Abstract
Small, hydrophobic proteins whose synthesis is repressed by small RNAs (sRNAs), denoted type I toxin–antitoxin modules, were first discovered on plasmids where they regulate plasmid stability, but were subsequently found on a few bacterial chromosomes. We used exhaustive PSI-BLAST and TBLASTN searches across 774 bacterial genomes to identify homologs of known type I toxins. These searches substantially expanded the collection of predicted type I toxins, revealed homology of the Ldr and Fst toxins, and suggested that type I toxin–antitoxin loci are not spread by horizontal gene transfer. To discover novel type I toxin–antitoxin systems, we developed a set of search parameters based on characteristics of known loci including the presence of tandem repeats and clusters of charged and bulky amino acids at the C-termini of short proteins containing predicted transmembrane regions. We detected sRNAs for three predicted toxins from enterohemorrhagic Escherichia coli and Bacillus subtilis, and showed that two of the respective proteins indeed are toxic when overexpressed. We also demonstrated that the local free-energy minima of RNA folding can be used to detect the positions of the sRNA genes. Our results suggest that type I toxin–antitoxin modules are much more widely distributed among bacteria than previously appreciated.
INTRODUCTION
Plasmid maintenance in many bacteria is attributed to the presence of toxin–antitoxin loci on the plasmids. These loci consist of two genes: one encodes a stable toxic protein, and the second an unstable antitoxin. If the plasmid is lost from the cell upon division, the unstable antitoxin is degraded, and the stable toxin is able to kill the cell. This phenomena, referred to as ‘post-segregational killing’ or ‘plasmid addiction’ has been described for plasmids in both Gram negative and Gram positive bacteria. The toxin–antitoxin loci are categorized into two broad classes based on the type of antitoxin: the antitoxin of type I systems is a small RNA (sRNA) which base pairs with the toxin mRNA to prevent protein synthesis, whereas the antitoxin of the type II systems is a protein that binds to and inhibits the toxin protein. Generally, type I toxins are small (under 60 amino acids in length), highly hydrophobic proteins, while type II toxins are slightly larger (∼100 amino acids) and less hydrophobic. The best-studied type I toxin–antitoxin systems include the hok-sok locus of plasmid R1, and the par locus of plasmid pAD1 of Enterococcus faecalis (1,2).
Although the toxin–antitoxin loci were initially described on plasmids, recent studies have shown that many of these gene pairs are also present on bacterial chromosomes. The type II toxin–antitoxin systems, in which the antitoxin is a protein, have been documented in diverse bacteria with many genomes carrying dozens of distinct toxin–antitoxin pairs (3). The type II toxins have been shown to degrade RNA or inhibit cellular enzymes such as DNA gyrase (4,5). The physiological role(s) of the type II systems remains a subject of debate; proposed functions include stress survival, protection of the bacteria against foreign DNA, and stabilization of chromosomal regions (6,7).
Several studies have shown that type I toxin–antitoxin systems, in which the antitoxin is an sRNA, are also present on some bacterial chromosomes. The hok-sok locus from plasmid R1 is encoded in the genomes of several enteric bacteria (8,9). In some strains, the sequences of these loci have degenerated and appear to be non-functional whereas in other cases, the systems are intact. Similarly, the par locus from plasmid pAD1 is present on the chromosomes of E. faecalis, Lactobacillus casei and a Staphylococcus saprophyticus strain (10). Additional type I toxin–antitoxin loci were found serendipitously on bacterial chromosomes (1). These include the ldr-rdl, ibs-sib, tisB-istR-1 and shoB-ohsC loci of Escherichia coli and the txpA-ratA locus of Bacillus subtilis. Interestingly, for these loci, there was no reported homology to known plasmid sequences. However, as for the plasmid-encoded systems, overproduction of the corresponding protein leads to cell death, and this toxicity is repressed by an antisense sRNA regulator. The exact biochemical activities of the small, hydrophobic toxin proteins are not known, although similarity to phage holin proteins has been noted, and overexpression of the proteins is associated with membrane depolarization and increased membrane permeability (1). As for the chromosomally-encoded type II toxin–antitoxin loci, the physiological function(s) of the chromosomally-encoded type I toxin–antitoxin systems remains unclear.
As mentioned above, type II toxin–antitoxin loci are broadly distributed among diverse bacteria. We hypothesized that type I systems are also widespread. To test this, we sought to identify homologs of the known type I toxins. Our computational approach identified many more putative toxins than have been previously reported. We experimentally validated a homolog of the par locus encoded in the chromosome of Streptococcus pneumoniae, the first report of a type I toxin–antitoxin system in this pathogen.
In addition to documenting the distribution of known type I systems in bacteria, we sought to identify new type I loci. Given the hydrophobicity and short length of type I toxins, and the difficulties in predicting sRNAs computationally, we developed search parameters based upon the characteristics of the known type I toxin–antitoxin systems. For example, given that the ibs-sib and ldr-rdl loci of E. coli are duplicated in the same intergenic region, we hypothesized that a short open reading frame (ORF) encoding a protein with a putative transmembrane domain and repeated in tandem could be a component of a type I toxin–antitoxin system. We also searched for amino acid sequences containing specific features derived from the analysis of known toxins, such as polar C-terminal residues. Finally, because the known antitoxin sRNAs form complex secondary structures, we developed a computational approach based upon the RNA folding energy profile of a putative type I locus to identify the location of the antisense sRNAs. Through these multiple approaches, we identified three new type I toxin–antitoxin loci which were experimentally validated. Our searches greatly expand the number of type I toxin–antitoxin systems known to be encoded in bacterial genomes.
MATERIALS AND METHODS
Computational approaches
Sequence data
All analyzed sequences were from the non-redundant protein sequence database at the NCBI. For the analysis of completed genomes, the RefSeq v.30 database was used for obtaining genome sequences and annotation (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/).
Exhaustive PSI-BLAST and TBLASTN searches
Methodological problems associated with a similarity search for short proteins are well-recognized. We tried different sets of parameters to improve the recognition of type I toxins by PSI-BLAST (Supplementary Table S1). The best results were obtained with the following set of parameters: matrix = PAM70; word size = 2, gap cost = existence 9, extension 2; PSI-BLAST inclusion threshold = 1; expect threshold = 100; no low complexity filtering; no composition based statistics (11). Searches at the NCBI server were performed for each protein family of the known type I toxins. After each run, all hits were clustered (with a sequence identity cutoff of 95%), and one representative from each cluster was used as a query for a new PSI-BLAST search until no new sequences were detected. Hits below the PSI-BLAST inclusion threshold were carefully inspected after each iteration, and if some of these hits were to a protein from a species closely related to those already detected and had other characteristic features of type I toxins (a predicted membrane protein under 70 amino acids in length), the respective sequences were manually included into the profile for the next search iteration. We denote this procedure exhaustive PSI-BLAST (Figure 1A). A similar approach was carried out with the TBLASTN program for those type I toxins that are often missed by ORF prediction programs (Figure 1B). The only difference is that TBLASTN cannot be used in an iterative mode, so the coverage of the respective families could be improved only by using multiple queries. Accordingly, all non-identical sequences from each toxin family were used as queries for TBLASTN searches.
Figure 1.
Computational approaches used to identify and predict type I toxins. (A) Exhaustive PSI-BLAST to identify homologs of known toxins not among previously annotated protein sequences. (B) Exhaustive TBLASTN search to identify homologs of known type I toxins not among previous annotated ORFs. (C) Tandem repeat search to identify new type I toxins encoded in the same intergenic region. (D) Search for new type I toxins based upon characteristics of known toxins.
Search for tandem repeats
Previous studies have shown that type I toxin–antitoxin loci have a tendency to be tandemly duplicated in the genomes of some bacteria (12,13). Many of these regions did not have annotated ORFs corresponding to the toxin. A pipeline of in-house Perl scripts was developed to detect tandem genes encoding proteins with characteristics similar to those of type I toxins, including genes missed by ORF prediction programs. The procedure includes the following steps (Figure 1C). First, the intergenic regions were extended by 30 nt into each of the two adjacent coding regions to allow for the possibility that the start (more common) or stop codons of the adjacent ORFs were annotated erroneously. Second, each intergenic region was translated in all 6 possible frames; predicted proteins longer than 70 amino acids and shorter than 16 amino acids were excluded. Third, the remaining proteins were searched for potential transmembrane helices using two approaches. For proteins longer than 50 amino acids, the TMHMM prediction method was employed (14). Since membrane protein prediction programs often perform poorly for short proteins, we had to apply a simpler approach to detect potential membrane proteins shorter than 50 amino acids. For such proteins we required at least one stretch of 15 amino acids with a minimum 10 hydrophobic amino acids (I, V, L, F, C, M, A) as an approximation for a transmembrane region. All predicted proteins that did not have a membrane region predicted by either approach were discarded. Fourth, to exclude protein fragments or pseudogenes, each remaining predicted protein was searched against the corresponding proteome using BLASTP. Predicted proteins with a highly significant match to previously annotated, longer proteins (e-value <10 and no more than two gaps) were excluded. The remaining proteins were also searched against the genomic DNA sequence by the TBLASTN program to find evidence that they might be located in a region which is likely to be non-coding. Those that matched a translated fragment containing one or more stop codons were considered as non-coding and discarded. Finally, to detect repeated sequences, the remaining predicted proteins in each intergenic region were grouped using BLASTCLUST (50% amino acid identity; length coverage 0.7 for at least one ORF). Since the small type I toxins generally do not have a variable length and internal gaps within a family, we required that no more than two internal and no more than one C-terminal gaps occur in the alignment of the proteins within a cluster.
Identification of characteristic features of type I toxins
We combined previously observed features of type I toxins with the new features identified in this work (for the set of type I toxins identified by PSI-BLAST and TBLASTN) in order to detect putative new toxin loci (Figure 1D, Supplementary Table S2). First, we took into account the observation that type I toxins are small (generally ≤ 70 amino acids in length) and secondly are membrane proteins. Third, type I toxin genes are separated from their neighboring genes by relatively long intergenic regions. We analyzed the up- and down-stream regions of the type I toxin genes and calculated the mean value for the up- and down-stream distances for all families (Supplementary Table S2). Based on the results of this analysis, we set the following thresholds for the distance between the putative toxin and its flanking genes: >400 nt between the toxin and the gene upstream, and >250 nt between the toxin and the gene downstream (Figure 1D). Finally, we noticed that many type I proteins have clusters of charged or bulky amino acids at their C-terminus. Therefore, in the selected genome set, we computed the absolute frequencies (number of occurrences) of non-hydrophobic amino acids within the 10 C-terminal amino acids for the combined set of all type I toxins identified by PSI-BLAST and TBLASTN in those genomes (Supplementary Table S2). We used these absolute frequencies to calculate a score for the 10 C-terminal amino acids for a protein. This was done by assigning a corresponding value of absolute frequency from the above estimate to each non-hydrophobic amino acid and calculating the sum of all such values.
Multiple sequence alignment and phylogenetic analysis methods
Multiple alignments of protein sequences were constructed using the MUSCLE program (15). Maximum likelihood (ML) phylogenetic trees were constructed from an alignment by using the MOLPHY program (16) with the JTT substitution matrix to perform local rearrangement of an original Fitch tree (17). The MOLPHY program was also used to compute RELL bootstrap values. Prediction of transmembrane helices was performed using TMHMM program (14) implemented in the web server (http://www.cbs.dtu.dk/services/TMHMM-2.0/).
Prediction of RNA secondary structure
Sequences of predicted and experimentally detected antisense sRNAs, random sequences from the same genomes and di-shuffled sequences were computationally folded, and the free energy of the most stable secondary structure was calculated using Afold and Mfold, as described previously (18,19). Energy minimization was performed by a dynamic programming method that employs nearest neighbor parameters to evaluate free energy and finds the secondary structures with the minimum free energy by summing up the contributions from stacking, loop length, and other structural features, using improved thermodynamic parameters (20–22). The sequence fold variant with the lowest secondary-structure energy was used in our analysis. The P-values for randomizations were calculated using paired t-tests (18). Results were presented as the free-energy profiles along the nucleotide sequences of interest with window lengths corresponding to the lengths of the antisense sRNAs. Starts and lengths of predicted antisense sRNAs were defined as the local minima of estimated free-energy profiles in the vicinity of predicted ORFs, taking into account the characteristic features (location and length) of known type I toxin families. The dinucleotide randomization procedure randomly shuffled all dinucleotides, retaining nucleotide composition of native RNAs (18,23).
Molecular approaches
Bacterial strains and plasmids
The strains and plasmids utilized in this study are listed in Supplementary Table S3, and the sequences of all oligonucleotides are given in Supplementary Table S4.
Growth conditions
E. coli strains were routinely grown in Luria–Burtani (LB) medium (10 g tryptone, 5 g yeast extract and 10 g NaCl per liter) or M9 minimal glucose medium (1 mM MgSO4, 0.1 mM CaCl, 1 µg/ml thiamine and 0.2% glucose) at 37°C with shaking. Arabinose was added as indicated to a final concentration 0.2%. Bacillus subtilis strains were grown in LB at 37°C with shaking. IPTG was supplemented to a 1 mM final concentration as indicated. Enterococcus faecalis OG1RF was grown in BHI medium (Difco) at 37°C. Streptococcus pneumoniae R6 was grown in BHI at 37°C in an atmosphere containing 5% (vol/vol) CO2/95% air. Antibiotics were added as needed at the following concentrations: 25 µg/ml chloramphenicol, 100 µg/ml spectinomycin, 100 µg/ml ampicillin.
RNA extraction
For E. coli, total RNA was harvested from cells grown in LB or M9 + 0.2% glucose media harvested at OD600 ≈ 0.4 and from overnight cultures (OD600 ≈ 5.0 in LB; OD600 ≈ 2.2 in M9) by the method of hot acid phenol as previously described (12). For B. subtilis, S. pneumoniae and E. faecalis strains, RNA was isolated as described (24) with some modifications. Briefly, 12-ml aliquots of culture were harvested by centrifugation at 4°C at OD600 ≈ 0.3, 1.0, 1.5 for E. faecalis and S. pneumoniae, and at OD600 ≈ 0.3, 2.0, 3.5 for B. subtilis ssp. subtilis str. 168 and B. subtilis PY79. Pellets were resuspended in 600 µl of Solution GP (50 mM Tris–HCl, 10 mM EDTA, 1% SDS, 30 mM sodium acetate), and transferred to tubes containing 0.5 g sterile glass beads (average diameter ≤106 µm; Sigma) and 650 µl of acid phenol:chloroform. The mixture was bead beated twice for 45 s at 4°C. The samples were separated by centrifugation, and the aqueous layer was transferred to tubes containing 500 µl of acid phenol: chloroform, and incubated at 65°C for 10 min. The supernatant was extracted two more times with phenol: chloroform, and once with chloroform. RNA was then ethanol precipitated, and resuspended in RNase-free water.
Northern analysis
For all antitoxin sRNAs, total RNA (10 µg) was separated on a denaturing 8% polyacrylamide–8 M urea gel. For detection of the z3289/z3290 mRNAs, total RNA (10 µg) was separated on a denaturing 6% polyacrylamide–8 M urea gel. RNA was then transferred to a Zeta-Probe Genomic GT membrane (Bio-Rad). The membranes were incubated with specific oligonucleotide probes labeled with 32P by T4 polynucleotide kinase and washed as previously described (25).
Primer extension analysis
Total RNA (5 µg) was used for primer extension analysis as previously described, and cDNA products were separated on a denaturing 8% polyacrylamide–8 M urea gel (25). Gene specific primers are found in Supplementary Table S4.
Overproduction of toxic proteins
For the toxicity studies in E. coli MG1655, potential toxins were cloned behind the PBAD promoter of the pAZ3 vector (26). As the ends of the potential toxin mRNA were unknown, a region containing ∼50 nt upstream of the predicted ribosome binding site and 100 nt downstream of the stop codon was amplified from genomic DNA, digested with EcoRI and HindIII, and cloned into the corresponding sites of pAZ3. For yhzE-2, the amplified fragment and pAZ3 were digested with EcoRI and XbaI.
To overproduce the toxins in B. subtilis PY79, the same regions were amplified from genomic DNA, digested with NheI and SphI, and cloned behind the Plac promoter of pDR111 (27). The resulting plasmids were then used for recombination into the amyE locus of B. subtilis PY79. Integration was confirmed by PCR and sequencing.
RESULTS
Identification of additional members of previously-characterized type I toxin–antitoxin families
Several studies have used sequence similarity searches, and in particular TBLASTN with default parameters, to identify chromosomally-encoded type I toxins (9,10). However, given the short lengths of these proteins and the strict parameters of such similarity searches, we suspected that a substantial fraction of homologs might have been missed in these studies. Thus, we performed a comprehensive analysis using customized, exhaustive PSI-BLAST and TBLASTN searches for 774 complete bacterial genomes (Figure 1A and B, and ‘Materials and Methods’ section). The results, presented in full in Supplementary Tables S5 and S6, and as a condensed list in Table 1 (for multiple alignments, see Supplementary Figure S1), substantially expand the number of detected type I toxin homologs, especially when compared with results that would be obtained if default BLAST parameters were used (Supplementary Table S1).
Table 1.
Type I toxins in selected completely sequenced genomes
| Species | Taxonomy | Total number of proteins | Number of different families | Total count | Ibs | Ldr/Fst | TxpA | Hok | TisB | ShoB | EHEC | YhzE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacillus halodurans C-125 | Bacilli, Bacillales, Bacillaceae | 4066 | 2 | 10 | 1 | 9 | ||||||
| Bacillus pumilus SAFR-032 | Bacilli, Bacillales, Bacillaceae | 3681 | 2 | 7 | 1 | 6 | ||||||
| Bacillus subtilis ssp. subtilis str. 168 | Bacilli, Bacillales, Bacillaceae | 4105 | 2 | 11 | 2 | 9 | ||||||
| Listeria monocytogenes str. 4b F2365 | Bacilli, Bacillales, Listeriaceae | 2821 | 1 | 1 | 1 | |||||||
| Listeria welshimeri serovar 6b str. SLCC5334 | Bacilli, Bacillales, Listeriaceae | 2774 | 1 | 2 | 2 | |||||||
| Staphylococcus aureus ssp. aureus NCTC 8325 | Bacilli, Bacillales, Staphylococcaceae | 2892 | 2 | 5 | 3 | 2 | ||||||
| Staphylococcus epidermidis RP62A | Bacilli, Bacillales, Staphylococcaceae | 2526 | 1 | 5 | 5 | |||||||
| Staphylococcus haemolyticus JCSC1435 | Bacilli, Bacillales, Staphylococcaceae | 2676 | 1 | 5 | 5 | |||||||
| Staphylococcus saprophyticus ssp. saprophyticus ATCC 15305 | Bacilli, Bacillales, Staphylococcaceae | 2514 | 2 | 4 | 3 | 1 | ||||||
| Enterococcus faecalis V583 | Bacilli, Lactobacillales, Enterococcaceae | 3265 | 2 | 7 | 1 | 6 | ||||||
| Lactobacillus casei ATCC 334 | Bacilli, Lactobacillales, Lactobacillaceae | 3044 | 2 | 6 | 2 | 4 | ||||||
| Leuconostoc citreum KM20 | Bacilli, Lactobacillales, Leuconostocaceae | 1820 | 1 | 2 | 2 | |||||||
| Leuconostoc mesenteroides ssp. mesenteroides ATCC 8293 | Bacilli, Lactobacillales, Leuconostocaceae | 2005 | 1 | 2 | 2 | |||||||
| Streptococcus pyogenes MGAS10270 | Bacilli, Lactobacillales, Streptococcaceae | 1986 | 1 | 1 | 1 | |||||||
| Streptococcus pneumoniae CGSP14 | Bacilli, Lactobacillales, Streptococcaceae | 2206 | 1 | 3 | 3 | |||||||
| Streptococcus suis 05ZYH33 | Bacilli, Lactobacillales, Streptococcaceae | 2186 | 1 | 1 | 1 | |||||||
| Streptococcus agalactiae 2603V/R | Bacilli, Lactobacillales, Streptococcaceae | 2124 | 1 | 1 | 1 | |||||||
| Streptococcus thermophilus CNRZ1066 | Bacilli, Lactobacillales, Streptococcaceae | 1915 | 1 | 1 | 1 | |||||||
| Aeromonas salmonicida ssp. salmonicida A449 | Gammaproteobacteria, Aeromonadales | 4437 | 1 | 2 | 2 | |||||||
| Alteromonas macleodii ‘Deep ecotype’ | Gammaproteobacteria, Alteromonadales | 4072 | 1 | 1 | 1 | |||||||
| Escherichia coli O157:H7 EDL933 | Gammaproteobacteria, Enterobacteriales | 5411 | 6 | 26 | 6 | 4 | 12 | 1 | 1 | 2 | ||
| Salmonella enterica ssp. enterica serovar Paratyphi B str. SPB7 | Gammaproteobacteria, Enterobacteriales | 5592 | 3 | 7 | 2 | 4 | 1 | |||||
| Shigella flexneri 2a str. 301 | Gammaproteobacteria, Enterobacteriales | 4440 | 4 | 11 | 4 | 5 | 1 | 1 | ||||
| Klebsiella pneumoniae 342 | Gammaproteobacteria, Enterobacteriales | 5777 | 2 | 4 | 3 | 1 | ||||||
| Shigella boydii CDC 3083-94 | Gammaproteobacteria, Enterobacteriales | 4557 | 5 | 20 | 7 | 4 | 7 | 1 | 1 | |||
| Citrobacter koseri ATCC BAA-895 | Gammaproteobacteria, Enterobacteriales | 5008 | 2 | 2 | 1 | 1 | ||||||
| Enterobacter sp. 638 | Gammaproteobacteria, Enterobacteriales | 4240 | 2 | 3 | 2 | 1 | ||||||
| Enterobacter sakazakii ATCC BAA-894 | Gammaproteobacteria, Enterobacteriales | 4420 | 2 | 3 | 2 | 1 | ||||||
| Escherichia fergusonii ATCC 35469 | Gammaproteobacteria, Enterobacteriales | 4269 | 6 | 18 | 7 | 3 | 4 | 1 | 1 | 2 | ||
| Photorhabdus luminescens ssp. laumondii TTO1 | Gammaproteobacteria, Enterobacteriales | 4683 | 1 | 1 | 1 | |||||||
| Proteus mirabilis HI4320 | Gammaproteobacteria, Enterobacteriales | 3662 | 1 | 1 | 1 | |||||||
| Salmonella enterica ssp. enterica serovar Typhi str. CT18 | Gammaproteobacteria, Enterobacteriales | 4758 | 4 | 8 | 2 | 2 | 3 | 1 | ||||
| Salmonella typhimurium LT2 | Gammaproteobacteria, Enterobacteriales | 4527 | 3 | 5 | 2 | 2 | 1 | |||||
| Serratia proteamaculans 568 | Gammaproteobacteria, Enterobacteriales | 4942 | 1 | 4 | 4 | |||||||
| Shigella dysenteriae Sd197 | Gammaproteobacteria, Enterobacteriales | 4503 | 5 | 10 | 3 | 2 | 3 | 1 | 1 | |||
| Shigella sonnei Ss046 | Gammaproteobacteria, Enterobacteriales | 4471 | 5 | 22 | 7 | 10 | 3 | 1 | 1 | |||
| Haemophilus influenzae 86-028NP | Gammaproteobacteria, Pasteurellales | 1792 | 1 | 1 | 1 | |||||||
| Haemophilus somnus 2336 | Gammaproteobacteria, Pasteurellales | 1980 | 1 | 7 | 7 | |||||||
| Vibrio vulnificus CMCP6 | Gammaproteobacteria, Vibrionales | 4472 | 1 | 1 | 1 |
Some families, such as the Hok (also denoted Gef), TxpA, Ldr and Fst families, were well represented in protein databases; the best-annotated group is the Hok family in which 72% are correctly named proteins. By contrast, others, such as the Ibs, TisB and ShoB families, were often missed by ORF-calling programs. The majority of the putative type I toxins that we identified with this approach are currently annotated as ‘hypothetical proteins’ or are unannotated.
To date, type I toxin–antitoxin loci have been experimentally characterized only in a few lineages of Enteroproteobacteria (Enterobacteria and Vibrionales) and Firmicutes (Bacillus and Enterococcus genus) (1,2). Our searches failed to detect any homologs of the known type I toxins outside these taxa; however, we identified previously unnoticed representatives of these families in many additional lineages of Enteroproteobacteria and Firmicutes (Supplementary Tables S5 and S6). For example, Fst-like sequences were detected in several Listeriaceae, Staphylococcaceae and Clostridiales species, and TxpA-like sequences were detected in Lactobacillales, Staphylococcaceae and Clostridiales species (Supplementary Table S5). The number of type I toxin loci varies greatly between different species and strains. So far the largest number was identified in E. coli O157:H7 str. Sakai. This genome carries 26 toxin–antitoxin loci of six distinct families including 14 Hok/Gef genes and seven Ibs genes. Taking into account our previous estimates of the number of type II toxin–antitoxin loci (3) (given in Supplementary Table S6) on a genome-wide scale we can conclude that type I toxin–antitoxin system are even more abundant in some genomes.
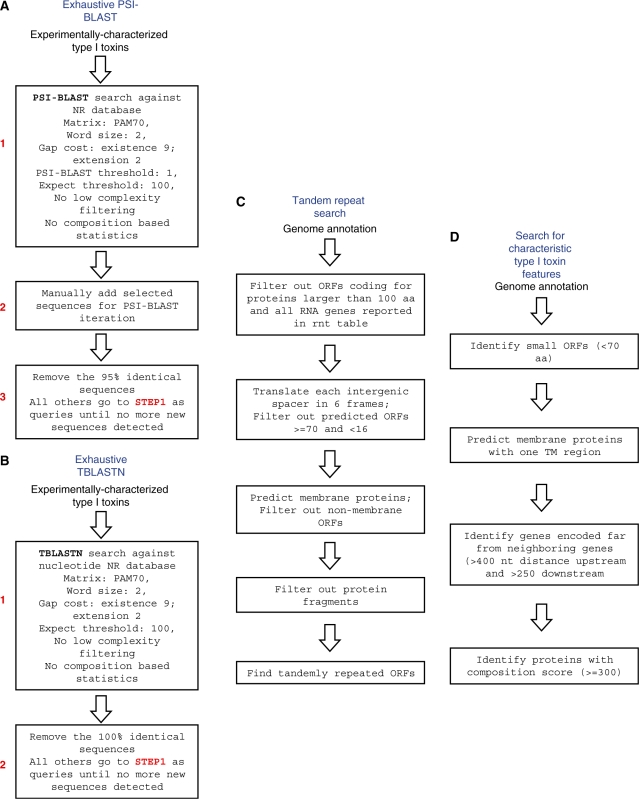
This approach also allows for the discovery of non-trivial links between families. Thus, using exhaustive PSI-BLAST, we detected a previously unnoticed connection between the Ldr and Fst families. The multiple amino acid sequence alignment shows considerable conservation between the two families including an apparent superfamily signature, a highly conserved tryptophan after a predicted transmembrane helix followed by a cluster of charged amino acids (Supplementary Figure S1A). This finding implies that the two families are probably homologous. The Ldr and Fst toxins are widely distributed across both Firmicutes and Enterobacteria. Given that representatives of these families are found in potential vectors for horizontal gene transfer, such as phages and plasmids, we were interested in potential evidence of horizontal gene transfer, and reconstructed a phylogenetic tree of the Ldr/Fst sequences. Despite limitations in tree construction because the sequences are so short, we were surprised to find that the topology of the tree matched the taxonomy of the respective bacteria (Supplementary Figure S2). There was no evidence of recent horizontal gene transfer events between distantly related bacteria for this family of type I toxins. Instead, we infer that there was a duplication of the ancestral toxin–antitoxin locus in the common ancestor of enterobacteria (LdrD- and LdrB-group) and in at least two distinct clades in Staphylococcaceae. These duplications were followed by other lineage-specific duplication events and a few losses in some species.
We also constructed a tree for the Ibs family, for which duplications in several genomes of enterobacteria were detected as well (Supplementary Figure S3). The analysis of this tree revealed the same trends as those seen in the Ldr/Fst tree. At least three copies of the ibs gene could have been present in the common ancestor of Enterobacteriaceae, and two in the Pasteurellaceae and the Haemophilus clades each. Subsequent duplications occurred independently in Haemophilus somnus and Shigella boydii lineages. These observations suggest that duplications of the type I toxin–antitoxin loci are relatively stable in evolution, with the implication that either these loci are prone to duplication and subject to relaxed selection, as in the case of transposons, or that the duplications are functionally important, possibly for stress resistance (28,29), and accordingly are maintained by purifying selection.
Experimental validation of the predicted Fst homologs in S. pneumoniae
Two Fst homologs (referred to herein as Fst-A and Fst-B), predicted by the exhaustive PSI-BLAST searches, are encoded in tandem in 27 out of the 29 S. pneumoniae genomic sequences deposited in the Microbial genome database at NCBI. These genes were missed by ORF prediction programs used for genome annotation in several strains including S. pneumoniae R6 (Figure 2A). The ORFs in S. pneumoniae R6 are flanked by fcsR, which encodes an annotated regulator of the fucose operon, and adcA, an ABC-transporter. We selected one of these ORFs, Fst-B (genomic coordinates 1 965 747–1 965 842), to test whether an antisense sRNA is expressed from the same locus and whether the product of the ORF is indeed toxic.
Figure 2.
(A) Multiple alignment of selected representatives of the Ldr/Fst family. Proteins studied in this work are denoted by asterisk, experimentally characterized type I toxins are shown in bold; the predicted transmembrane region is shaded; conserved small amino acids are colored blue; the conserved tryptophan is colored magenta; charged amino acids (RKDE) are colored red. The consensus was built using CONSENSUS program (http://coot.embl.de/Alignment//consensus.html) for a larger set of Ldr/Fst proteins (see Supplementary Figure S1A). The sequences are denoted by both the abbreviated species name and the GI number or the coordinates in the corresponding genome in parentheses. Species abbreviations: SP, Streptococcus pneumoniae R6; CB, Clostridium bolteae; EC, Escherichia coli K-12 substr. MG1655; SB, Shigella boydii CDC 3083-94; SE, Salmonella enterica arizonae z4z23; ECO, Escherichia coli O127:H6 str. E2348/69; SA, Staphylococcus aureus ssp. aureus Mu50; LG, Lactobacillus gasseri MV-22; EF, Enterococcus faecalis plasmid pAD1. (B) Northern blot showing expression of an sRNA antisense to S. pneumoniae fst-B-homolog. Total RNA (10 µg) isolated from S. pneumoniae R6 cells grown to OD600 ≈ 0.3 (E), OD600 ≈ 1.0 (L) and OD600 ≈ 1.5 (S) in BHI medium was loaded in each lane. (C) Overproduction of the S. pneumoniae Fst-B homolog in E. coli. MG1655 harboring pAZ3-fst-B was grown in LB medium to OD600 ≈ 0.3. The culture was split (indicated by the arrow); half was left untreated (blue) while arabinose (0.2% final concentration) was added to the other half (red). Cell dilutions were plated 0 (T0) and 60 (T60) min following arabinose induction.
If the S. pneumoniae protein is functionally analogous to Fst, there should be a corresponding antisense sRNA regulator. The Fst protein is the toxin component of the par locus of the plasmid pAD1. The organization of the par locus has been well-characterized, and the antisense sRNA regulator (RNA II) overlaps the 3′-end of the mRNA encoding the toxic protein (10,30,31). Total RNA was isolated from S. pneumoniae R6 and used for northern analysis. A strong signal corresponding to an RNA species of ∼65–75 nt was detected using an end-labeled oligonucleotide complementary to the 3′ end of fst-B (Figure 2B). This signal is in agreement with the previously characterized size and location of the antisense sRNA from pADI, as well as RNA II expressed from copies of the par locus encoded in the chromosomes of other bacteria (10).
To test the toxicity of the protein, fst-B from S. pneumoniae R6 was cloned on a plasmid behind an arabinose-inducible promoter (PBAD) and overproduced in E. coli MG1655. As shown in Figure 2C, induction of the protein halted cell growth, and there was a significant decrease in colony forming units over time, confirming that high levels of this protein are indeed toxic.
Finding new type I toxin–antitoxins: tandem repeats
Among the previously identified chromosomally encoded type I toxin–antitoxin systems, the ibs-sib and ldr-rdl loci are repeated multiple times in the same intergenic region (12,13). Therefore we developed a computational procedure to identify tandem repeats encoding potential type I toxins in intergenic regions of bacterial genomes. We then examined a selected set of sequenced genomes in order to identify new type I toxin families (see Figure 1C, and ‘Materials and Methods’ section). The complete search results for Proteobacteria and Firmicutes are given in Supplementary Table S7.
This approach reproduced some findings obtained in our exhaustive BLAST search, including the S. pneumoniae Fst toxins described above. The search also led to the identification of a duplication of the apparent Ibs homologs in the genome of Helicobacter pylori that were missed by TBLASTN but recently were identified experimentally (32). We were particularly interested in further analyzing E. coli and B. subtilis toxin–antitoxin candidates predicted by this approach.
Experimental analysis of new candidate type I toxins from E. coli strain O157:H7
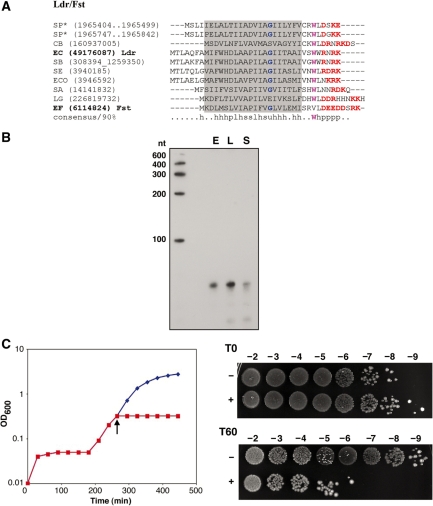
One repeat family was observed between yehI and yehL in the enterohemmoragic E. coli strain O157:H7 EDL933. Two genes, z3289 and z3290, encoding proteins 29 amino acids in length each, are encoded in tandem and share extensive sequence similarity that extends beyond the coding region (Figure 3A). The same loci are present in most E. coli and Shigella strains, and in Escherichia fergusonii ATCC 35469, either as tandem repeats or as single genes (Supplementary Table S7), but are not found in the laboratory strain E. coli MG1655. In fact, the length of the entire yehI-yehL intergenic region in MG1655 is 0.3 kb compared to 1.2 kb in O157:H7 EDL933.
Figure 3.
(A) Genomic arrangement of EHEC Z3289 and Z3290. The ORFs are indicated by the black regions of the leftward arrows and the regions of complementarity are indicated by the white boxes. The sequences capable of base pairing are shown below the gene arrangement. (B) Multiple alignment of selected representatives of EHEC family. Most designations are the same as in the Figure 2A. The predicted transmembrane regions is shaded (predicted for E. coli O157:H7 EDL933 proteins and extended for other sequences): small amino acids are colored blue; charged amino acids (RKDE) are colored red. Species abbreviations (strains are also indicated for E. coli species): EC, E. coli; SB, Shigella boydii CDC 3083-94; SF, Shigella flexneri 2a str. 2457T. (C) Expression of the antitoxin RNAs for Z3289 (sRNA-1) and Z3290 (sRNA-2). Total RNA (10 µg) isolated from E. coli O157:H7 EDL933 cells grown to OD600 ≈ 0.4 (E) and OD600 ≈ 5.0 (overnight, S) in LB medium and from cells grown to OD600 ≈ 0.4 (E) and OD600 ≈ 2.2 (overnight, S) in M9 media supplemented with 0.2% glucose was loaded in each lane. (D) Overproduction of Z3290 in MG1655. MG1655 harboring pAZ3-z3290 was grown in LB medium to OD600 ≈ 0.3. The culture was split (indicated by the arrow); half was left untreated (blue) while arabinose (0.2% final concentration) was added to the other half (red). Cell dilutions were plated 0 (T0) and 60 (T60) min following arabinose induction.
The antitoxin RNAs described to date are encoded directly opposite the coding sequence of the toxin, opposite the 5′ untranslated region (5′ UTR), or opposite the 3′ UTR of the toxin mRNA, or even divergent to the toxin gene but with long stretches of complementarity to the toxin mRNA (1). We were unable to detect antisense sRNAs using oligonucleotide probes corresponding to the coding sequence, 5′ or 3′ UTR of z3289 and z3290 (data not shown). To test for the presence of an sRNA in the region around z3289 and z3290, we carried out northern analysis using three riboprobes, which together would span a 1 kb segment encompassing the two small ORFs. We observed a strong band of ∼80 nt with the probe that spanned the intergenic region between the two genes (data not shown). To further refine the position of the putative sRNA, we calculated the predicted free-energy profile of the yehI-yehL intergenic region (see below). This analysis revealed two local minima of predicted free-energy, corresponding to regions of complex secondary structure, 240–300 nt upstream of z3289 and z3290. Upon further examination of these regions, we identified potential terminators and promoter sequences (Supplementary Figure 4A). Using oligonucleotides complementary to these predicted sRNAs, we detected two transcripts of ∼85 nt in length each. Interestingly, these transcripts were abundant during the exponential phase in both rich and minimal media, but decreased during stationary phase (Figure 3B).
As these sRNA genes were encoded divergent from the toxin genes, we were interested in whether they had the potential to base pair with the toxin mRNAs. There is perfect complementarity between the sRNA (sRNA-1) encoded divergent to z3289 and the sequence 72–92 nt upstream of the start codon of the toxin (Figure 3A). Similar complementarity is also observed between z3290 and the sRNA (sRNA-2) encoded divergent from this gene (Figure 3A). We carried out primer extension analysis to map the transcription start sites of the z3289 and z3290 mRNAs and the newly discovered sRNAs (Supplementary Figure S4). The results indicate that both toxin mRNAs contain long 5′ UTRs (180 nt), similar to what has been reported with other toxins (1). The gene orientations and base pairing potentials are very reminiscent of the tisB-istR and shoB-ohsC toxin loci. In these pairs, the sRNA is encoded divergent, and distant from the toxin, but has the potential for extended base pairing with the 5′ UTR of the toxin mRNA.
The putative toxin genes were cloned with their native ribosome binding sites, behind the PBAD promoter on a multicopy plasmid to measure toxicity. Overproduction of both small proteins (Figure 3C and data not shown) in the laboratory strain E. coli MG1655 led to cell stasis and a mild decrease in colony forming units, indicating that the proteins are toxic at high levels.
Experimental analysis of new candidate type I toxins from B. subtilis
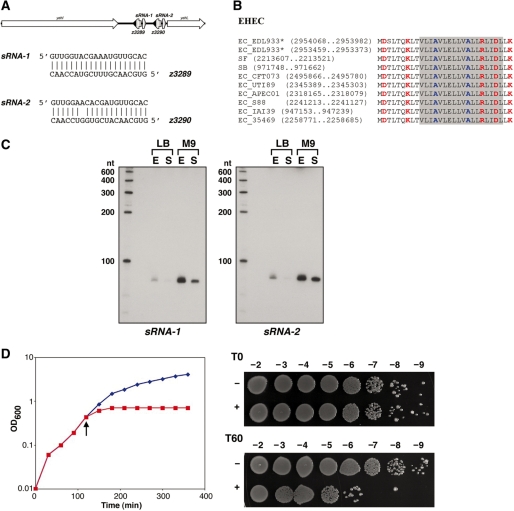
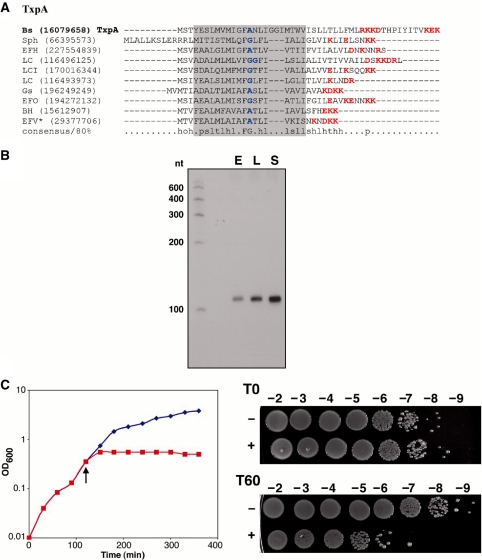
A separate duplication was identified in B. subtilis ssp. subtilis str. 168 genome. The duplicated gene encodes a 28 amino acid hydrophobic protein that is conserved across multiple species of Firmicutes, but it is not similar to any of the known type I toxins (Figure 4A and Supplementary Figure S1C). One of these duplicated genes has been annotated as yhzE; herein, we will refer to the annotated gene as yhzE-1 and the second copy in the same intergenic region as yhzE-2. The genes encoding proteins of this family are highly abundant in Firmicutes. For instance, the Bacillus subtilis ssp. subtilis str. 168 genome contains eight genes for these proteins (Supplementary Figure S1C and Figure 4A). Analysis of an alignment of this family reveals a distinct feature: both the N- and C-termini are highly variable in length but are rich in glycines and aromatic residues (Supplementary Figure S1C). Genes encoding these proteins are tandemly duplicated in the genomes of several other bacteria and are also present in several phages and plasmids. Combined with the sequence features of this proteins, such a distribution makes them possible type I toxin candidates.
Figure 4.
(A) Multiple alignment of selected representatives of YhzE family. Most designations are the same as in the Figure 2A. The consensus was built using CONSENSUS program for a larger set of YhzE family proteins (see Supplementary Figure S1C). The predicted transmembrane regions is shaded (predicted for B. subtilis YhzE protein and extended for other sequences); small amino acids are colored blue; aromatic residues are colored magenta. Species abbreviations: Bs, B. subtilis str. 168; Bph, Bacillus phage SPBc2; Gsp, Geobacillus sp. G11MC16; BH, B. halodurans C-125; BA, B. anthracis str. Ames; Psp, Paenibacillus sp. JDR-2; BP, B. pumilus SAFR-032. (B) Expression of an sRNA antisense to yhzE-2. Total RNA (10 µg) isolated from B. subtilis PY79 cells grown to OD600 ≈ 0.3 (E), OD600 ≈ 2.0 (L) and OD600 ≈ 3.5 (S) in LB medium was loaded in each lane. (C) Overproduction of YhzE-2 and TxpA in B. subtilis PY79. YhzE-2 (graph on the left) or TxpA (right) under the control of the Plac promoter was integrated into the amyE locus of PY79. The cultures were grown in LB medium to OD600 ≈ 0.3. The cultures were split; (indicated by the arrow); half was left untreated (blue) while IPTG (1 mM final concentration) was added to the other half (red).
To test whether this region has features of a type I toxin–antitoxin locus, we isolated RNA from B. subtilis PY79 and carried out northern blot analysis. As the B. subtilis ratA antitoxin RNA base pairs at the 3′-end of the toxin mRNA, we used an oligonucleotide probe that overlaps the 3′-end of the yhzE-2 ORF. A strong signal, of ∼110–120 nt in length was detected throughout growth in rich media using this probe (Figure 4B).
We initially overexpressed the YhzE-2 protein from a PBAD plasmid in E. coli MG1655 but observed no effects on growth (data not shown). Given that the protein is native to B. subtilis and not E. coli, we next measured its toxicity in B. subtilis. The yhzE-2 gene was cloned behind the Plac promoter of the plasmid pDR111, and the construct was integrated into the amyE gene of B. subtilis PY79 (27). As a control, we similarly examined the toxicity of TxpA, a known type I toxin found in B. subtilis (33). TxpA was highly toxic to B. subtilis whereas there were no obvious growth defects upon overproduction of YhzE-2 (Figure 4C). The lack of YhzE-2 mediated toxicity could be due to insufficient levels of protein production, possibly because of repression by endogenous antisense sRNAs expressed from the multiple paralogous copies of the locus. Alternatively, the protein may not function as a toxin, even at high levels.
Finding new type I toxin–antitoxins: characteristic protein features
In addition to being encoded in tandem repeats, there are other characteristics shared by many type I protein toxins. The described type I toxins are under 70 amino acids in length, contain a transmembrane region and a small C-terminal region rich in polar or aromatic residues. The toxin–antitoxin loci also are often encoded distant from their flanking genes. We combined these observations into a set of search parameters (see ‘Materials and Methods’ section) taking into account data obtained by the analysis of all known and new toxins described here (Figure 1D and Supplementary Table S2). Briefly, we identified all proteins under 70 amino acids in length that were predicted to contain at least one transmembrane region. We then selected those ORFs that were separated by at least 400 nt from the upstream flanking gene and by at least 250 nt from the downstream gene. From this set of proteins, we selected those that contained a C-terminus rich in polar or aromatic residues. Results for the selected genomes are presented in Supplementary Table S8. Using these parameters, we identified, among other putative novel type I toxins, the 27 amino-acid protein BH0344 from Bacillus halodurans C-125, which has homologs in several L. monocytogenes strains and in E. faecalis V583 (protein EF3263), as well as YonT encoded in the B. subtilis ssp. subtilis str. 168 genome. The EF3263 and YonT proteins were chosen for further analysis.
Experimental validation of a candidate type I toxin in E. faecalis V583
The use of exhaustive PSI-BLAST and TBLASTN searches with the E. faecalis EF3263 protein as a starting query led us to link this group of sequences with the TxpA family (Supplementary Figure S1D, Figure 5A), a connection we did not make with our preceding analysis. This finding demonstrates that BLAST searches are highly sensitive to query sequences and database content (which had changed since we obtained the first results for the TxpA family described above). Accordingly, it seems likely that we are still underestimating the number of type I toxin genes even for known families and that the development of new, customized computational approaches, some of which are presented here, can help find homologs not identified by standard BLAST searches.
Figure 5.
(A) Multiple alignment of selected representatives of the TxpA family. The consensus was built using CONSENSUS program for a larger set of the TxpA family proteins (see Supplementary Figure S1D). Most designations are the same as in the Figure 2A. The predicted transmembrane regions is shaded (predicted for Enterococcus faecalis V583 protein and extended for other sequences). Species abbreviations: Bs, B. subtilis sub. subtilis str. 168; Sph, Staphylococcus phage 42E; EFH, E. faecalis HH22; LC, Lactobacillus casei ATCC 334; LCI, Leuconostoc citreum KM20; LC, Lactobacillus casei ATCC 334; Gs, Geobacillus sp. G11MC16; EFO, E. faecalis OG1RF; BH, B. halodurans C-125; EFV, E. faecalis V583. (B) Northern blot showing expression of an sRNA antisense to EF3263 in E. faecalis OG1RF. Total RNA (10 µg) isolated from E. faecalis OG1RF cells grown to OD600 ≈ 0.3 (E), OD600 ≈ 1.0 (L) and OD600 ≈ 1.5 (S) in BHI medium was loaded in each lane. (C) Overproduction of EF3263 in E. coli. MG1655 harboring pAZ3-ef3263 was grown in LB medium to OD600 ≈ 0.3. The culture was split (indicated by the arrow); half was left untreated (blue) while arabinose (0.2% final concentration) was added to the other half (red). Cell dilutions were plated 0 (T0) and 60 (T60) min following arabinose induction.
As EF3263 is distantly related to TxpA, we predicted that the organization of the EF3263 toxin–antitoxin gene pair would be similar to that of TxpA-RatA (33). We isolated RNA from E. faecalis OG1RF (which contains EF3263) grown in BHI. As shown in Figure 5B, a transcript of ∼110 nt in length was detected using a probe that would overlap the 3′ UTR of the EF3263 transcript. This RNA, although readily detected under all growth conditions examined here, appeared to accumulate as the cells entered stationary phase, suggesting an increase in either its transcription or stability under these conditions.
Toxicity of this protein was tested by overproduction in E. coli, as described above. Upon induction of the small protein, cell growth stopped and a mild decrease in colony forming units was observed (Figure 5C). These results show that EF3263 is toxic upon overexpression and support the hypothesis that EF3263 is a divergent member of the TxpA toxin family. Interestingly, overproduction of TxpA in E. coli had no effects on growth [(33) and data not shown], suggesting that, despite their relatedness, there are differences in the levels of the small proteins and/or the functions of TxpA and EF3263.
Experimental validation of a candidate type I toxin in B. subtilis
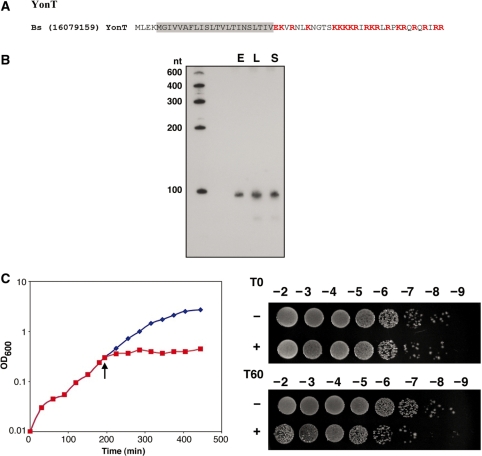
An additional protein identified using the search parameters derived from the characteristic features of type I toxins was YonT encoded in the genome of B. subtilis ssp. subtilis str. 168 (Figure 6A). Given that the yonT gene resides within the SPβ prophage of B. subtilis, it appeared to be a plausible toxin candidate.
Figure 6.
(A) Amino acid sequence of yonT gene product of Bacillus subtilis ssp. subtilis str. 168 (Bs). Charged amino acids (EKR) are colored red and the predicted transmembrane regions is shaded. (B) Expression of an sRNA antisense to yonT. Total RNA (10 µg) isolated from B. subtilis ssp. subtilis str. 168 cells grown to OD600 ≈ 0.3 (E), OD600 ≈ 2.0 (L) and OD600 ≈ 3.5 (S) in LB medium was loaded in each lane. A smaller band of ∼80 nt can be seen upon overexposure as the cells enter the stationary phase of growth. (C) Overproduction of YonT in E. coli. MG1655 harboring pAZ3-yonT was grown in LB medium to OD600 ≈ 0.3. The culture was split (indicated by the arrow); half was left untreated (blue) while arabinose (0.2% final concentration) was added to the other half (red). Cell dilutions were plated 0 (T0) and 60 (T60) min following arabinose induction.
Since the yonT region is absent in B. subtilis PY79, which has been cured of SPβ, we examined whether there is an sRNA encoded in the antisense strand of yonT in B. subtilis ssp. subtilis str. 168 (34). Northern analysis of RNA isolated from cells grown in rich media revealed the presence of a transcript, just under 100 nt in length, encoded opposite the 3′ end of yonT. Upon longer exposures, a smaller, ∼80 nt band also could be seen, and this transcript accumulates as the culture exits log phase and enters stationary phase (Figure 6B).
To examine the toxicity of YonT, the gene with its predicted ribosome binding site was cloned behind the PBAD promoter as described above. Induction of YonT led to a decrease in the growth rate of E. coli although this effect was milder than the effects of the other toxins examined in this study, with the exception of YhzE-2 (Figure 6C).
Computational prediction of regulatory antisense sRNAs using thermodynamic parameters of RNA folding
The known type I antitoxin RNAs are predicted to fold into complex secondary structures (1,2). Thus, we analyzed RNA secondary structures and RNA folding characteristics to determine whether it is possible to predict the location of antisense sRNA genes. Using computer algorithms to predict RNA folding and to estimate the free energy for optimal and suboptimal secondary structures (see ‘Materials and Methods’ section), we first created free-energy profiles for the previously characterized antitoxin RNA regions. We found that the transcriptional starts for all known antitoxin RNAs (IstR1, Sok, SibA, SibB, RdlD, RatA) are located in the local minima of predicted free-energy profiles (Supplementary Figure S5). Specifically, the differences in the local minima and the average free-energy levels in the thermodynamic profiles for known antitoxin RNAs compared to those calculated for di-shuffled sequences and random sequences of comparable lengths from the same genome were statistically significant (P < 0.001).
To validate this approach, we compared the distribution of free-energy values for predicted antitoxin RNA regions for known type I loci identified using BLAST (Supplementary Table S9) with those for random sequences of comparable lengths taken from elsewhere in the same genomes, and with randomly shuffled sequences with the same dinucleotide content as the RNA antitoxin sequences (‘Materials and Methods’ section). The starts and lengths of the predicted antitoxin RNA were defined based on the characteristic features (location and length) of known type I toxin families. Again the folding free energies for the predicted antisense sRNAs were substantially lower than those for the di-shuffled sequences (P = 9.2E−32; Supplementary Figure S6). Notably, mRNA folding energies for the random sequences were distributed differently, as compared to those of the antitoxin RNAs that contain numerous domains capable of folding into highly stable secondary structures. The results show that the predicted antisense sRNA regions generally have a propensity to form more stable secondary structures and possess lower free-energy values that the random genomic sets (P = 2.62E−12; Supplementary Figure S6).
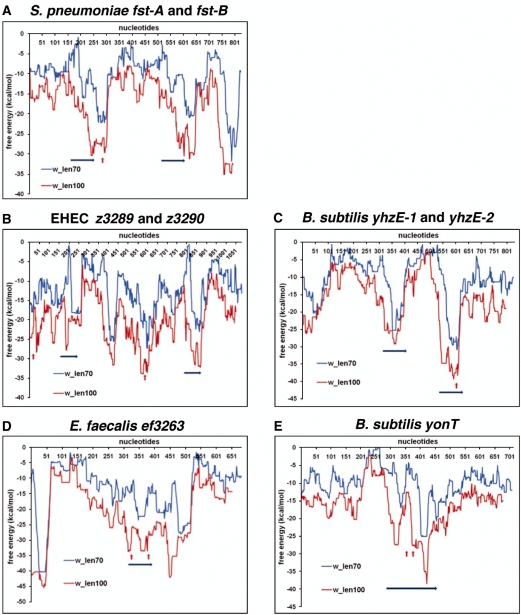
Using this approach, we predicted the locations of the genes encoding the antitoxin RNAs identified in E. coli O157:H7, B. subtilis and E. faecalis, and experimentally analyzed in this study (Figure 7). The predicted energy minima coincided perfectly with the sequences of the oligonucleotides used to detect the sRNAs by northern analysis. The analysis was particularly helpful for z3289 and z3290 of E. coli O157:H7, where antisense sRNAs were not detectable with oligonucleotide probes corresponding to the coding sequence, 5′ or 3′ UTR of the genes. For these toxins the locations of putative sRNA genes were predicted based on the analysis of the free-energy profile of the yehI-yehL intergenic region. This analysis revealed two local minima of free energy, which corresponded to the regions of complex secondary-structure upstream of z3289 and z3290 (Figure 7B) and were confirmed experimentally to express sRNAs.
Figure 7.
Prediction of antitoxin sRNAs using free-energy profiles for RNA local secondary structures. Free-energy profiles for RNA local secondary structures along nucleotide sequences in experimentally tested RNA antitoxin systems in S. pneumoniae (A), E. coli (B), B. subtilis (C and E) and E. faecalis (D). The lengths of the sliding window used for free-energy estimations (70 and 100 nt) corresponded to common lengths of the previously described sRNA antitoxins. Blue arrows show location of predicted ORFs. Red arrows show the positions of the oligonucleotides used to detect the antisense sRNAs. Other local free-energy minima correspond to annotated terminators or unrelated short ORFs. x-axis: nucleotide positions; y-axis: free energy of RNA folding.
Whenever possible, the lengths of sliding windows were chosen on the basis of the characteristic location and length of known type I toxins. For new type I toxin families, we performed a more extensive analysis with sliding windows of varying length. Some of the local free-energy minima observed outside of predicted ORFs corresponded to annotated transcription terminators or unrelated short ORFs and were excluded from consideration. Most of the remaining stable free-energy minima, readily detectable with different window lengths, were candidates for experimental evaluation. Our results support the observations that the antitoxins are encoded by highly structured RNAs, which justifies the use of this parameter to predict new type I loci.
DISCUSSION
Several recent studies have focused on the identification and characterization of the many type II toxin–antitoxin gene pairs in which both the toxin and antitoxin are proteins. These loci are broadly distributed across bacteria and archaea, and the numbers of loci vary extensively between species. In contrast, little is known about the distribution of type I toxin–antitoxin loci in which the antitoxin is an antisense sRNA. We thus set out to screen for homologs of known type I toxin–antitoxin pairs as well as to identify new loci.
Approaches to identify type I toxin–antitoxin systems
Prior to these studies, identification of homologs of known type I toxins relied solely upon TBLASTN and PSI-BLAST searches carried out using default parameters. These searches revealed few homologs (9,10), and consequently suggested that the distribution of these toxins was limited. However, using customized, exhaustive PSI-BLAST and TBLASTN searches, we predicted numerous type I toxin–antitoxin loci in a wide range of bacteria including homologs of the Fst toxins in S. pneumoniae, which we experimentally validated. The combined results from our searches greatly increased the number of known toxins across many different bacterial lineages (see Supplementary Tables S1, S5 and S6). The majority of the putative type I toxins that we identified with this approach are currently annotated as ‘hypothetical proteins’ or are not represented in protein databases at all (missed by gene prediction methods). For example, for the TxpA family, previously represented by the single, originally identified type I toxin from B. subtilis (33), we describe 118 representatives, of which only eight were annotated as holin-like toxins.
The development of computational approaches to identify novel type I toxin–antitoxin systems is challenging due to the short, hydrophobic character of the toxin proteins and the difficulty in predicting the antitoxin sRNAs. By examining common features of known toxins, we were able to establish a list of potential features to use in searches for new families. Here we described two computational approaches that led to discovery of new type I toxin families. One nucleotide-based approach identified tandemly repeated ORFs encoding potential type I toxins in intergenic regions. The second approach is based on the characteristics of known protein sequences of type I toxins and was aimed at searching the protein content of sequenced genomes. Both approaches identified candidates that could be further analyzed in silico and in vivo to test our predictions. We experimentally examined three new putative toxin–antitoxin loci: the Z3289/Z3290 family of EHEC and the YhzE and YonT proteins of B. subtilis. All three loci were confirmed to have associated sRNAs and the Z3289, Z3290 and YonT proteins were found to be toxic or partially toxic at high levels.
We additionally found that the results could be refined by applying RNA folding predictions to the potential type I loci. The characterized antisense sRNA antitoxins have extensive secondary structures and are located in regions that have very low predicted free-energies. By incorporating this observation, we were able to predict the location of the type I antitoxin RNA genes. This was especially useful in locating the sRNA regulators of the EHEC toxins. Given that these sRNAs are not encoded directly opposite the toxin genes, they would have been missed in our searches. Overall, the regions of predicted low free-energy corresponded well to the chromosomal location of the antitoxins (Figure 7).
The computational approaches described here certainly require further improvement. As a case in point, our exhaustive PSI-BLAST search failed to identify the divergent TxpA toxin in E. faecalis. Thus, even this approach underestimates the number of such loci for known families. In addition, our computational approaches based upon toxin characteristics produced a considerable number of apparent false positives. At present, there is no single, universal criterion to identify false-positives among the predictions. However, case-by-case analysis for features such as the presence of a conserved ribosome binding site, and start and stop codons allowed us to dismiss a considerable fraction of detected ORFs as inconsistent with a type I toxin function (see Supplementary Table S7). The novel type I toxin–antitoxin gene pairs reported in this work should be helpful in the further refinement of the computational parameters and methods described here.
It is also expected that new pairs of type I toxin–antitoxins will be identified in the transcriptomes of the many bacteria that are being studied by whole genome expression analysis with tiling arrays or deep sequencing. Indeed, homologs of the Ibs genes were detected by deep sequencing of H. pylori (32). Similarly, a deep sequencing study of Prochlorococcus, a marine cyanobacterium, revealed two distinct loci encoding overlapping RNAs, in which one gene in each pair is predicted to encode a short protein (35) although further studies are required to test the hypothesis that these are toxin–antitoxin pairs.
Overall, the computational part of this work pursued two major goals. The first was to achieve the maximum coverage of the known type I toxin families and evaluate the number of representatives of each family in sequenced genomes. The second goal was to develop computational approaches to predict new families of toxins that could not be identified with sequence similarity searches. The latter approach did not aim to find all representatives of putative new toxin families but rather to pinpoint a small number of plausible candidates for further case-by-case analysis using both computational and experimental techniques. Once a new family is confirmed as a true positive, similarity search methods are the best way to identify homologs of the respective proteins in genomes of interest.
Distribution and evolution of type I toxin–antitoxin systems
The distribution of the previously characterized type I toxin–antitoxin loci, as well as the new ones identified here, can vary greatly. For example, homologs of ShoB are found mainly in E. coli and Shigella, whereas Ldr/Fst homologs are detected in multiple Firmicutes and enteric bacteria (Supplementary Tables S5 and S6). Our analysis of the evolution of type I toxins unexpectedly showed that, unlike type II toxins, the type I toxin–antitoxin systems are not prone to horizontal gene transfer, but instead have evolved by lineage-specific duplication. The duplicated copies are stable in evolution suggesting a possible functional role of these loci in the respective organisms. Thus, it appears that ShoB emerged later in evolution as it is not present outside a group of closely related organisms. In contrast, the genes of the Ldr/Fst family were probably present in the ancestors of both Firmicutes and enterobacteria taxa and retained by many lineages after their divergence.
Interestingly, the sRNAs associated with the E. coli Ldr proteins are encoded opposite the 5′-ends of these genes, while the sRNAs associated with the proteins that are more Fst-like are encoded opposite the 3′-ends of these genes. This raises questions about the evolution of the toxin and the antisense sRNA; when and how did the sRNA arise? Related to this, there could be a difference in the distribution of the ‘traditional’ type I toxin–antitoxin loci where the sRNA is encoded opposite the toxin gene versus the families such as ShoB-OhsC, TisB-IstR-1, Z3289-sRNA-1 and Z3290-sRNA-2 pairs, where the sRNA is encoded divergent from the toxin gene, but possesses extensive base pairing potential. Thus far these loci have only been found in E. coli and closely related bacteria. However, predicting new families of this subset of type I toxin–antitoxin modules is difficult, and consequently it remains unknown whether other bacteria possess toxin–antitoxin loci with this divergent gene arrangement.
It is likely that still other permutations of type I toxin–antitoxin loci as well as combinations of type I and II toxin–antitoxin modules will be found. In E. coli, the SymR antisense sRNA represses the synthesis of the toxic SymE protein (26). Interestingly, although SymE functions like a type II toxin, it actually resembles type II antitoxins. A number of ‘orphan’ type II toxin genes lacking the adjacent antitoxin gene have been found in searches for type II loci (3); it is quite possible that the synthesis of these toxins is repressed by antisense sRNAs. In another recent study, an RNA which carries a striking repeated sequence and is encoded upstream of the ToxN protein of Erwinia carotovora was reported to act as an antitoxin by binding to the ToxN protein rather than blocking synthesis as an antisense sRNA (36).
Until now type I toxin–antitoxin gene pairs have only been experimentally characterized in Firmicutes and γ-Proteobacteria, and our searches were based upon what is known about these few examples. Thus, the apparent absence of known type I toxins in the genomes of bacteria and archaea other than Firmicutes and γ-Proteobacteria may reflect the limits of our methods to detect new families or highly diverged members of known families. As the methods for computational prediction of type I toxin–antitoxin pairs are refined and more transcriptome information is obtained and validated from other bacteria, the number of type I toxin–antitoxin families is likely to expand.
Function of type I toxins
Some features of the type I toxin proteins point to parallels with phage holins despite the absence of obvious sequence similarity. Both type I toxins and holins are predicted small membrane proteins with charged or aromatic terminal regions. Holin family proteins are extremely diverse but all appear to retain the same mechanism of action, namely, the formation of pores in bacterial membranes. It is plausible that type I toxins function through the same mechanism as holins (37); killing the cell by forming pores. However, similarities between type I toxins and other small hydrophobic proteins, such as peptides that affect ribosome stalling (38), suggest other potential modes of action. It is also quite possible that different families of type I toxin proteins have unique biological activities.
The toxic phenotype of many chromosomally encoded type I toxins has only been reported upon overproduction from a multicopy plasmid. There is very little evidence for toxicity when these proteins are natively expressed from the chromosome (1). Thus, as it is unlikely the levels of the toxin would reach amounts high enough to cause lethality, the main function of these chromosomally encoded proteins might not be to kill the cell (calling into question the use of the term ‘toxin’). In addition, although we detected an sRNA encoded antisense to B. subtilis yhzE-2, we were unable to demonstrate that the protein was toxic, even in its native species. This lack of toxicity could be due to insufficient protein production, or it could suggest that the protein does not function to kill B. subtilis.
Support for a biological function other than toxicity comes from the apparent species specificity in the effects of type I toxins. For example, TxpA is toxic only upon overproduction in B. subtilis and is not toxic in E. coli [(33) and data not shown]. This observation could be due to differences in the amounts of the proteins overproduced by different bacteria but might also reflect the native target/function of TxpA. It has been suggested that the function of TxpA is to maintain the skin element, a chromosomal region excised during spore formation, within the B. subtilis genome (33). The E. faecalis TxpA homolog EF3263 is toxic to E. coli upon overproduction; possibly it has evolved a separate target, that is shared between Enterococcus and E. coli, from the B. subtilis protein.
We suggest that the distribution and evolutionary conservation of the type I toxins implies a genuine function in the bacterial cell. Despite the variation in the protein sequences across a toxin family, there are distinct sequence signatures that unite the families (Supplementary Figure S1), suggestive of conserved functions for the toxins. Such a proposed function does not contradict the selfish role of plasmid and phage-encoded type I toxin–antitoxin loci. Although we still lack a clear understanding of the role of the type I toxins, our data demonstrates their broad distribution across bacterial species. With the discovery of new families, and further experimentation with identified systems, the function of these loci will undoubtedly be revealed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Programs of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (E.M.F. and G.S.) and National Center for Biotechnology Information (K.S.M., S.A.S., N.Y. and E.V.K.) and a Research Associateship from the National Research Council (E.M.F.). Funding for open access charge: Intramural program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to D. Friedman, J. Lemos, W. Haas, E. Hobbs, K. Ramamurthy and Y. Wolf for strains and/or technical advice. Additionally, the authors thank K. Elkins and S. Gottesman for the use of equipment and S. Gottesman for comments on the manuscript.
REFERENCES
- 1.Fozo EM, Hemm MR, Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 2008;72:579–589. doi: 10.1128/MMBR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerdes K, Wagner E.GH. RNA antitoxins. Curr. Opin. Microbiol. 2007;10:117–124. doi: 10.1016/j.mib.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Makarova KS, Wolf YI, Koonin EV. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct. 2009;4:19. doi: 10.1186/1745-6150-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamaguchi Y, Inouye M. mRNA interferases, sequence-specific endoribonucleases from the toxin-antitoxin systems. Prog. Mol. Biol. Transl. Sci. 2009;85:467–500. doi: 10.1016/S0079-6603(08)00812-X. [DOI] [PubMed] [Google Scholar]
- 5.Gerdes K, Christensen SK, Løbner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 6.Van Melderen L, Saavedra De Bast M. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 2009;5:e1000437. doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buts L, Lah J, Dao-Thi MH, Wyns L, Loris R. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 2005;30:672–679. doi: 10.1016/j.tibs.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Pedersen K, Gerdes K. Multiple hok genes on the chromosome of Escherichia coli. Mol. Microbiol. 1999;32:1090–1102. doi: 10.1046/j.1365-2958.1999.01431.x. [DOI] [PubMed] [Google Scholar]
- 9.Faridani OR, Nikravesh A, Pandey DP, Gerdes K, Good L. Competitive inhibition of natural antisense Sok-RNA interactions activates Hok-mediated cell killing in Escherichia coli. Nucleic Acids Res. 2006;34:5912–5922. doi: 10.1093/nar/gkl750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weaver KE, Reddy SG, Brinkman CL, Patel S, Bayles KW, Endres JL. Identification and characterization of a family of toxin-antitoxin systems related to the Enterococcus faecalis plasmid pAD1 par addiction module. Microbiology. 2009;155:2930–2940. doi: 10.1099/mic.0.030932-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schäffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawano M, Oshima T, Kasai H, Mori H. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol. Microbiol. 2002;45:333–349. doi: 10.1046/j.1365-2958.2002.03042.x. [DOI] [PubMed] [Google Scholar]
- 13.Fozo EM, Kawano M, Fontaine F, Kaya Y, Mendieta KS, Jones KL, Ocampo A, Rudd KE, Storz G. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 2008;70:1076–1093. doi: 10.1111/j.1365-2958.2008.06394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 15.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adachi J, Hasegawa M. Computer Science Monographs No. 27. Tokyo: Institute of Statistical Mathematics; 1992. [Google Scholar]
- 17.Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 18.Shabalina SA, Ogurtsov AY, Spiridonov NA. A periodic pattern of mRNA secondary structure created by the genetic code. Nucleic Acids Res. 2006;34:2428–2437. doi: 10.1093/nar/gkl287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, Maixner W, Diatchenko L. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- 20.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 21.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogurtsov AY, Shabalina SA, Kondrashov AS, Roytberg MA. Analysis of internal loops within the RNA secondary structure in almost quadratic time. Bioinformatics. 2006;22:1317–1324. doi: 10.1093/bioinformatics/btl083. [DOI] [PubMed] [Google Scholar]
- 23.Kondrashov AS, Shabalina SA. Classification of common conserved sequences in mammalian intergenic regions. Hum. Mol. Genet. 2002;11:669–674. doi: 10.1093/hmg/11.6.669. [DOI] [PubMed] [Google Scholar]
- 24.Abranches J, Candella MM, Wen ZT, Baker HV, Burne RA. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J. Bacteriol. 2006;188:3748–3756. doi: 10.1128/JB.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawano M, Aravind L, Storz G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007;64:738–754. doi: 10.1111/j.1365-2958.2007.05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Ooij C, Losick R. Subcellular localization of a small sporulation protein in Bacillus subtilis. J. Bacteriol. 2003;185:1391–1398. doi: 10.1128/JB.185.4.1391-1398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unoson C, Wagner E.GH. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol. Microbiol. 2008;70:258–270. doi: 10.1111/j.1365-2958.2008.06416.x. [DOI] [PubMed] [Google Scholar]
- 29.Weel-Sneve R, Bjørås M, Kristiansen KI. Overexpression of the LexA-regulated tisAB RNA in E. coli inhibits SOS functions; implications for regulation of the SOS response. Nucleic Acids Res. 2008;36:6249–6259. doi: 10.1093/nar/gkn633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenfield TJ, Ehli E, Kirshenmann T, Franch T, Gerdes K, Weaver KE. The antisense RNA of the par locus of pAD1 regulates the expression of a 33-amino-acid toxic peptide by an unusual mechanism. Mol. Microbiol. 2000;37:652–660. doi: 10.1046/j.1365-2958.2000.02035.x. [DOI] [PubMed] [Google Scholar]
- 31.Weaver KE, Jensen KD, Colwell A, Sriram SI. Functional analysis of the Enterococcus faecalis plasmid pAD1-encoded stability determinant par. Mol. Microbiol. 1996;20:53–63. doi: 10.1111/j.1365-2958.1996.tb02488.x. [DOI] [PubMed] [Google Scholar]
- 32.Sharma CM, Hoffmann S, Darfeuille F, Findeiß S, Sittka A, Reignier J, Chabas S, Reiche K, Hackermüller J, Stadler PF, et al. The primary transcriptome of the major human pathogen. Helicobacter pylori. 2009 doi: 10.1038/nature08756. Nature, in press. [DOI] [PubMed] [Google Scholar]
- 33.Silvaggi JM, Perkins JB, Losick R. Small untranslated RNA antitoxin in Bacillus subtilis. J. Bacteriol. 2005;187:6641–6650. doi: 10.1128/JB.187.19.6641-6650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeigler DR, Prágai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, Bai R, Wyss M, Perkins JB. The origins of 168, W23, and other Bacillus subtilis legacy strains. J. Bacteriol. 2008;190:6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steglich C, Futschik ME, Lindell D, Voss B, Chisholm SW, Hess WR. The challenge of regulation in a minimal photoautotroph: non-coding RNAs in Prochlorococcus. PLoS Genet. 2008;4:e1000173. doi: 10.1371/journal.pgen.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl Acad. Sci. USA. 2009;106:894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang IN, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 38.Ramu H, Mankin A, Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Mol. Microbiol. 2009;71:811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.