Abstract
A number of studies have shown that the structure and composition of bacterial nucleoid influences many a processes related to DNA metabolism. The nucleoid-associated proteins modulate not only the DNA conformation but also regulate the DNA metabolic processes such as replication, recombination, repair and transcription. Understanding of how these processes occur in the context of Mycobacterium tuberculosis nucleoid is of considerable medical importance because the nucleoid structure may be constantly remodeled in response to environmental signals and/or growth conditions. Many studies have concluded that Escherichia coli H-NS binds to DNA in a sequence-independent manner, with a preference for A-/T-rich tracts in curved DNA; however, recent studies have identified the existence of medium- and low-affinity binding sites in the vicinity of the curved DNA. Here, we show that the M. tuberculosis H-NS protein binds in a more structure-specific manner to DNA replication and repair intermediates, but displays lower affinity for double-stranded DNA with relatively higher GC content. Notably, M. tuberculosis H-NS was able to bind Holliday junction (HJ), the central recombination intermediate, with substantially higher affinity and inhibited the three-strand exchange promoted by its cognate RecA. Likewise, E. coli H-NS was able to bind the HJ and suppress DNA strand exchange promoted by E. coli RecA, although much less efficiently compared to M. tuberculosis H-NS. Our results provide new insights into a previously unrecognized function of H-NS protein, with implications for blocking the genome integration of horizontally transferred genes by homologous and/or homeologous recombination.
INTRODUCTION
The bacterial nucleoid is a dynamic entity whose structure and composition is governed by a delicate balance between a plethora of nucleoid-associated proteins (NAPs), global superhelicity and general transcription status of the cell (1–4). The NAPs are ‘architectural’ proteins that profoundly affect not only the DNA conformation but also regulate the DNA metabolic processes such as replication, recombination, repair and transcription; however, their precise roles in these processes remain poorly understood (4–10). The most abundant NAPs, which exist often at micromolar concentrations, are HU, IHF, H-NS, Fis and Dps proteins (3–6). The NAP pool in Escherichia coli consists of 10–20 DNA binding proteins (7). However, H-NS (histone nucleoid structuring protein) and its paralogue, StpA (suppressor of td mutant phenotype A) are major protein components of the nucleoid structure in Escherichia coli and Salmonella enterica serovar Typhimurium (11). It has also been proposed that these two proteins form part of a global regulation network (8,12,13) and, an integral part of the protein scaffold responsible for DNA condensation in these organisms (5,8). Although their mechanism of conversion of linear DNA into supramolecular structure is beginning to be understood, little is known about how the NAPs engage their DNA substrates.
Genetic studies in both Salmonella and E. coli have shown that mutations in the hns gene display pleiotropic phenotypes, many of which are linked to adaptation to environmental stress such as increased resistance to osmotic and cold shock in Salmonella (14), carbon source utilization (15), homologous recombination and genome stability in E. coli (16,17). Studies in E. coli and other enterobacteria have revealed that H-NS plays a dual role of architectural organization of the nucleoid and regulator of gene expression of about 5% of the total chromosomal genes (see refs 8–10 and references therein). Consistent with the pleiotropic effects of hns mutations, transcriptomic studies suggest that 1439 genes were regulated by H-NS in S. typhimurium (18–20). Likewise, in uropathogenic E. coli strain, 536, H-NS regulates the expression of more than 500 genes, including many virulence factors such as fimbriae, cytotoxins and siderophores (21). Consequently, H-NS has been regarded as a paradigm to understand the role(s) of NAPs as a global regulator of gene expression, environmental adaptation and virulence (8–15). H-NS is responsible for silencing the expression of horizontally acquired DNA (19,20). A combination of in vivo and in vitro approaches have demonstrated that binding of H-NS to linear duplex DNA to be sequence non-specific; however, with a preference for A/T rich tracts embedded in curved DNA (19,22–27). Recently, specific high-affinity DNA binding sites have been identified (28), and these sites may serve as initiation sites for supra-structuring via H-NS oligomerization (29–31). Although, oligomerization is not required for interactions of H-NS with DNA, this property has been suggested to be essential for its architectural function (5). Like in E. coli, hns mutations in other types of bacteria display pleiotropic effects (15,32–36). Interestingly, hns mutations show an increase in the frequency of illegitimate recombination and reduced intra-chromosomal recombination (37,38). Several studies have shown that H-NS recognizes and transcriptionally represses horizontally transferred sequences in enteric bacteria in a process known as xenogeneic silencing (39,40). The repression mechanisms include promoter exclusion and RNA polymerase entrapment, both depend on the ability of H-NS to bind DNA and undergo oligomerization (see refs 8–12 and references therein). Although silencing of horizontally acquired DNA sequences may avoid potential toxic effects, the acquired genes must be expressed if they are to contribute to the organism to change its phenotype. This means that bacteria must have evolved means to counteract the H-NS silencing effects and to transcribe the acquired genes when their products are needed (39,40). It is unknown whether H-NS can also function as a repressor of other DNA metabolic processes such as replication, recombination or repair.
The progress in sequencing of entire microbial genomes has led to the discovery of several genes encoding homologues or interaction partners for H-NS (see refs 8–12 and references therein). Two mycobacterial NAPs have been studied in some detail: histone like protein (with homology to eubacterial HU protein) from Mycobacterium tuberculosis, Mycobacterium smegmatis and Mycobacterium bovis, and Lsr2 from M. tuberculosis and M. smegmatis (41–45). However, much less is known about the nucleoid structure and roles of NAPs in the tubercle bacillus. To insure the stability of its genome, adaptation of M. tuberculosis to persistence, latency and drug tolerance (as conferred by the state of non-replication) most likely requires changes in the expression pattern of nucleoid proteins (1–4). To gain insights into the mechanism(s) by which hns mutations affect the recombination events, we have studied the biochemical activities of M. tuberculosis H-NS protein. Here, we show that the M. tuberculosis H-NS protein binds in a more structure-specific manner to DNA replication/repair intermediates, but with substantially higher affinity to the Holliday junction (HJ), and abolished the three-strand exchange promoted by its cognate RecA protein. Consistent with M. tuberculosis H-NS, E. coli H-NS was able to bind the HJ and suppress DNA strand exchange, although much less efficiently than M. tuberculosis H-NS. Our results provide new insights into a previously unrecognized mechanism of H-NS function, with implications for blocking the genome integration of horizontally transferred genes by homologous and/or homeologous recombination.
EXPERIMENTAL PROCEDURES
Enzymes, DNA, bacterial strains and biochemicals
Fine chemicals were purchased from GE biosciences and Sigma. Restriction endonucleases, T4 DNA ligase, T4 polynucleotide kinase and Taq polymerase were purchased from New England Biolabs (NEB). Escherichia coli strain BL-21(DE3)pLysS strain and plasmid pET17b were purchased from Novagen. [γ-32P]ATP was purchased from Bhabha Atomic Research Center, Mumbai. Fast performance liquid chromatography columns were purchased from GE Biosciences. Escherichia coli and M. tuberculosis RecA was purified as described (46).
ODNs were synthesized by Sigma-Genosys and their sequences are shown in Table 1. The ODNs were labeled at the 5′-end by using [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs). The HJ, DNA replication fork and Y-shaped DNA substrates were prepared and characterized as described (47). Briefly, stoichiometric amounts of four purified ODNs were annealed by incubation in 0.3 M sodium citrate buffer (100 µl), pH 7, containing 3 M NaCl at 95°C for 3 min, followed by at 20°C for 4 h. The HJ was prepared by mixing ODN1, ODN2, ODN3 and 4; DNA replication fork from ODN2, 3, 5 and 6; Y-shaped junction from ODN2 and 3; double-stranded DNA with 70% GC base pairs from ODN7 and 8, and double-stranded DNA with 40% GC base pairs from ODN9 and 10. The annealing mixture was electrophoresed on a 6% polyacrylamide gel in 45 mM Tris–borate buffer (pH 8.3) containing 1 mM EDTA at 10 V/cm for 3 h. The bands corresponding to the annealed substrates were excised from the gel, eluted into TE buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA).
Table 1.
Sequences of oligonucleotides used in this study
| ODN 1 | 5′-GCCGTGATCACCAATGCAGATTGACGAACCTTTGCCCACGT-3′ |
| ODN 2 | 5′-GACGTGGGCAAAGGTTCGTCAATGGACTGACAGCTGCATGG-3′ |
| ODN 3 | 5′-GCCATGCAGCTGTCAGTCCATTGTCATGCTAGGCCTACTGC-3′ |
| ODN 4 | 5′-GGCAGTAGGCCTAGCATGACAATCTGCATTGGTGATCACGG-3′ |
| ODN 5 | 5′-GCAGTAGGCCTAGCATGA-3′ |
| ODN 6 | 5′-CGAACCTTTGCCCACGTC-3′ |
| ODN 7 70% GC (+Strand) | 5′GCGGTGGACGGCTGGGTCGGGTGGTGAGTGGGTTGCGATGGAGGTGGTGGGCTAGGGGGCTTAGGGGGGAGTCGGTGGTGG-3′ |
| ODN 8 70% GC (−Strand) | 5′-CCACCACCGACTCCCCCCTAAGCCCCCTAGCCCACCACCTCCCATCGCCAACCCACTCACCACCCGACCCAGCCGTCCACCGC-3′ |
| ODN 9 40% GC (+Strand) | 5′-GTACTATACGGTTGTACAGTGTTGTTAGTGAGTTGAAGATTGGAAGTAGTTTGCTAGGTGTCTTAGGAGAGAATCGTTAGTGT-3′ |
| ODN 10 40% GC (−Strand) | 5′-ACACTAACGATTCTCTCCTAAGACACCTAGCAAACTACTTCCAATCTTCAACTCACTAACAACACTGTACAACCGTATAGATC-3′ |
| ODN 11 50% GC (+Strand) | 5′-GTACTGTACGGCTGGACAGTGTTGTGAGTGAGTTGAAGATGGGAGGTAGTGTGCTAGGTGGCTTAGGAGAGAGTCGTTAGTGT-3′ |
| ODN 12 50% GC (−Strand) | 5′-ACACTAACGACTCTCTCCTAAGCCACCTAGCACACTACCTCCCATCTTCAACTCACTCACAACACTGTCCAGCCGTACAGATC-3′ |
Construction of an M. tuberculosis hns expression plasmid
The coding sequence corresponding to the hns gene of M. tuberculosis H37Rv (Rv3852) was PCR-amplified from its genomic DNA using the following ODNs (forward primer, 5′-GAGGGCCATATGCCAGACCCGCAGGATCG-3′ and reverse primer, 5′-CGCATTGAGCTCAGCGGCGGCGCAGTTGC-3′) carrying the sites for NdeI and SacI. The PCR product was gel purified and digested with restriction enzymes. The DNA was extracted with phenol–chloroform, precipitated by ethanol and ligated into pET17b (Novagen) expression vector. A portion of the ligation mixture was used to transform E. coli DH5α cells. The identity of the recombinant plasmid was ascertained by restriction analysis and DNA sequencing. These analyses revealed a sequence, which is identical to that of the M. tuberculosis hns gene (Rv3852) as annotated by the genome project (http://genolist.pasteur.fr/TubercuList/index.html) (48). The resultant plasmid was designated pMTHNS.
Overexpression and purification of M. tuberculosis Rv3852 in E. coli
H-NS protein was overexpressed in E. coli strain BL21(DE3)pLysS harboring the plasmid pMTHNS. Bacteria were grown in LB broth supplemented with antibiotics (100 μg/ml ampicillin and 34 μg/ml chloramphenicol) at 37°C to A600 of 0.6. H-NS was induced by the addition of 0.5 mM IPTG, and the cultures were incubated for overnight at 18°C. Cells were collected by centrifugation, washed in STE buffer [10 mM Tris–HCl (pH 8), 100 mM NaCl and 1 mM EDTA], resuspended in buffer A [10 mM Tris–HCl (pH 7.5), 4 mM EDTA, 5 mM DTT, 150 mM NaCl, and 5% (v/v) glycerol], and stored at −80°C. Cells were thawed and lysed by sonication (Model No. GEX-750, Ultrasonic Processor) on ice at 60% duty cycles in a pulse mode. The sonicated suspension was centrifuged in a Beckman Ti-45 rotor at 30000 r.p.m. for 1 h at 4°C. To the supernatant, solid ammonium sulfate was added to 40% saturation at 4°C and the precipitate was removed by centrifugation. To the supernatant, ammonium sulfate was added to 60% saturation at 4°C, and the precipitated proteins were recovered by centrifugation. The precipitate was resuspended and dialyzed overnight against buffer A at 4°C. The dialyzed sample was then loaded onto a dsDNA-cellulose column that had been previously equilibrated with buffer A. H-NS was eluted with a 50 → 1 M linear gradient of NaCl in buffer A. Peak fractions were pooled and dialyzed against buffer B [10 mM Tris–HCl (pH 7.5), 4 mM EDTA, 5 mM DTT, 500 mM NaCl and 10% (v/v) glycerol]. The dialyzed sample was loaded onto a Superdex-75 gel-filtration column. Peak fractions were pooled, dialyzed against storage buffer C (20 mM Tris–HCl, pH 7.5, 0.5 mM EDTA, 0.5 mM DTT, 50 mM NaCl and 20% glycerol). The purity of H-NS was assessed by SDS–PAGE and found to be >98%. Aliquots of M. tuberculosis H-NS were stored at −80°C.
Overexpression and purification of E. coli H-NS
Escherichia coli H-NS was overexpressed in E. coli UT5600 cells containing hns plasmid, pPLc2833 (a kind gift of Drs R. Spurio and C. O. Gualerzi, University of Rome-Tor Vergata, 00133 Rome, Italy) as described (49). The expressed protein was purified to homogeneity by chromatography on a double-stranded DNA–cellulose column, which retained H-NS, whereas many other proteins flowed through the column. The bound proteins were eluted by a linear gradient of NaCl (150 mM →1 M) in buffer A. Fractions containing H-NS were pooled and subjected to gel filtration through a Superdex S-75 column. The H-NS containing fractions were pooled and dialyzed into storage buffer C and stored at −80°C. The resulting E. coli H-NS protein was >95% pure by SDS–PAGE.
Glutaraldehyde crosslinking of M. tuberculosis H-NS
Reaction mixtures (25 µl) contained 16 mM Tris–HCl (pH 7.5), 16% glycerol, 0.4 mM EDTA, 0.4 mM DTT, 40 mM NaCl, 6.5 µM H-NS and the indicated amounts of freshly diluted glutaraldehyde. After incubation at 37°C for 20 min, the samples were diluted into gel loading buffer, incubation was extended for 3 min at 95°C. Samples were loaded onto 15% SDS–PAGE. The products were visualized by staining with silver nitrate.
Gel mobility shift assay
Reaction mixtures (20 μl) contained 20 mM Tris–HCl (pH 8), 20 mM KCl, 1 mM DTT, 1 mM potassium phosphate, 5% glycerol, 20 mM EDTA, 32P-labeled DNA substrate and M. tuberculosis or E. coli H-NS at concentrations as specified in the figure legends. After incubation at 37°C for 20 min, 2.5 μl of 10 × loading dye [0.42% (w/v) bromophenol blue and xylene cyanol in 50% glycerol] was added to each sample, and were electrophoresed on 6% polyacrylamide gel in 18 mM Tris–borate (pH 8.3) containing 0.4 mM EDTA at 80 V at 4°C. The duration of electrophoresis was as follows: in the case of M. tuberculosis H-NS, 16 h for complexes with HJ, replication fork or Y-shaped DNA structures, and 8 h with ssDNA and dsDNA, respectively. In case of E. coli H-NS, 15 h for complexes with HJ and 2.5 h for replication fork and dsDNA, respectively. The gels were dried, and the bands were visualized using a Fuji FLA-9000 phosphorimager. The bands were quantified using UVI-BAND MAP and the data was plotted in GraphPad Prism version 4.0.
H-NS constrains negative supercoiling in DNA
One microgram of pBend3 form I DNA was incubated in a buffer (25 μl) containing 50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 1 mM DTT, 100 μg/ml bovine serum albumin with indicated amounts of H-NS at 37°C for 20 min. Wheat germ topoisomerase I (2 U) (Promega) was then added and incubation was continued at 37°C for 1 h. Samples were deproteinized with proteinase K (0.4 mg/ml), ETDA (5 mM) and SDS (0.1%) at 37°C for 15 min. The reaction was terminated by the addition of 2.5 μl of 10 × gel loading dye [0.42% (w/v) bromophenol blue and xylene cyanol in 50% glycerol]. The samples were electrophoresed on 0.8% agarose gel in 89 mM Tris–borate buffer (pH 8.3), at 3 V/cm at 4°C for 20 h. The gel was soaked in 300 ml of electrophoresis buffer containing 0.5 μg/ml ethidium bromide for 1 h, rinsed thoroughly with water, and captured using UVI gel documentation system.
Three-strand exchange assay
The assay was performed as described (46). Briefly, reaction mixtures (15 µl) containing 20 mM Tris–HCl (pH 7.5), 3 mM dATP, 8 mM MgCl2, 2 µM ssDNA (ODN11) was incubated with 1 µM M. tuberculosis or E. coli RecA in the presence of ATP regeneration system at 37°C for 5 min. In parallel, 83-bp (0.4 µM) duplex DNA (32P-labeled ODN11 annealed to ODN12) was incubated with increasing concentrations (0.5–4 µM) of M. tuberculosis H-NS at 37°C for 20 min. The strand transfer reaction was initiated by mixing the contents of both the reactions, followed by incubation at 37°C for 10 min. Reaction was stopped by the addition of 1.5 µl of 1% SDS, followed by 1 µl proteinase K (10 mg/ml). After incubation at 37°C for 15 min, 2 µl 10 × gel loading dye (50% glycerol, 0.42% bromophenol blue, 0.42% xylene cyanol) was added to each reaction. Samples were loaded onto 10% polyacrylamide gel and electrophoresed in 45 mM Tris–borate buffer (pH 8.3), 1 mM EDTA at 120 V for 10 h. The 32P-labeled DNA substrates and products were visualized by phosphorimaging analysis of the dried gel.
Bioinformatics
In the published genome sequence of M. tuberculosis strain H37Rv, Rv3852, which has been annotated as hns, has been proposed to encode HU-histone protein (48). The sequences of M. tuberculosis H-NS and E. coli H-NS homologues were retrieved from the database searches of microbial genome sequences from the TIGR Comprehensive Microbial Resource (CMR) database site (http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi). Sequence alignments were performed using the ClustalW ver. 2 program (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and viewed using Jalview 2.4.0.b2.
RESULTS
Mycobacterium tuberculosis nucleoid protein-encoding genes
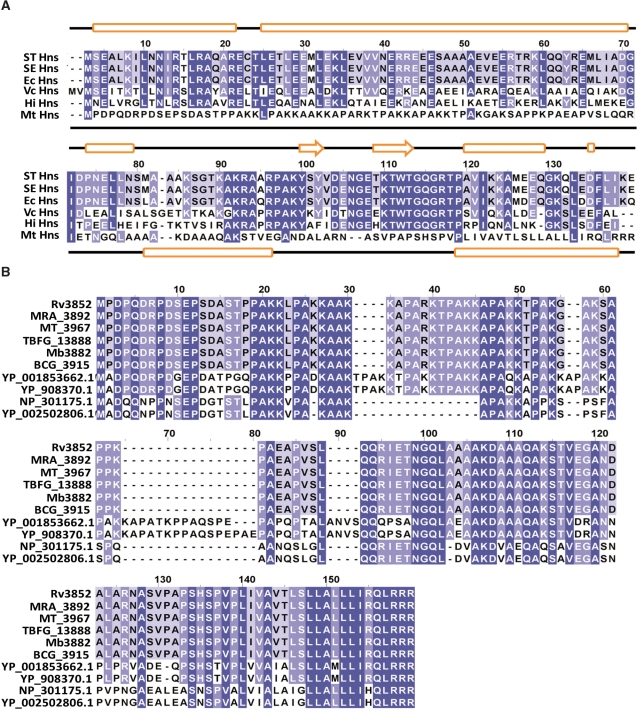
To identify H-NS homologues, the sequence of the prototype E. coli H-NS was used in database searches. The original sequence and annotation of M. tuberculosis strain H37Rv identified Rv3852 as the hns gene (www.sanger.ac.uk/Projects/M_tuberculosis). Consequently, the protein sequence of Rv3852 was used to search the database for H-NS homologues in other mycobacterial species. The searches in the TIGR CMR database identified H-NS homologues from a diverse set of prokaryotes including S. typhimurium, S. enterica, Vibrio cholerae and Haemophilus influenzae. Direct inspection of the complete sequence alignment revealed striking conservation as well as some notable differences. A remarkable conservation of sequence (95% identity) was found between E. coli H-NS and its homologues in Salmonella species (Figure 1A). When more distant species were compared, the sequence divergence was clearly visible. V. cholerae and H. influenzae share 54% and 47% sequence identity with E. coli H-NS, respectively. Sequence identity decays further in the case of M. tuberculosis H-NS: it shares 6% sequence identity with its counterpart in E. coli (Figure 1A). Pair-wise comparison revealed that M. tuberculosis H-NS is devoid of amino acid residues 84 and 106–107 compared to the E. coli H-NS. Notably, M. tuberculosis H-NS is a conserved protein (100%) across M. tuberculosis H37Ra, M. tuberculosis CDC1551, M. bovis and M. bovis BCG. However, protein conservation and sequence length was less noticeable between M. tuberculosis H-NS and its counterpart in the other mycobacterial species such as Mycobacterium marinum, Mycobacterium ulcerans and Mycobacterium leprae. In contrast to E. coli, Salmonella, V. cholerae and H. influenzae, the mycobacterial H-NS homologues contain varying number of tetrapeptide repeats (PAKK, KAAK), which are crucial for high affinity DNA-binding activity of H1/H5 histone family of proteins. Interestingly, the numbers of PAKK repeats vary within mycobacterial species: four each in M. tuberculosis and M. bovis, 6 in M. marinum and M. ulcerans and 1 in M. lepare. Taken together, these results suggest that M. tuberculosis protein is a non-canonical H-NS and is structurally distinct from its counterpart in E. coli.
Figure 1.
ClustalW alignment of H-NS homologues from various bacterial species. (A) Alignment of amino acid sequences of H-NS homologues include the following: STHns, S. typhimurium LT2 SGSC1412 Hns (STM1751), SEHns, S. enterica serovar Typhi Ty2 Hns (t1662), EcHns, Escherichia coli Hns (b1237), VcHns, V. cholerae El Tor N16961 Hns (VC1130), HiHns, Haemophilus influenzae 86028NP (NTHI1464) and MtHns, Mycobacterium tuberculosis H37Rv Hns (Rv3852). Dark blue shading indicates identical amino acid residues; whereas, light blue indicates identical residues among the specified species. Secondary structure assignments for E. coli and M. tuberculosis H37Rv Hns proteins were carried out using the Jpred program (75) (http://www.compbio.dundee.ac.uk/∼www-jpred/) and are displayed above and below the alignments, respectively. The predicted secondary structures (cylinders represent alpha helices, arrows represent beta strands and lines represent loops, respectively) of E. coli and M. tuberculosis H-NS are shown at the top and bottom of the plot, respectively. (B) Amino acid sequences of M. tuberculosis H37Rv Hns (Rv3852), M. tuberculosis H37Ra Hns (MRA_3892), M. tuberculosis CDC1551 Hns (MT_3967), M. tuberculosis F11 Hns (TBFG_13888), M. bovis subsp. bovis AF2122/97 Hns (Mb3882), M. bovis BCG str. Pasteur 1173P2 Hns (BCG_3915), M. marinum M Hns(YP_001853662.1), M. ulcerans Agy99 Hns (YP_908370.1), M. leprae TN Hns (NP_301175.1) and M. leprae Br4923 Hns (YP_002502806.1) were aligned using ClustalW2 and the resulting alignment was viewed using Jalview 2.4.0.b2 (76). Dark blue shading indicates identical amino acid residues in all the species; whereas, light blue indicates identical residues among the specified species.
Purification of M. tuberculosis H-NS
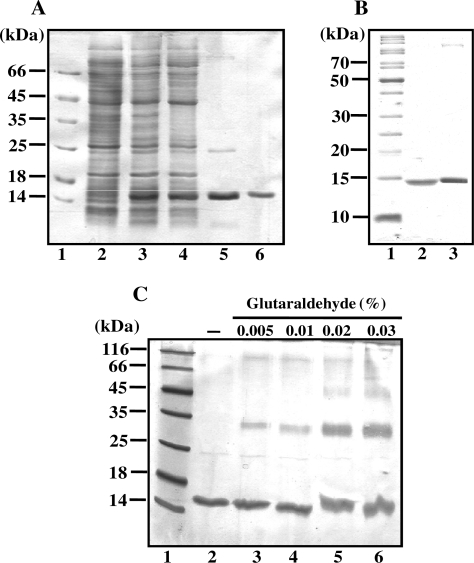
Mycobacterium tuberculosis Rv3852 gene is predicted to encode a small 134 amino acid protein with a predicted molecular mass of 13.8 kDa. Because so little is known about the function of mycobacterial NAPs, and considering the sequence dissimilarities between E. coli and M. tuberculosis H-NS, we wished to explore the biochemical activities of M. tuberculosis H-NS. Escherichia coli BL-21(DE3)pLysS, a lonA mutant, was used as host strain for expression of the recombinant H-NS protein. Both supernatant and pellet from cell-free lysates were examined for the presence of recombinant H-NS protein. The expressed recombinant H-NS was found to be in the soluble fraction of the induced cell-free lysates. Purification of the recombinant H-NS was performed as described under Experimental procedures. H-NS was purified to (>98%) homogeneity as estimated from polyacrylamide gels stained with Coomassie brilliant blue R-250. A representative SDS–PAGE pattern of the sample at each purification step is shown in Figure 2A. Side-by-side comparison of E. coli and M. tuberculosis H-NS proteins, after separation by SDS–PAGE and staining with Coomassie brilliant blue R-250, is shown in Figure 2B.
Figure 2.
(A) SDS–PAGE analysis showing induced expression of M. tuberculosis H-NS and at various stages during its purification. Ten micrograms of proteins from the indicated sample was resolved on SDS–PAGE and visualized by staining with Coomassie blue. Lanes: 1, SDS–PAGE standards molecular mass markers; 2, uninduced cell-free lysate; 3, induced cell-free lysate; 4, (NH4)2SO4 precipitate; 5, chromatography on dsDNA cellulose; 6, chromatopraphy on Superdex S-75. (B) Side-by-side comparison of M. tuberculosis and E. coli H-NS proteins by SDS–PAGE. Lanes: 1, SDS–PAGE standards molecular mass markers; 2, E. coli H-NS (1 µg); 3, M. tuberculosis H-NS (1 µg). (C) Glutaraldehyde crosslinking of M. tuberculosis H-NS. The reactions were performed as described under ‘Experimental Procedures’ section. Lane 1, molecular weight standards; 2, H-NS incubated in the absence of glutaraldehyde; 3-6, H-NS incubated with concentrations of glutaraldehyde as indicated at the top of the gel image.
The oligomerization of H-NS was investigated by cross-linking with glutaraldehyde. Glutaraldehyde reacts with the amino group of the Lys side chain and has been extensively used to cross-link oligomeric proteins. H-NS was incubated with increasing concentrations of glutaraldehyde, and then the oligomeric state was analyzed by SDS–PAGE. As shown in Figure 2C, we observed a slower migrating band corresponding to a dimer (lanes 3–6), whose intensity increased with increasing concentrations of glutaraldehyde, suggesting that M. tuberculosis H-NS exists in equilibrium between monomeric and dimeric forms in solution.
Mycobacterium tuberculosis H-NS binds poorly to DNA containing high GC base pairs
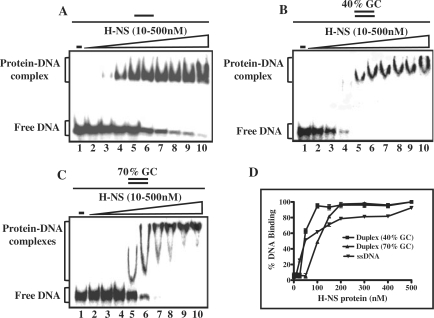
Much effort has been devoted in understanding the DNA-binding specificity of E. coli H-NS. The sequences to which it displays high-affinity are usually A+T-rich and are often associated with regions of intrinsic curvature in DNA (1–4). Analysis of the M. tuberculosis genome has disclosed that it possess a high (65.5%) GC content across its genome; however, a group of genes that belong to the family of PE or PPE proteins contain a very high GC (>80%) content (50). In addition, it has been demonstrated that Salmonella H-NS selectively silences genes that possess substantially lower GC content than the overall genome (20). We have therefore investigated the DNA-binding activities of purified M. tuberculosis H-NS by electrophoretic mobility shift assay (EMSA) using different DNA substrates. Reactions were performed with a fixed amount of 32P-labeled (83-mer) single- or double-stranded DNA (83-bp) containing either 40% GC (formed by annealing ODN 9 with 10, Table 1) or 70% GC-base pairs (formed by annealing ODN 7 with 8, Table 1) and increasing concentrations of M. tuberculosis H-NS. The amounts of H-NS required for complex formation showed a wide range of concentrations. Like the E. coli homologue, M. tuberculosis H-NS does not need magnesium or ATP under conditions where the protein binds efficiently. As shown in Figure 3A and B, M. tuberculosis H-NS formed a distinct protein–DNA complex with ssDNA or dsDNA containing 40% GC base pairs. On the other hand, with DNA containing 70% GC base pair and at low concentrations of H-NS, a less well-defined protein–DNA complex was formed as evidenced by the smeared distribution of radioactivity between the retarded complex and free DNA, indicating that the complex is unstable and dissociates on electrophoresis (Figure 3C, lanes 5–8). However, the much retarded protein–DNA complex progressively increased with increasing concentrations of M. tuberculosis H-NS (Figure 3A–C, lanes 9–10). Figure 3D shows quantification of DNA in the shifted complex, relative to free DNA, with various DNA substrates as a percentage of total amount of DNA bound over a range of protein concentrations. Notably, the concentration of H-NS required for 50% binding for duplex DNA containing 70% GC base pairs is ∼3-fold higher, compared to a similar substrate containing 40% GC base pairs. These results are consistent with robust binding of M. tuberculosis H-NS to ssDNA or dsDNA containing 40% GC base pairs, but substantially reduced binding to dsDNA containing 70% GC base pairs (Figure 3D and see below).
Figure 3.
Mycobaterium tuberculosis H-NS binds poorly to double-stranded DNA containing high GC base pairs. Reactions were performed with 5 nM of the indicated 32P-labeled DNA substrate in the absence (lane1) or presence of 10, 25, 50,100, 150, 200, 250, 300 or 500 nM H-NS (lanes 2–0), respectively. A single or two parallel lines on the top of each panel of the Figure denote single- or double-stranded DNA, respectively. The open triangle on the top of the gel image denotes increasing concentrations of H-NS. Reaction products were separated as described under ‘Experimental Procedures’ section. (A) ssDNA; (B) dsDNA (40% GC); (C) dsDNA (70% GC). The positions of free DNA and protein–DNA complexes are shown in the left-hand side of each panel. (D) Graphical representation of binding of H-NS to different DNA substrates. The extent of formation of H-NS–DNA complexes in (A–C) is plotted versus varying concentrations of H-NS. Error bars indicate SEM.
Mycobacterium tuberculosis H-NS binds to the HJ with high affinity
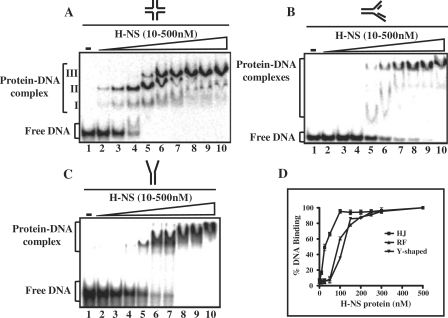
We next examined the ability of M. tuberculosis H-NS to bind DNA replication and recombination intermediates. For these assays, we assembled a HJ containing four 20-bp arms, DNA replication fork with three 20-bp arms and Y-shaped junction containing a 20-bp duplex region with two 20-nt heterologous single strands and characterized them as described (47). Gel mobility shift assays were performed to separate the protein–DNA complex from free DNA. Significantly, M. tuberculosis H-NS displayed robust binding to the HJ relative to the DNA replication intermediates (compare Figure 4A with B and C). At low concentrations, binding of H-NS to the HJ led to the formation of two protein–DNA complexes, complex I and II (Figure 4A, lanes 2–4). As the amount of H-NS was increased, these complexes were replaced by a progressively slower-migrating species, complexes III, resulting in a single well-defined complex. The stoichiometric ratio of H-NS/HJ complexes is currently under investigation.
Figure 4.
Mycobacterium tuberculosis H-NS binds to the Holliday junction with high affinity. Reaction mixtures contained 5 nM of 32P-labeled HJ (A), 32P-labeled DNA replication fork (B) or 32P-labeled Y-shaped junction (C) in the absence (lane1) or presence of 10, 25, 50,100, 150, 200, 250, 300 or 500 nM H-NS (lanes 2–10), respectively. The open triangle on the top of the gel denotes increasing concentrations of H-NS. Reaction products were separated as described under ‘Experimental Procedures’ section. The positions of free DNA and protein–DNA complexes are shown in the left-hand side of each panel. (D) Graphical representation of the extent of binding of H-NS to different DNA substrates in (A–C) is plotted versus varying concentrations of H-NS.
Mobility shift assays were also performed to determine the relative affinity of H-NS to two different DNA replication intermediates: replication fork and Y-shaped junctions. Although H-NS showed efficient binding to both the replication intermediates, we found striking differences (compare Figure 4B with C). At lower concentrations of H-NS with the DNA replication fork, we observed smeared distribution of radioactivity between the retarded nucleoprotein complex and free DNA, suggesting the formation of weak protein–DNA complexes which may have dissociated in the gel. However, the much slower migrating band was greatly retarded with increasing concentration of H-NS (Figure 4B, lanes 8–10). On the other hand, at low concentrations, H-NS formed a distinct complex with the radiolabeled Y-shaped junction. Increasing amounts of H-NS led to an increase in the formation of a large macromolecular complex that positioned near the well of the gel (Figure 4C, lane 10). These results suggest the occurrence of complexes containing different amounts of H-NS bound to the DNA. Figure 4D shows quantification of DNA in the shifted complexes with various DNA substrates as a percentage of total amounts of DNA bound over a range of protein concentration. The binding isotherm for DNA replication fork and Y-shaped junction are almost superimposable with each other. Notably, the concentration of H-NS required for 50% binding for Holliday junction is ∼6-fold lower, compared to the DNA replication fork or Y-shaped structure.
The equilibrium dissociation constant (Kd) for binding of H-NS to various DNA substrates was determined by EMSA. The reaction mixtures contained fixed amounts of the indicated DNA substrate and varying concentrations of H-NS. The concentration of DNA was much lower than required for half-maximal binding, so that the protein concentration at half-maximal binding was very close to Kd. We note that H-NS exhibited a higher apparent binding constant for the Holliday junction (Kd≈ 32 nM) (Table 2). The Kd value for H-NS binding to the Holliday junction is comparable to those of the high-affinity (proU) sites with E. coli H-NS (28). Furthermore, the apparent Kd values for H-NS binding varies with the nature of the substrate: it is 6- and 7-fold higher for the DNA replication fork and Y-shaped junction, respectively, compared with the HJ. Noteworthy, these results suggest that H-NS binding to the HJ is cooperative, whereas H-NS- binding to dsDNA, DNA replication fork and Y-shaped structure is not.
Table 2.
Equilibrium association constants of H-NS DNA complexes
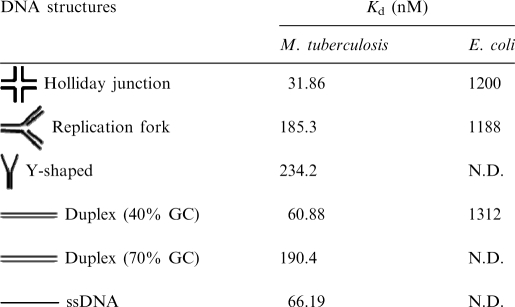 |
N.D. = Not determined.
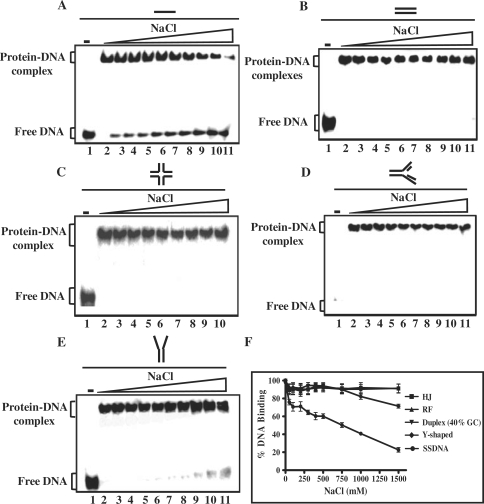
A number of studies have shown that the sensitivity of protein–DNA complex to salt is a relative measure of the affinity of the protein to DNA. The effect of salt on H-NS binding to various DNA substrates was measured by varying the concentration of NaCl in the incubation buffer from 0 to 1.5 M (Figure 5A–E). Intriguingly, quantification of protein–DNA complexes disclosed that the binding of M. tuberculosis H-NS to the HJ, replication fork or double-stranded DNA, as construed from the salt stability curves, were superimposable with each other, whereas the stability curve for H-NS-Y shaped DNA complex diverged at 0.75 M NaCl (Figure 5F). In contrast, we note that H-NS-ssDNA complexes were unstable and the salt titration mid-point was 0.75 M, indicating the binding affinity of H-NS for ssDNA is weak (Figure 5F).
Figure 5.
Effect of NaCl on the stability of H-NS–DNA complexes. Reaction mixtures contained 5 nM of indicated 32P-labeled DNA substrate and 500 nM of M. tuberculosis H-NS. After incubation for 30 min, NaCl was added to the final concentration of 50 100, 200, 300, 400, 500, 750, 1000 or 1500 mM (lanes 3–11), respectively. After 10 min with NaCl, samples were electrophoresed on polyacrylamide gel, and this was followed by autoradiography as described under ‘Experimental Procedures’ section. (A) ssDNA; (B) dsDNA (40% GC); (C) HJ; (D) DNA replication fork; (E) Y-shaped junction; (F) the extent of dissociation of H-NS–DNA complex containing the indicated recombination intermediate is plotted versus varying concentrations of NaCl. Error bars indicate SEM.
Comparison with E. coli H-NS protein
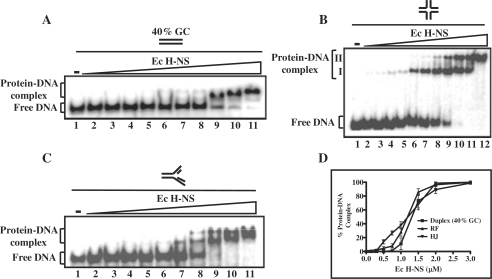
In light of significant differences between the M. tuberculosis and E. coli H-NS proteins in their primary sequence, we asked whether E. coli H-NS can bind structures similar to those bound by its M. tuberculosis counterpart. Using the experimental conditions described for the M. tuberculosis H-NS protein, the DNA-binding activity of E. coli H-NS was first measured with linear duplex DNA containing 40% GC content. A distinct protein–DNA complex was seen with linear duplex DNA (Figure 6A, lanes 9–11), although with apparently lower affinity than M. tuberculosis H-NS (Table 2). On the other hand, E. coli H-NS was able to bind the HJ in a stepwise manner resulting in the formation of two distinct protein–DNA complexes (Figure 6B, lanes 6–12), indicating that E. coli H-NS may bind as a dimer or higher multimer, and that the substrate provides more than one binding site. These results are consistent with a mechanism in which H-NS interaction with DNA results in the formation of DNA–H-NS–DNA bridges that are implicated in the regulation transcription initiation and organization of bacterial chromatin (4–6). A similar result was also seen with the three-way junction (Figure 6C, lanes 7–11). All the DNA substrates were shifted completely by E. coli H-NS at the concentrations shown; however, it displayed somewhat higher affinity for the four-way junction (Figure 6D, Table 2). These experiments were performed several times with the same substrates with similar results. We note that, in contrast to M. tuberculosis H-NS, the complexes formed by E. coli H-NS were relatively unstable during the extended periods of electrophoresis, and under these conditions the complexes were less well defined (data not shown).
Figure 6.
Binding of E. coli H-NS to the HJ and to other DNA substrates. Reactions were performed with 1 nM of 32P-labeled DNA (duplex DNA having 40% GC content or replication fork) in the absence (lane 1) or presence of 0.1, 0.15, 0.25, 0.3, 0.5, 0.75, 1, 1.5, 2 and 3 µM E. coli H-NS (lanes 2–11), respectively. Similarly, 1 nM of 32P-labeled HJ was incubated in the absence (lane 1) or in the presence of 0.1, 0.15, 0.25, 0.3, 0.5, 0.75, 1, 1.5, 2, 2.5 and 3 µM E. coli H-NS (lanes 2–12), respectively. Reaction products were separated and visualized as described under ‘Experimental Procedures’ section. The open triangle on the top of the gel image denotes increasing concentrations of E. coli H-NS. (A) Linear duplex DNA; (B) HJ; (C) replication fork. The positions of free DNA and protein–DNA complexes are indicated on the left-hand side of the gel. (D) Graphical representation of E. coli H-NS binding to different DNA substrates. The extent of protein–DNA complexes in (A–C) is plotted versus varying protein concentrations. Standard deviations are derived from three independent experiments.
M. tuberculosis H-NS constrains negative supercoiling in DNA
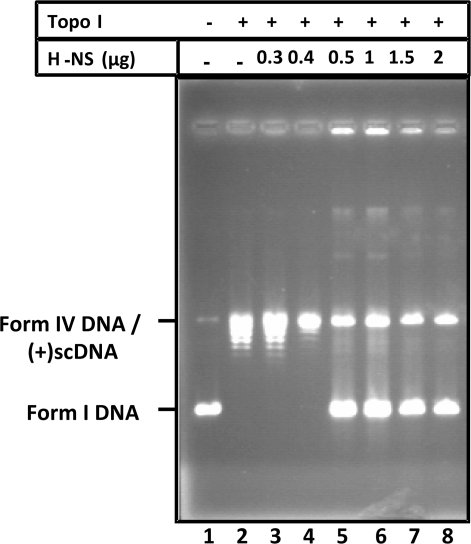
To investigate the type of structure formed by M. tuberculosis H-NS, we examined whether this protein modifies the topological state of the DNA substrate. This assay which was carried out in the presence of eukaryotic topoisomerase I traps DNA supercoils by protein binding (51,52). The relaxation of form I DNA was assessed using wheat germ topoisomerase I (Figure 7, lane 2). Incubation of form I DNA with increasing concentrations of H-NS, and then with topoisomerase I, produced a species of DNA which migrated to the same position as the initial form I DNA (Figure 7, lanes 5–8). Interestingly, at the low concentrations employed, M. tuberculosis H-NS by itself was unable to constrain the negative supercoils in DNA (Figure 7, lanes 3–4). As the concentration increased, H-NS could constrain negative supercoils in >70% of form I DNA (Figure 7, lanes 5–8). However, H-NS failed to convert all the DNA into form I DNA. We therefore considered the possibility that high concentrations of H-NS may have converted form I DNA into positively supercoiled DNA, which migrates at the same position as form IV DNA on one-dimensional agarose gels. To test the above possibility, we performed two-dimensional agarose gel electrophoresis in the presence of chloroquine. We observed that there were no bands corresponding to positively supercoiled topoisomers at the indicated concentrations of H-NS protein (data not shown). We conclude from these results that inside the H-NS–DNA complex the DNA is negatively supercoiled and protected from relaxation by topoisomerase I.
Figure 7.
H-NS constrains DNA supercoils in vitro. Form-I DNA (1 μg) was incubated with the indicated amounts of purified H-NS protein and then treated with topoisomerase I. After deproteinization, DNA samples were electrophoresed on 0.8% agarose gel to resolve the topoisomers. Lane 1, the positions of form I and form IV (relaxed) DNA and positively supercoiled [(+) SC DNA] are shown in the left-hand side. Lane: 2, DNA incubated with topoisomerase I in the absence of H-NS; 3–8, contained topoisomerase I and indicated amounts of H-NS.
H-NS proteins act as barriers in RecA protein-promoted DNA strand exchange
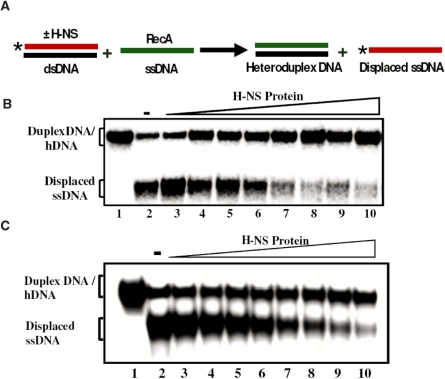
To gain insights into homologous recombination in the context of bacterial nucleoid, we focused on the ability of M. tuberculosis RecA to catalyze three-strand exchange with linear duplex DNA bound by increasing amounts of H-NS. The function of H-NS is of particular interest because: (i) E. coli hns mutations display pleiotropic effects, including increase in the frequency of illegitimate recombination and reduced intra-chromosomal recombination, and (ii) its role in xenogeneic silencing, where it represses the transcription/integration of foreign genes acquired by horizontal transfer. In this assay, 32P-labeled 83-bp linear duplex DNA and its complementary 83-mer ssDNA were used as DNA substrates. In this combination of DNA substrates, complete strand exchange will culminate in the displacement of a faster migrating 83-mer 32P-labeled ssDNA. Active nucleoprotein filaments of E. coli or M. tuberculosis RecA-ssDNA were prepared in the presence of dATP as described previously (46). In parallel, 83-bp linear duplex DNA was incubated with increasing concentrations of H-NS protein. As a control, H-NS was omitted from the reaction (Figure 8, lane 2). The contents of the two reaction mixtures were directly combined and the strand-exchange reaction was performed as described under the ‘Experimental Procedures’ section. After halting the reaction by deproteinization, the samples were resolved by PAGE and analyzed by autoradiography. In the absence of H-NS, E. coli or M. tuberculosis RecA proteins were able to execute significant levels of strand transfer. In contrast, under similar conditions, M. tuberculosis H-NS, in a concentration-dependent manner, dramatically reduced the amount of displaced ssDNA with concomitant increase in the DNA corresponding to the 32P-labeled duplex DNA. On the other hand, E. coli H-NS also was able to suppress DNA strand exchange promoted by its cognate RecA, but at relatively high concentrations. Nonetheless, these results suggest that H-NS proteins act as ‘roadblocks’ to strand exchange promoted by their respective RecA proteins.
Figure 8.
H-NS proteins suppress DNA strand exchange promoted by RecA protein. Assay was performed as described under ‘Experimental Procedures’ section. (A) Schematic depicting the experimental design. (B) Effect of M. tuberculosis H-NS on strand exchange by its cognate RecA. (C) Effect of E. coli H-NS on strand exchange by its cognate RecA. The positions of 32P-labeled displaced ssDNA, 32P-labeled duplex DNA or unlabeled heteroduplex DNA (hDNA) generated by RecA promoted strand transfer are shown in the left-hand side of the gel images. The open triangle on top of the gel images denotes increasing concentrations of H-NS. Lane1, control reactions lacking H-NS and RecA; lane 2, complete reaction in the absence of H-NS; lanes 3–10, complete reaction in the presence of 0.5, 1, 1.5, 2, 2.5, 3, 3.5 and 4 μM H-NS, respectively. An asterisk represents the labeled phosphate at the 5′ end.
DISCUSSION
Analysis of the biochemical activities of M. tuberculosis H-NS protein leads to two main conclusions. First, H-NS possess high-affinity HJ-binding activity, compared to DNA replication intermediates and GC-rich double-stranded DNA; and second, the H-NS binding to duplex DNA impedes the three-strand exchange promoted by the cognate RecA. When viewed in the context of the role(s) of E. coli hns in homologous recombination and repression of the integration of the horizontally transferred genes into the genome, these observations are indeed striking. While this manuscript was in preparation, Werlang and colleagues (53) reported the purification of M. tuberculosis H-NS and its ability to bind DNA derived from its proU promoter region. Taken together, these studies suggest that the product of the Rv3852 gene in M. tuberculosis as being the H-NS protein.
In an effort to relate the sequence of H-NS proteins to their function, we have performed sequence alignment of H-NS proteins in a range of eubacterial species, including various species of mycobacteria. From the results of sequence analyses perhaps the most intriguing aspect that emerges is the lack of homology among E. coli/Salmonella H-NS and mycobacterial H-NS proteins. Mycobacterium tuberculosis H-NS showed a very low sequence identity (<10% with E. coli/Salmonella HN-S) with H-NS proteins from several other eubacterial species (Figure 1A). On the other hand, the size and sequence of H-NS is strongly conserved among several mycobacterial genomes sequenced so far, with 100% identity between the M. bovis, M. bovis BCG and M. tuberculosis H37Rv and H37Ra (Figure 1B). It is noteworthy that the M. tuberculosis H-NS sequence is more divergent compared to the M. marinum, M. ulcerns and M. leprae H-NS proteins. However, all mycobacterial H-NS proteins possess varying numbers of PAKK and KAKK tetrapeptide motifs. The low percentage of sequence identity between M. tuberculosis and E. coli/Salmonella proteins is not unprecedented. For example, M. tuberculosis chorismate mutase shares ∼20% amino acid sequence identity with the E. coli enzyme, while the level of identity with yeast enzyme is ∼10% (54,55). Despite such a low degree of sequence identity, these proteins possess the catalytic activity and also display significant similarities in their tertiary structures (56).Other examples include, CeoB/TrkA (Rv2691) shares 21% sequence identity with the Escherichia coli TrkA protein, which codes for the NAD+-binding subunit of the potassium transport system (57). Taken collectively, these results suggest that primary sequence alone is a poor discriminator of the biological function of proteins, and the three-dimensional structure might provide a more reliable guide.
Following the isolation and characterization of the prototype H-NS from E. coli, similar proteins have been identified in other enteric or related bacteria through purification and DNA binding assays or in silico analysis of bacterial genomes. All these proteins possess amino acid sequence identity in the range of 40–70% with the E. coli H-NS (58). In Pseudomonas strains, the hns gene product lacks the conserved consensus motif, in particular the conserved amino acid residues in the C-terminal domain of H-NS-like proteins (58). Interestingly, MvaT protein of Pseudomonas sp strain Y1000, which contains 18% amino acid identity with the E. coli H-NS protein and shows no significant homology with any known H-NS-related proteins (58,59), fully reversed the various phenotypes of E. coli related to hns mutations (59). These observations suggest that, despite low amino acid conservation, proteins functionally related to E. coli H-NS are likely to be widespread than thought previously.
The M. tuberculosis NAPs, Lsr2 and H-NS, encoded by Rv3597 and Rv3852 genes, respectively, have been overexpressed and purified from E. coli. Biochemical studies have shown that Lsr2 is a DNA binding protein (60), a function also shared by H-NS protein (53). Mycobacterium tuberculosis Lsr2 protein shares 4% amino acid identity with E. coli H-NS. Intriguingly, genetic studies have revealed that M. tuberculosis lsr2 was able to complement the phenotypes related to hns mutations in E. coli (60). Consequently, the authors have stated ‘unequivocally that Lsr2 is an H-NS-like protein’. Although M. tuberculosis Rv3852 encodes a strongly divergent H-NS compared to E. coli/Salmonella H-NS, it contains the KAAK and PAKK motifs found in the vertebrate histone H1/H5 family of proteins (61–63), thereby suggesting that M. tuberculosis H-NS may be positioned between the E. coli H-NS and histone H1 family of proteins. Likewise, M. tuberculosis 32 kDa HupB protein, a close homologue to bacterial HU-type DNA-binding protein, is similar in primary amino acid sequence to the human H1/H5 histones (64). Mycobacterium tuberculosis is a human pathogen and cannot survive outside eukaryotic cells. In the course of evolution and adaptation to the host environment, it is possible that gene duplication might have led to the formation of an hns gene with a sequence that is analogous similarity to the vertebrate histone H1/H5 family of proteins. Nonetheless, our results suggest that the Rv3852 gene product shares the same functional properties as members of H-NS family of proteins despite the unusually low amino acid conservation.
In Gram-negative bacteria, H-NS functions as a transcriptional repressor for a large number of genes, many of which encode proteins involved in stress response and virulence pathways (15,42). Although several lines of evidence suggest that H-NS binds DNA in a sequence-independent manner, the binding region coincides with the A/T-rich tracts (6,8–10). However, medium- and low-affinity binding sites have been mapped in the vicinity of the curved DNA (negative regulatory element of proU promoter) (28,65,66). Binding of H-NS to DNA traps RNA polymerase at the initiation stage of transcription; therefore, is conceivable that it can repress transcription through its DNA-binding and bridging activities (8–10,35). Characterization of the binding sites has revealed an unusual property. The binding sites could still be recognized by H-NS outside of their natural context. The insertion of the 10-bp site into a (G+C)-rich fragment led to base opening and the affinity of H-NS for this fragment increased, and the in vivo binding of H-NS was essentially restricted to (A+T)-rich DNA (20). These results are consistent with our findings that H-NS binds robustly to double-stranded DNA containing low percentage of GC base pairs (40%), but poorly to duplex DNA containing high GC content (70%). Mycobacterium tuberculosis H-NS, like E. coli H-NS (67), constrains negative superhelicity, suggesting a role for H-NS in DNA organization in the bacterial nucleoid and maintenance of genomic integrity in vivo.
The ability of E. coli and M. tuberculosis H-NS proteins to bind such a diverse array of DNA structures is rather unusual. Escherichia coli HU binds to a variety of DNA substrates much more avidly under stringent conditions (68). Specifically, binding of HU protein to the HJ has been shown to be 1000-fold stronger than linear DNA (68). However, there is limited knowledge about the structural and kinetic aspects of H-NS DNA interactions. To identify the precise structures recognized by M. tuberculosis and E. coli H-NS proteins, we used a series of DNA substrates likely to be present within the nucleoid structure. Notably, M. tuberculosis H-NS displayed higher affinity for the HJ. Similar experiments showed relatively weak binding to the DNA replication intermediates. It is indeed striking that H-NS displays high-affinity for the HJ, which points to its role in the repair of some specific DNA lesions during DNA replication and recombination. Interaction with ssDNA was much weaker, and the stability of ssDNA–H-NS complexes was significantly reduced in low salt, which can be attributed to the electrostatic screening of protein–protein and/or protein–DNA interactions. Nevertheless, the capacity to bind ssDNA can also be necessary for the function of H-NS in DNA replication and recombination mechanisms.
For our perspective, the mobility shift assay was especially useful for measuring protein–DNA complexes formed by E. coli and M. tuberculosis H-NS proteins in terms of discerning differences in the nature of the protein–DNA complexes. The specificity of E. coli H-NS for the DNA structures contrasts with M. tuberculosis H-NS under similar conditions. Some aspects of the complexes formed by both E. coli and M. tuberculosis H-NS with the HJ are especially intriguing. The formation of stepwise protein–DNA complexes suggests the recognition of a common binding motif between these two proteins. It is conceivable that in the stepwise manner of binding of H-NS to the HJ, the first complex may represent the binding of a dimer to two arms of the junction. If this proposal is correct, the second complex may result in binding of two dimers each to a pair of arms of the junction.
The horizontal transfer of DNA sequences provides bacteria with the opportunity for diversification and evolution leading to the development of new traits (69,70). To this end, bacteria must select the desired genes among the acquired DNA sequences for integration into the host genome. It has been estimated that a new sequence is acquired and stably maintained by enteric bacteria on an average of only once every several hundred thousand years (70), suggesting that only rarely is a new sequence favorably selected, and thus the integration of newly transferred genes has to be regulated. Our results directly address the question of how H-NS may contribute to the regulation underlying the integration of horizontally transferred genes into the host genome. Notably, our results show that H-NS may negatively regulate at two stages in the pathway of homologous recombination: (i) impede strand exchange promoted by RecA, and (ii) inhibit branch migration and/or compete for binding with the components of the HJ migration and/or resolution. A role for E. coli HU in homologous recombination has been demonstrated: the binding of HU to linear duplex DNA resulted in the inhibition of homologous paring promoted by its cognate RecA (71). Likewise, Hc1 nucleoid condensation protein from Chlamydia trachomatis selectively inhibited some of the RecA functions (72). However, we note that the ability of H-NS to act as a modulator of HR, by inhibiting strand exchange promoted by its cognate RecA, is different from its effect on site-specific recombination. Interestingly, H-NS stimulates Tn10 transposition in vitro by acting at the transition point between transposon excision and integration (73). H-NS also functions in four other transposition systems including IS903, Tn552, IS1 and bacteriophage Mu (74). Escherichia coli H-NS promotes transposition of IS903 and Tn552; whereas, it is inhibitory in the Mu system (74). These results suggest that H-NS acts on a number of transposition systems in different ways because of its ability to interact with other proteins and/or via its DNA structure-specific binding activity.
In summary, our findings have broad implications for the mechanism of H-NS action. In particular, the dual mode of action, that is, high-affinity binding to the HJ and suppression of strand exchange promoted by RecA can play a crucial role in the control or accuracy of DNA replication and recombination mechanisms. The ability of H-NS to inhibit strand exchange, together with specific binding to the HJ, may have synergistic effects in the overall pathway of homologous recombination.
FUNDING
European Community (CSI_LTB LSHP-CT-2007-037235); the Department of Biotechnology, New Delhi, under the ‘Centre of Excellence’ in research on mycobacteria; J. C. Bose National Fellowship (to K. M.). Funding for open access charge: Projects by the European Community (CSI_LTB LSHP-CT-2007-037235) and the Department of Biotechnology, New Delhi.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank the anonymous reviewers whose insightful comments have helped to improve the article, and members of our lab for critical and constructive suggestions. We appreciate the help of Dr K. Neelakanteshwar Patil with some of the experiments.
REFERENCES
- 1.Travers A, Muskhelishvili G. Bacterial chromatin. Curr. Opin. Genet. Dev. 2005;15:507–514. doi: 10.1016/j.gde.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Rouviere-Yaniv J, Gros F. Characterization of a novel, low-molecular-weight DNA-binding protein from Escherichia coli. Proc. Natl Acad. Sci. USA. 1975;72:3428–3432. doi: 10.1073/pnas.72.9.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drlica K, Rouviere-Yaniv J. Histone-like proteins of bacteria. Microbiol. Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dame RT. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol. Microbiol. 2005;56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 5.Luijsterburg MS, White MF, van Driel R, Dame RT. The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2008;43:393–418. doi: 10.1080/10409230802528488. [DOI] [PubMed] [Google Scholar]
- 6.Dorman CJ, Deighan P. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 2003;13:179–184. doi: 10.1016/s0959-437x(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 7.Azam TA, Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- 8.Dorman CJ. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 9.Dorman CJ. H-NS, the genome sentinel. Nat. Rev. Microbiol. 2007;5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- 10.Rimsky S. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 2004;7:109–114. doi: 10.1016/j.mib.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Luijsterburg MS, Noom MC, Wuite GJ, Dame RT. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J. Struct. Biol. 2006;156:262–272. doi: 10.1016/j.jsb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Williams RM, Rimsky S. Molecular aspects of the E. coli nucleoid protein, H-NS: a central controller of gene regulatory networks. FEMS Microbiol Lett. 1997;156:175–185. doi: 10.1111/j.1574-6968.1997.tb12724.x. [DOI] [PubMed] [Google Scholar]
- 13.Dorman CJ, Hinton JC, Free A. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 1999;7:124–128. doi: 10.1016/s0966-842x(99)01455-9. [DOI] [PubMed] [Google Scholar]
- 14.Hinton JC, Santos DS, Seirafi A, Hulton CS, Pavitt GD, Higgins CF. Expression and mutational analysis of the nucleoid-associated protein H-NS of Salmonella typhimurium. Mol. Microbiol. 1992;6:2327–2337. doi: 10.1111/j.1365-2958.1992.tb01408.x. [DOI] [PubMed] [Google Scholar]
- 15.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 16.Lejeune P, Danchin A. Mutations in the bglY gene increase the frequency of spontaneous deletions in Escherichia coli K-12. Proc. Natl Acad. Sci. USA. 1990;87:360–363. doi: 10.1073/pnas.87.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dri AM, Moreau PL, Rouvière-Yaniv J. Role of the histone-like proteins OsmZ and HU in homologous recombination. Gene. 1992;120:11–16. doi: 10.1016/0378-1119(92)90003-8. [DOI] [PubMed] [Google Scholar]
- 18.Ono S, Goldberg MD, Olsson T, Esposito D, Hinton JC, Ladbury JE. H-NS is a part of a thermally controlled mechanism for bacterial gene regulation. Biochem. J. 2005;391:203–213. doi: 10.1042/BJ20050453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JCD. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:746–752. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 21.Müller CM, Dobrindt U, Nagy G, Emödy L, Uhlin BE, Hacker J. Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J. Bacteriol. 2006;188:5428–5438. doi: 10.1128/JB.01956-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dame RT, Wyman C, Goosen N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 2000;28:3504–3510. doi: 10.1093/nar/28.18.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rimsky S. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 2004;7:109–114. doi: 10.1016/j.mib.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Oshima T, Ishikawa S, Kurokawa K, Aiba H, Ogasawara N. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 2006;13:141–153. doi: 10.1093/dnares/dsl009. [DOI] [PubMed] [Google Scholar]
- 25.Amit R, Oppenheim AB, Stavans J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys. J. 2003;84:2467–2473. doi: 10.1016/S0006-3495(03)75051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dame RT, Wuite GJL. On the role of H-NS in the organization of bacterial chromatin: from bulk to single molecules and back. Biophys. J. 2003;85:4146–4148. doi: 10.1016/S0006-3495(03)74826-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dame RT, Noom MC, Wuite GJL. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444:387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- 28.Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 2007;14:441–448. doi: 10.1038/nsmb1233. [DOI] [PubMed] [Google Scholar]
- 29.Esposito D, Petrovic A, Harris R, Ono S, Eccleston JF, Mbabaali A, Haq I, Higgins CF, Hinton JC, Driscoll PC, et al. H-NS oligomerization domain structure reveals the mechanism for high order self-association of the intact protein. J. Mol. Biol. 2002;324:841–850. doi: 10.1016/s0022-2836(02)01141-5. [DOI] [PubMed] [Google Scholar]
- 30.Badaut C, Williams R, Arluison V, Bouffartigues E, Robert B, Buc H, Rimsky S. The degree of oligomerization of the HNS nucleoid structuring protein is related to specific binding to DNA. J. Biol. Chem. 2002;277:41657–41666. doi: 10.1074/jbc.M206037200. [DOI] [PubMed] [Google Scholar]
- 31.Dame RT, Luijsterburg MS, Krin E, Bertin PN, Wagner R, Wuite GJ. DNA bridging: a property shared among H-NS like proteins. J. Bacteriol. 2005;187:1845–1848. doi: 10.1128/JB.187.5.1845-1848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barth M, Marschall C, Muffler A, Fischer D, Hengge-Aronis R. Role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of σS and many σS-dependent genes in Escherichia coli. J. Bacteriol. 1995;177:3455–3464. doi: 10.1128/jb.177.12.3455-3464.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dersch P, Kneip S, Bremer E. The nucleoid-associated DNA-binding protein H-NS is required for the efficient adaptation of Escherichia coli K-12 to cold environment. Mol. Gen. Genet. 1994;245:255–259. doi: 10.1007/BF00283274. [DOI] [PubMed] [Google Scholar]
- 34.Hommais F, Krin E, Laurent-Winter C, Soutourina O, Malpertuy A, Le Caer JP, Danchin A, Bertin P. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 2001;40:20–36. doi: 10.1046/j.1365-2958.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- 35.McGovern V, Higgins NP, Chiz RS, Jaworski A. H-NS over-expression induces an artificial stationary phase by silencing global transcription. Biochimie. 1994;76:1019–1029. doi: 10.1016/0300-9084(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh A, Paul K, Chowdhury R. Role of the histone-like nucleoid structuring protein in colonization, motility, and bile-dependent repression of virulence gene expression in Vibrio cholerae. Infect. Immun. 2006;74:3060–3064. doi: 10.1128/IAI.74.5.3060-3064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashino T, Ueguchi C, Mizuno T. Quantitative control of the stationary phase-specific sigma factor, sigma S, in Escherichia coli: involvement of the nucleoid protein, H-NS. EMBO J. 1995;14:594–602. doi: 10.1002/j.1460-2075.1995.tb07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shanado Y, Hanada K, Ikeda H. Suppression of gamma ray-induced illegitimate recombination in Escherichia coli by the DNA-binding protein H-NS. Mol. Gen. Genet. 2001;265:242–248. doi: 10.1007/s004380000399. [DOI] [PubMed] [Google Scholar]
- 39.Navarre WW, McClelland M, Libby SJ, Fang FC. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- 40.Stoebel DM, Free A, Dorman CJ. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology. 2008;154:2533–2545. doi: 10.1099/mic.0.2008/020693-0. [DOI] [PubMed] [Google Scholar]
- 41.Prabhakar S, Annapurna PS, Jain NK, Dey AB, Tyagi JS, Prasad HK. Identification of an immunogenic histone-like protein (HLPMt) of Mycobacterium tuberculosis. Tuber. Lung Dis. 1998;79:43–53. doi: 10.1054/tuld.1998.0004. [DOI] [PubMed] [Google Scholar]
- 42.Lee BH, Murugasu-Oei B, Dick T. Upregulation of a histone-like protein in dormant Mycobacterium smegmatis. Mol. Gen. Genet. 1998;260:475–479. doi: 10.1007/s004380050919. [DOI] [PubMed] [Google Scholar]
- 43.Mukherjee A, Bhattacharyya G, Grove A. The C-terminal domain of HU-related histone-like protein Hlp from Mycobacterium smegmatis mediates DNA end-joining. Biochemistry. 2008;47:8744–8753. doi: 10.1021/bi800010s. [DOI] [PubMed] [Google Scholar]
- 44.Shires K, Steyn L. The cold-shock stress response in Mycobacterium smegmatis induces the expression of a histone-like protein. Mol Microbiol. 2001;39:994–1009. doi: 10.1046/j.1365-2958.2001.02291.x. [DOI] [PubMed] [Google Scholar]
- 45.Chen JM, Ren H, Shaw JE, Wang YJ, Li M, Leung AS, Tran V, Berbenetz NM, Kocíncová D, Yip CM, et al. Lsr2 of Mycobacterium tuberculosis is a DNA-bridging protein. Nucleic Acids Res. 2008;36:2123–2135. doi: 10.1093/nar/gkm1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaze MB, Muniyappa K. RecA protein of Mycobacterium tuberculosis possesses pH-dependent homologous DNA pairing and strand exchange activities: implications for allele exchange in mycobacteria. Biochemistry. 1999;38:3175–3186. doi: 10.1021/bi9819125. [DOI] [PubMed] [Google Scholar]
- 47.Tripathi P, Anuradha S, Ghosal G, Muniyappa K. Selective binding of meiosis-specific yeast Hop1 protein to the Holliday junctions distorts the DNA structure and its implications for junction migration and resolution. J. Mol. Biol. 2006;364:599–611. doi: 10.1016/j.jmb.2006.08.096. [DOI] [PubMed] [Google Scholar]
- 48.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry C.E., III, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 49.Sette M, Spurio R, Trotta E, Brandizi C, Brandi A, Pon CL, Barbato G, Boelens R, Gualerzi CO. Sequence-specific recognition of DNA by the C-terminal domain of nucleoid-associated protein H-NS. J. Biol. Chem. 2009;284:30453–30462. doi: 10.1074/jbc.M109.044313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Del Portillo P, Reyes A, Salazar L, del Carmen Menéndez M, García MJ. Genomics and proteomics, Chapter 4. In: Ritacco V, Leão SC, Palomino JC, editors. Tuberculosis 2007, A Medical Text Book. London: Pitman Medical and Scientific Publishing Co. Ltd; 2007. pp. 113–156. [Google Scholar]
- 51.Rould E, Muniyappa K, Radding CM. Unwinding of heterologous DNA by RecA protein during the search for homologous sequences. J. Mol. Biol. 1992;226:127–139. doi: 10.1016/0022-2836(92)90129-8. [DOI] [PubMed] [Google Scholar]
- 52.Mukherjee A, Sokunbi AO, Grove A. DNA protection by histone-like protein HU from the hyperthermophilic eubacterium Thermotoga maritime. Nucleic Acids Res. 2008;36:3956–3968. doi: 10.1093/nar/gkn348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Werlang ICR, Schneider CZ, Mendonc JD, Palma MS, Luiz A, Basso LA, Santos DS. Identification of Rv3852 as a nucleoid-associated protein in Mycobacterium tuberculosis. Microbiology. 2009;155:2652–2663. doi: 10.1099/mic.0.030148-0. [DOI] [PubMed] [Google Scholar]
- 54.Prakash P, Aruna B, Sardesai AA, Hasnain SE. Purified recombinant hypothetical protein coded by open reading frame Rv1885c of Mycobacterium tuberculosis exhibits a mono-functional AroQ class of periplasmic chorismate mutase activity. J. Biol. Chem. 2005;280:19641–19648. doi: 10.1074/jbc.M413026200. [DOI] [PubMed] [Google Scholar]
- 55.Sasso S, Ramakrishnan C, Gamper M, Hilvert D, Kast P. Characterization of the secreted chorismate mutase from the pathogen Mycobacterium tuberculosis. FEBS Lett. 2005;272:375–389. doi: 10.1111/j.1742-4658.2004.04478.x. [DOI] [PubMed] [Google Scholar]
- 56.Qamra R, Prakash P, Aruna B, Hasnain SE, Mande SC. The 2.15 A crystal structure of Mycobacterium tuberculosis chorismate mutase reveals an unexpected gene duplication and suggests a role in host-pathogen interactions. Biochemistry. 2006;45:6997–700. doi: 10.1021/bi0606445. [DOI] [PubMed] [Google Scholar]
- 57.Argyrides Argyrou A, Jin L, Siconilfi-Baez L, Angeletti RH, Blanchard JS. Proteome-wide profiling of isoniazid targets in Mycobacterium tuberculosis. Biochemistry. 2006;45:13947–13953. doi: 10.1021/bi061874m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tendeng C, Bertin PN. H-NS in Gram-negative bacteria: a family of multifaceted proteins. Trends Microbiol. 2003;11:511–518. doi: 10.1016/j.tim.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 59.Tendeng C, Soutourina OA, Danchin A, Bertin PN. MvaT proteins in Pseudomonas spp.: a novel class of H-NS-like proteins. Microbiology. 2003;149:3047–3050. doi: 10.1099/mic.0.C0125-0. [DOI] [PubMed] [Google Scholar]
- 60.Gordon BRG, Imperial R, Wang L, Navarre WW, Liu J. Lsr2 of Mycobacterium represents a novel class of H-NS-like proteins. J. Bacteriol. 2008;190:7052–7059. doi: 10.1128/JB.00733-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Erard M, Lakhdar-Ghazal J, Amalric J. Repeat peptide motifs which contain p-turns and modulate DNA condensation in chromatin. Eur. J. Biochem. 1990;191:19−25. doi: 10.1111/j.1432-1033.1990.tb19088.x. [DOI] [PubMed] [Google Scholar]
- 62.Khadake JR, Rao MRS. Condensation of DNA and chromatin by an SPKK-containing octapeptide repeat motif present in the C-terminus of histone H1. Biochemistry. 1997;36:1041–1051. doi: 10.1021/bi961617p. [DOI] [PubMed] [Google Scholar]
- 63.Kasinsky HE, Lewis JD, Dacks JB, Ausio J. Origin of H1 linker histones. FASEB J. 2001;15:34–42. doi: 10.1096/fj.00-0237rev. [DOI] [PubMed] [Google Scholar]
- 64.Cohavy O, Harth G, Horwitz M, Eggena M, Landers C, Sutton C, Targan SR, Braun J. Identification of a novel mycobacterial histone H1 homologue (HupB) as an antigenic target of pANCA monoclonal antibody and serum immunoglobulin A from patients with Crohn's; disease. Infect. Immun. 1999;67:6510–6517. doi: 10.1128/iai.67.12.6510-6517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gowrishankar J, Manna D. How is osmotic regulation of transcription of the Escherichia coli proU operon achieved: a review and a model. Genetica. 1996;97:363–378. doi: 10.1007/BF00055322. [DOI] [PubMed] [Google Scholar]
- 66.Rajkumari K, Gowrishankar J. In vivo expression from the RpoS-dependent P1 promoter of the osmotically regulated proU operon in Escherichia coli and Salmonella enterica serovar Typhimurium: activation by rho and hns mutations and by cold stress. J. Bacteriol. 2001;183:6543–6550. doi: 10.1128/JB.183.22.6543-6550.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Owen-Hughes TA, Pavitt GD, Santos DS, Sidebotham JM, Hulton CS, Hinton JC, Higgins CF. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell. 1992;71:255–265. doi: 10.1016/0092-8674(92)90354-f. [DOI] [PubMed] [Google Scholar]
- 68.Kamashev D, Rouviere-Yaniv J. The histone-like protein HU binds specifically to DNA recombination and repair intermediates. EMBO J. 2000;19:6527–6535. doi: 10.1093/emboj/19.23.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Groisman EA, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 70.Smets BF, Barkay T. Horizontal gene transfer: perspectives at a crossroads of scientific disciplines. Nat. Rev. Microbiol. 2005;3:675–678. doi: 10.1038/nrmicro1253. [DOI] [PubMed] [Google Scholar]
- 71.Ramdas J, Mythili E, Muniyappa K. RecA protein promoted homologous pairing in vitro. Pairing between linear duplex DNA bound to HU Protein (nucleosome cores) and nucleoprotein filaments of recA protein-single-stranded DNA. J. Biol. Chem. 1989;264:17395–17400. [PubMed] [Google Scholar]
- 72.Ennis DG, Woodgate R, Shi M. Selective inhibition of RecA functions by the Hc1 nucleoid condensation protein from Chlamydia trachomatis. FEMS Microbiol. Lett. 2006;182:279–283. doi: 10.1111/j.1574-6968.2000.tb08908.x. [DOI] [PubMed] [Google Scholar]
- 73.Wardle SJ, O’Carroll M, Derbyshire KM, Haniford DB. The global regulator H-NS acts directly on the transpososome to promote Tn10 transposition. Genes Dev. 2005;19:2224–2235. doi: 10.1101/gad.1338905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Falconi M, McGovern V, Gualerzi C, Hillyard D, Higgins NP. Mutations altering chromosomal protein H-NS induce mini-Mu transposition. New Biol. 1991;3:615–625. [PubMed] [Google Scholar]
- 75.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36(Web Server issue):W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview version 2: a multiple sequence alignment and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]