms. First, Hu proteins may bind to specific AU-rich elements in the intron and recruit U1 snRNP to the 5′ splice site downstream of exon 6. In our previous study, we found that when Hu proteins bind to an AU-rich sequence 15 nt downstream of exon 23a of the NF1 pre-mRNA, they antagonize the function of TIA-1/TIAR to block interaction between U1 and U6 snRNP and the 5′ splice site (6). Recent studies have demonstrated that the same element can have opposite splicing regulatory effect to inhibit or enhance exon inclusion when located in different positions relative to the alternative exon. Two well-studied examples are Nova and Fox proteins. Nova-1 binding to an exonic YCAY cluster changes the protein complexes assembled on pre-mRNA, blocking U1 binding and exon inclusion, whereas Nova-1 binding to an intronic YCAY cluster enhances spliceosome assembly and exon inclusion (32). The position of the binding sites determines either activation or repression of exon recognition by Fox-1/2 (36). It appears that a simple rule based on the location of AU-rich sequences relative to the regulated exon does not apply to Hu proteins. For example, our previous study showed that Hu proteins negatively regulate inclusion of exon 23a of the NF1 pre-mRNA. In that case, the major AU-rich sequence is also located downstream of the exon (6). However, it is possible that the distance of AU-rich sequences to the regulated exon is important in their regulatory role. The AU-rich sequence is 56 and 13 nt downstream of Hu exon 6 and NF1 exon 23a, respectively. Future studies will be carried out to address if and how this potential position-dependent effect is achieved.
Second, Hu proteins may increase exon 6 inclusion by competing with splicing repressors. The results of the truncated and mutant HuD exon 6 reporters showing different levels of exon 6 inclusion in cells suggest that very complex regulatory mechanisms may control the splicing of HuD exon 6. Exon 6 inclusion levels change with different truncated or mutant reporters in HeLa cells (Figures 4 and 5). The result that the E6-T6 reporter shows a higher exon 6 inclusion than the E6-T5 reporter suggests that some splicing silencer motifs exist in this minimal sequence required for Hu-mediated inclusion of exon 6. By searching the Human Splicing Finder (http://www.umd.be/HSF/), we found several putative splicing silencer motifs located in this region. In the future, more detailed analysis of the mechanism of Hu-mediated HuD exon 6 inclusion will be carried out to address these issues.
FUNDING
The National Institutes of Health (NS-049103 to H.L.); the Department of Defense (NF060083 to H.L.); and the American Heart Association (0415086B to H.Z.). The GE Healthcare Typhoon Trio Variable Mode Imager was made available through a National Center for Research Resources (NCRR) Shared Instrumentation Grant (#1 S10 RR024536). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGMENTS
We would like to acknowledge the following individuals for providing plasmids and cells: Nora Perrone-Bizzozero at the University of New Mexico Health Sciences Center (pcHuD), Xincheng Lu in Guangbin Luo’s lab at Case Western Reserve University (pU6P), Alison Hall at Case Western Reserve University and Andrew Russo at the University of Iowa (CA77 cell line). We thank Robert Hasman for generating the GST-HuC sv4 construct. We thank members of the Lou laboratory, Victoria Barron, Melissa Hinman and Hua-Lin Zhou, for helpful discussions and critical reading and editing of the manuscript.
REFERENCES
- 1.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 3.Xu Q, Modrek B, Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002;30:3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 5.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu H, Hinman MN, Hasman RA, Mehta P, Lou H. Regulation of neuron-specific alternative splicing of neurofibromatosis type 1 pre-mRNA. Mol. Cell. Biol. 2008;28:1240–1251. doi: 10.1128/MCB.01509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izquierdo JM. Hu antigen R (HuR) functions as an alternative pre-mRNA splicing regulator of Fas apoptosis-promoting receptor on exon definition. J. Biol. Chem. 2008;283:19077–19084. doi: 10.1074/jbc.M800017200. [DOI] [PubMed] [Google Scholar]
- 8.Bellavia D, Mecarozzi M, Campese AF, Grazioli P, Talora C, Frati L, Gulino A, Screpanti I. Notch3 and the Notch3-upregulated RNA-binding protein HuD regulate Ikaros alternative splicing. EMBO J. 2007;26:1670–1680. doi: 10.1038/sj.emboj.7601626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu H, Hasman RA, Barron VA, Luo G, Lou H. A nuclear function of Hu proteins as neuron-specific alternative RNA processing regulators. Mol. Biol. Cell. 2006;17:5105–5114. doi: 10.1091/mbc.E06-02-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szabo A, Dalmau J, Manley G, Rosenfeld M, Wong E, Henson J, Posner JB, Furneaux HM. HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell. 1991;67:325–333. doi: 10.1016/0092-8674(91)90184-z. [DOI] [PubMed] [Google Scholar]
- 11.Okano HJ, Darnell RB. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J. Neurosci. 1997;17:3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wakamatsu Y, Weston JA. Sequential expression and role of Hu RNA-binding proteins during neurogenesis. Development. 1997;124:3449–3460. doi: 10.1242/dev.124.17.3449. [DOI] [PubMed] [Google Scholar]
- 13.Gao FB, Keene JD. Hel-N1/Hel-N2 proteins are bound to poly(A)+ mRNA in granular RNP structures and are implicated in neuronal differentiation. J. Cell. Sci. 1996;109(Pt 3):579–589. doi: 10.1242/jcs.109.3.579. [DOI] [PubMed] [Google Scholar]
- 14.Antic D, Keene JD. Messenger ribonucleoprotein complexes containing human ELAV proteins: interactions with cytoskeleton and translational apparatus. J. Cell. Sci. 1998;111(Pt 2):183–197. doi: 10.1242/jcs.111.2.183. [DOI] [PubMed] [Google Scholar]
- 15.Akamatsu W, Fujihara H, Mitsuhashi T, Yano M, Shibata S, Hayakawa Y, Okano HJ, Sakakibara S, Takano H, Takano T, et al. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc. Natl Acad. Sci. USA. 2005;102:4625–4630. doi: 10.1073/pnas.0407523102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorospe M. HuR in the mammalian genotoxic response: post-transcriptional multitasking. Cell Cycle. 2003;2:412–414. [PubMed] [Google Scholar]
- 17.Lal A, Kawai T, Yang X, Mazan-Mamczarz K, Gorospe M. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha. EMBO J. 2005;24:1852–1862. doi: 10.1038/sj.emboj.7600661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Figueroa A, Cuadrado A, Fan J, Atasoy U, Muscat GE, Munoz-Canoves P, Gorospe M, Munoz A. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol. Cell. Biol. 2003;23:4991–5004. doi: 10.1128/MCB.23.14.4991-5004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherry J, Karschner V, Jones H, Pekala PH. HuR, an RNA-binding protein, involved in the control of cellular differentiation. In Vivo. 2006;20:17–23. [PubMed] [Google Scholar]
- 20.Katsanou V, Papadaki O, Milatos S, Blackshear PJ, Anderson P, Kollias G, Kontoyiannis DL. HuR as a negative posttranscriptional modulator in inflammation. Mol. Cell. 2005;19:777–789. doi: 10.1016/j.molcel.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Yaman I, Fernandez J, Sarkar B, Schneider RJ, Snider MD, Nagy LE, Hatzoglou M. Nutritional control of mRNA stability is mediated by a conserved AU-rich element that binds the cytoplasmic shuttling protein HuR. J. Biol. Chem. 2002;277:41539–41546. doi: 10.1074/jbc.M204850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMullen MR, Cocuzzi E, Hatzoglou M, Nagy LE. Chronic ethanol exposure increases the binding of HuR to the TNFalpha 3′-untranslated region in macrophages. J. Biol. Chem. 2003;278:38333–38341. doi: 10.1074/jbc.M304566200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilusz CJ, Wilusz J. HuR-SIRT: the hairy world of posttranscriptional control. Mol. Cell. 2007;25:485–487. doi: 10.1016/j.molcel.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Chung S, Jiang L, Cheng S, Furneaux H. Purification and properties of HuD, a neuronal RNA-binding protein. J. Biol. Chem. 1996;271:11518–11524. doi: 10.1074/jbc.271.19.11518. [DOI] [PubMed] [Google Scholar]
- 25.Ren XR, Zhou LJ, Luo GB, Lin B, Xu A. Inhibition of hepatitis B virus replication in 2.2.15 cells by expressed shRNA. J. Viral. Hepat. 2005;12:236–242. doi: 10.1111/j.1365-2893.2005.00587.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhu H, Hasman RA, Young KM, Kedersha NL, Lou H. U1 snRNP-dependent function of TIAR in the regulation of alternative RNA processing of the human calcitonin/CGRP pre-mRNA. Mol. Cell. Biol. 2003;23:5959–5971. doi: 10.1128/MCB.23.17.5959-5971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothrock CR, House AE, Lynch KW. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 2005;24:2792–2802. doi: 10.1038/sj.emboj.7600745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu H, Zhou HL, Hasman RA, Lou H. Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. J. Biol. Chem. 2007;282:2203–2210. doi: 10.1074/jbc.M609349200. [DOI] [PubMed] [Google Scholar]
- 29.Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, Darnell RB. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 30.Dredge BK, Darnell RB. Nova regulates GABA(A) receptor gamma2 alternative splicing via a distal downstream UCAU-rich intronic splicing enhancer. Mol. Cell. Biol. 2003;23:4687–4700. doi: 10.1128/MCB.23.13.4687-4700.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 32.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 33.Akusjarvi G, Stevenin J. Remodelling of the host cell RNA splicing machinery during an adenovirus infection. Curr. Top. Microbiol. Immunol. 2003;272:253–286. doi: 10.1007/978-3-662-05597-7_9. [DOI] [PubMed] [Google Scholar]
- 34.Huang TS, Nilsson CE, Punga T, Akusjarvi G. Functional inactivation of the SR family of splicing factors during a vaccinia virus infection. EMBO Rep. 2002;3:1088–1093. doi: 10.1093/embo-reports/kvf217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barreau C, Paillard L, Mereau A, Osborne HB. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 36.Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou HL, Lou H. Repression of prespliceosome complex formation at two distinct steps by Fox-1/Fox-2 proteins. Mol. Cell. Biol. 2008;28:5507–5516. doi: 10.1128/MCB.00530-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou HL, Baraniak AP, Lou H. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Mol. Cell. Biol. 2007;27:830–841. doi: 10.1128/MCB.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ponthier JL, Schluepen C, Chen W, Lersch RA, Gee SL, Hou VC, Lo AJ, Short SA, Chasis JA, Winkelmann JC, et al. Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. J. Biol. Chem. 2006;281:12468–12474. doi: 10.1074/jbc.M511556200. [DOI] [PubMed] [Google Scholar]
- 40.Akamatsu W, Okano HJ, Osumi N, Inoue T, Nakamura S, Sakakibara S, Miura M, Matsuo N, Darnell RB, Okano H. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems. Proc. Natl Acad. Sci. USA. 1999;96:9885–9890. doi: 10.1073/pnas.96.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao FB, Carson CC, Levine T, Keene JD. Selection of a subset of mRNAs from combinatorial 3′ untranslated region libraries using neuronal RNA-binding protein Hel-N1. Proc. Natl Acad. Sci. USA. 1994;91:11207–11211. doi: 10.1073/pnas.91.23.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson KD, Morin MA, Beckel-Mitchener A, Mobarak CD, Neve RL, Furneaux HM, Burry R, Perrone-Bizzozero NI. Overexpression of HuD, but not of its truncated form HuD I + II, promotes GAP-43 gene expression and neurite outgrowth in PC12 cells in the absence of nerve growth factor. J. Neurochem. 2000;75:1103–1114. doi: 10.1046/j.1471-4159.2000.0751103.x. [DOI] [PubMed] [Google Scholar]
- 43.Anderson KD, Sengupta J, Morin M, Neve RL, Valenzuela CF, Perrone-Bizzozero NI. Overexpression of HuD accelerates neurite outgrowth and increases GAP-43 mRNA expression in cortical neurons and retinoic acid-induced embryonic stem cells in vitro. Exp. Neurol. 2001;168:250–258. doi: 10.1006/exnr.2000.7599. [DOI] [PubMed] [Google Scholar]
- 44.Kasashima K, Terashima K, Yamamoto K, Sakashita E, Sakamoto H. Cytoplasmic localization is required for the mammalian ELAV-like protein HuD to induce neuronal differentiation. Genes Cells. 1999;4:667–683. doi: 10.1046/j.1365-2443.1999.00292.x. [DOI] [PubMed] [Google Scholar]
- 45.Manley GT, Smitt PS, Dalmau J, Posner JB. Hu antigens: reactivity with Hu antibodies, tumor expression, and major immunogenic sites. Ann. Neurol. 1995;38:102–110. doi: 10.1002/ana.410380117. [DOI] [PubMed] [Google Scholar]
- 46.Dredge BK, Stefani G, Engelhard CC, Darnell RB. Nova autoregulation reveals dual functions in neuronal splicing. EMBO J. 2005;24:1608–1620. doi: 10.1038/sj.emboj.7600630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanchette M, Chabot B. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 1999;18:1939–1952. doi: 10.1093/emboj/18.7.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spellman R, Rideau A, Matlin A, Gooding C, Robinson F, McGlincy N, Grellscheid SN, Southby J, Wollerton M, Smith CW. Regulation of alternative splicing by PTB and associated factors. Biochem. Soc. Trans. 2005;33:457–460. doi: 10.1042/BST0330457. [DOI] [PubMed] [Google Scholar]