Abstract
Mammalian mitochondria contain their own genome that is almost fully transcribed from both strands, generating polycistronic RNA units that are processed and matured. The mitochondrial mRNA is modified by oligo- or polyadenylation at the 3′ termini, but the exact function of this post-transcriptional addition is unclear. Current debate focuses on the role of polyadenylation in transcript stability. An equally likely function that has received little attention is that, as in the cytosol of eukaryotes, polyadenylation facilitates translation in the mitochondrion. To address this issue, we have targeted cytosolic proteins to the mitochondrion, a poly(A) specific 3′ exoribonuclease, mtPARN, and a poly(A)binding protein, mtPABP1. Removal of the 3′ adenylyl extensions had a variable effect on mt-mRNA steady-state levels, increasing (MTND1, 2, 5) or decreasing (MTCO1, 2, RNA14) certain species with minimal effect on others (RNA7, MTND3). Translation was markedly affected, but interpretation of this was complicated by the concomitant 3′ truncation of the open reading frame in most cases. Coating of the poly(A) tail by mtPABP1, however, did not lead to transcript decay but caused a marked inhibition of mitochondrial translation. These data are consistent with endogenous RNA-binding factor(s) interacting with the poly(A) to optimize mitochondrial protein synthesis.
INTRODUCTION
Every nucleated human cell contains many hundreds or thousands of copies of the mitochondrial genome, mtDNA. This small molecule is almost fully transcribed from both strands, producing polycistronic RNA units that are co-transcriptionally processed and matured into the three standard classes, mitochondrial (mt)-ribosomal, -transfer and -messenger RNA (1,2). Transcripts that encode polypeptides (mt-mRNAs) are normally poly- or oligoadenylated, a process that is required to produce the UAA termination codon in 7 out of the 11 major mt-mRNA species. Beyond this stop codon formation, the function of this simple maturation event in mammalian mitochondria is elusive (3,4). As has been well documented, polyadenylation is required to promote both stability and translatability of mRNAs in the cytosol of eukaryotes. In contradistinction, however, in plant organelles and prokaryotes the addition of 3′ adenylates is a harbinger of RNA degradation. There is no such maturation of mt-mRNAs in the budding yeast Saccharomyces cerevisiae, although all species contain a dodecamer sequence towards their 3′ termini (5). Finally, to confound the issue further, studies over the recently identified Trypanosoma brucei kinetoplast poly(A) polymerase KPAP1, revealed that short poly(A) tails are required and sufficient to maintain the steady-state levels of partially edited, fully edited and never-edited mRNAs, whereas they are not involved in stabilizing pre-edited mRNAs (6). Upon completion of the editing process, the short poly(A) tails are extended as (A/U) heteropolymers into structures previously thought to be long poly(A) tails. Authors of this report implied that these A/U structures could be essential for generating translationally competent, fully edited mRNAs.
Analysis of human fibroblast RNA from a patient with a microdeletion in the termination codon of one species of mt-mRNA, RNA14, showed this species to be less stable and carried a shortened poly(A) tail (7). Although this appeared to support a role for the poly(A) tail in stability of mt-mRNA, it was noted that the deadenylation was translation dependent, suggesting that the lack of stability may have been due to run through of the mitoribosome into the tail, promoting the degradation of the unprotected transcript. Conflicting results have been reported using siRNA-based approaches to deplete the natural human mitochondrial poly(A) polymerase (8,9). In the former example, shortened poly(A) extensions were reported not to affect transcript stability, whilst the latter reported decreased steady state levels of analysed mt-mRNAs with a concomitant decrease in translation. It is possible these contradictory results have been complicated by the retention of at least a minor population of polyadenylated mt-mRNA species, possibly implicating a second poly(A) polymerase activity (10).
A second function that could be modulated by polyadenylation is mitochondrial translation. In the cytosol, a protein bridge is formed between poly(A) binding factors and proteins that interact with the 5′7-methylguanosine cap (11). Mitochondrial mRNA do not carry any 5′ modification and to date, there has been no report of a mitochondrial poly(A) binding protein. Bioinformatic analyses reveal no obvious candidate, although several metabolic mitochondrial enzymes have been shown to be capable of binding RNA and of poly(A) sequences (12–14).
In an attempt to resolve whether oligo- or polyadenylation of mt-mRNAs does indeed affect the stability or translatability of transcripts, we have engineered and mitochondrially targeted two human proteins that are normally involved in mRNA expression in the cytosol: PARN, a cytosolic 3′–5′ exoribonuclease with reported specificity to poly(A) sequences and the cytosolic poly(A) binding protein, PABPC1, reasoning that if a known poly(A) binding protein could be re-directed to the mitochondrial matrix it may be possible to compete against endogenous proteins for the poly(A) sequence and inhibit translation.
MATERIALS AND METHODS
Preparation of constructs
To initiate construction of an inducible FLAG-tagged mitochondrially targeted PABP1 (mtPABP1), the open reading frame of PABPC1 (clone MGC: 43692, IMAGE 5271298) was PCR amplified. PARN constructs were polymerase chain reaction (PCR)-amplified from a cDNA source equivalent to NM_002582 to generate an amplicon containing the entire PARN open reading frame (639 aa). Subsequently, mtPARNΔC (430 aa) was produced. Both proteins also contained a C-terminal FLAG tag. All cloning steps, vectors and primers used for PCR amplication are given in Supplementary Data.
Site-directed mutagenesis of mtPABP1
Mutagenesis was performed with the QuikChange II Site-Directed Mutagenesis Kit (Stratagene), essentially following manufacturer’s guidelines. F337V mutant was generated with the following pair of primers; F 5′-gca aag ggt ttg gtg ttg tat gtt tct cc-3′, R 5′-gga gaa aca tac aac acc aaa ccc ttt gc-3′ and pcDNA5/FRT/TOmtPABP1 as the template. Y56V/F142V was generated by two rounds of the mutagenesis. First with the following pair of primers; F 5′-ctt ggg cta cgc ggt tgt gaa ctt cca g-3′, R 5′-ctg gaa gtt cac aac cgc gta gcc caa g-3′ and pcDNA5/FRT/TO mtPABP1 as template. Second round, with the primers introducing F142V (F 5′-caa ggg cta tgg agt tgt aca ctt tga gac-3′, R 5′-gtc tca aag tgt aca act cca tag ccc ttg-3′) and pcDNA5/FRT/TO Y56V as template. All mutations were confirmed by sequencing of both DNA strands.
Generating Flp-In™ T-Rex™ expression cell lines
HEK293 Flp-In™ T-Rex™ (HEK293T; Invitrogen) cells were grown as a monolayer in DMEM (Sigma, D6429) supplemented with 10% (v:v) FCS, 50 µg/ml uridine and non-essential amino acids (standard growth medium). Untransfected HEK293T cells were maintained in presence of blasticidin S (10 µg/ml) and zeocin (100 µg/ml). The cultures were grown in vented flasks at 37°C in a 5% CO2 humidified atmosphere. Medium was replaced every 2–3 days.
To generate stable, inducible expression cell lines, HEK293T were co-transfected with pcDNA5/FRT/TO harbouring the mtPARN, mtPABP1 or mtLUC derivatives, and pOG44 (Invitrogen) according to the manufacturer’s recommendations, using SuperFect (QIAGEN) transfection reagent following the manufacturer’s guidelines.
After transfection, cells were cultured in DMEM (Sigma) with hygromycin B (100 µg/ml) and blasticidin S (10 µg/ml). After approximately 3–4 weeks of selection clonal foci were identified, transferred into separated wells. Expression was induced by addition of tetracycline (1 µg/ml) to the growth medium for the indicated time.
Cell lysate and mitochondrial preparation
Production of mitochondria and cell lysates were as described in (15). Proteinase K treatment was carried out in a 50 µl volume of isolation buffer lacking bovine serum albumun (BSA) containing 50 µg freshly isolated mitochondria and proteinase K (20 ng/µl final). Reactions were incubated on ice for 30 min, stopped by addition of phenylmethylsulphonyl fluoride (PMSF; 1 mM), mitochondria were washed twice in 1 ml of the isolation buffer (-BSA) and re-suspended in the desired buffer. Where necessary, mitochondria were solubilized by addition of Triton X-100 to 1% v:v.
RNA isolation and northern blot procedure
RNA was prepared by TRIZOL (Invitrogen) extraction following manufacturer’s recommendations. Northerns were performed as described (16) and signals quantified by PhosphorImager analysis (GE Healthcare).
Analysis of proteins by western blot and blue native gel electrophoresis
After separation by standard sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE), protein species were immobilized on polyvinylidene difluoride (PVDF) membrane (Immobilon-P, Millipore Corporation) by wet transfer (500 mA, 2.5 h at 4°C) in Towbin transfer buffer. Membranes were blotted with appropriate primary antibodies [anti-FLAG M2 (Sigma); anti-β-actin (Sigma); anti-MRPL3 (Abcam); anti-MRPS18b (ProteinTech Group); anti-IF3mt; anti-porin (Molecular Probes)]; monoclonals against COX2, NDUFA9, NDUFB8, SDH 70 K, core 2 complex III (MitoSciences), followed by horseradish peroxidase (HRP)-conjugated secondary antibodies (DAKO) and visualised by ECL-plus (GE Healthcare). One-dimension blue native gel electrophoresis was performed as described in (17) with the modification that 50 µg protein was loaded per lane.
Immunoaffinity purification of FLAG tagged fusion proteins and RNA isolation
Immunoprecipitations were performed using the FLAG Tagged Protein Immunoprecipitation Kit (FLAGIPT-1 Sigma), essentially following manufacturer’s recommendations. Typically, 1.5 mg of proteinase-K-treated mitochondria isolated from HEK293T cells expressing mtPABP1 or mtLUC were re-suspended in 0.5 ml of lysis buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) supplemented with Complete EDTA free Protease Inhibitor Cocktail (Roche) and RNAguard (GE Healthcare). Lysis was performed for 20 min rotating at 4°C, followed by centrifugation at 12 000g for 10 min to remove insoluble material. Mitochondrial lysates were incubated with anti-FLAG M2-Agarose Affinity Gel (Sigma) and incubated for 30 min, rotating at 4°C. Unbound material was removed by series of washes in 50 mM Tris–HCl pH 7.4, 150 mM NaCl, 50 µg/ml heparin. Elution of bound FLAG tagged recombinant protein was performed with FLAG peptide (Sigma) for 45 min at 4°C with constant agitation.
To isolate RNA associated with FLAG tagged proteins, the eluted fractions were subjected to Trizol (Invitrogen) extraction. Equal volumes of extracted RNA were analysed by northern blot. To remove any possible DNA contaminations, RNA samples were treated with 1 U of RNase-Free DNase I (Epicentre Biotechnologies) for 30 min at 37°C, phenol–chloroform extracted and precipitated. Isolated RNA was reversed transcribed using random hexamers and Superscript II preamplication kit (Invitrogen) following manufacturer’s guidelines.
Mitochondrial poly(A)-tail length assay
The technique was performed as described in (7). The additional primers used in this paper to analyse the following transcripts were as follows: MTCO1, first primer, (nt 7151–7170), nested radiolabelled primer (nt 7488–7504); MTCO2, first primer (nt 8155–8172), nested radiolabelled primer (nt 8265-8284); MTND4 first primer (nt 11 717–11 736), nested radiolabelled primer (nt 12 108–12 125). An annealing temperature of 54°C was used for all reactions except with MTCO2 first primer, which was 50°C.
RNA circularization assays
The RNA ligation was adapted from (18). Reactions were performed in 12.5 µl 50 mM HEPES pH 8.3, 10 mM MgCl2, 3.5 mM DTT, 3.4 mM ATP, 1 mg/ml BSA, 10% DMSO, RNA (2 µg), 20 U T4 RNA Ligase (New England Biolabs). After 3 h at 37°C, reactions were phenol extracted, ethanol precipitated, washed and re-suspended in 25 µl of DEPC-treated water. Reverse transcription was effected with Superscript II (Gibco BRL) and cDNA generated with gene specific primers.
Primers for reverse transcription (RT) and subsequent PCR were: MTND3 RT-5′-aggccagacttagg and PCR F-5′-gagtgactacaaaaaggattagac/R-5′-ctaggatgatgattaataagaggg; MTCO2 RT-5′-gtaaaggatgcgtag and PCR F-5′-ctccttgacgttgacaatcgag/R-5′-gcaggatagttcagacggtttc; RNA14 RT-5′-gacgagtccgagg and PCR 5′-gcccacttcttaccacaaggc/R-5′-acttgttgggtggtgattagtc.
Cell proliferation assays
Untransfected cells were used to assess the effect of tetracycline on cell viability. To measure growth in standard growth conditions, HEK293T cells expressing mtPABP1 or mtLUC were induced for 5 days in standard growth conditions prior to the assay. To measure growth in conditions forcing oxidative respiration, cells were pre-induced with 1 µg/ml of tetracycline in standard medium for 5 days. Cells were then transferred to DMEM medium lacking glucose (Sigma) supplemented with 10% (v:v) dialysed FCS, 0.9 mg/ml galactose, 0.11 mg/ml sodium pyruvate and 1 µg/ml tetracycline for 24 h before the assay. Cells were then plated in six-well plates and grown with tetracycline in galactose (2 × 105/plate) or glucose containing medium (1 × 105/plate). Cells were harvested from replicate plates on alternate days and total cell counts made using a Neubauer haemocytometer.
35S-metabolic labelling of mitochondrial proteins
This technique was adapted from (20) with some major modifications. Stably transfected HEK293T cells expressing mtPABP1 or mtLUC were induced with tetracycline (1 µg/ml) for 3 or 5 days prior to 35S labelling. Growth medium was replaced with methionine/cysteine-free DMEM (Sigma) supplemented with 2 mM L-glutamine, 50 µg/ml uridine and non-essential amino acids. The cells were incubated for 2 × 10 min in this medium before transfer to fresh methionine/cysteine-free DMEM (Sigma) medium containing 10% (v:v) dialysed FCS and emetine dihydrochloride (100 µg/ml, final). Cells were incubated for 10 min before addition of 220 µCi of [35S]-methionine/cysteine mix (PerkinElmer EasyTag express protein labelling mix, 1175 Ci/mmol) per millililtre of medium. Labelling was performed for various amounts of time (10–45 min) before replacing the medium with chilled standard growth medium supplemented with 7.5 µg/ml methionine for two short washes. All protein samples (25–50 µg) were separated by 15% SDS-PAGE and products visualised and quantified with a PhosphorImager system with ImageQuant software (Molecular Dynamics, GE Healthcare).
RESULTS
Loss of poly(A) sequences has a variable effect on transcript stability
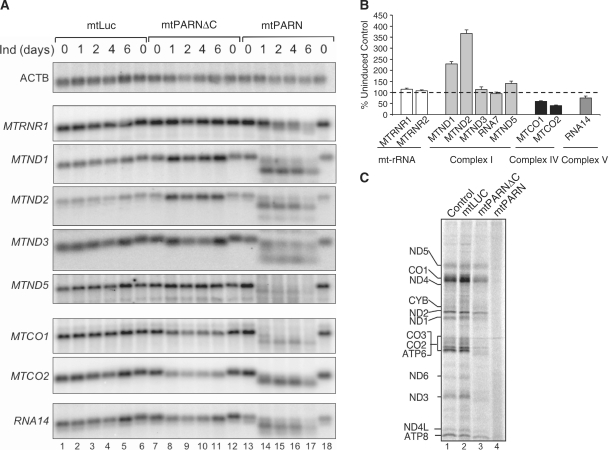
To determine the role of the 3′ poly(A) tail on mRNA stability, the 3′–5′ poly(A) specific exoribonuclease PARN (uniprot O95453) was engineered to target mitochondria in human HEK293T cells. The pcDNA5/FRT/TO vector was manipulated to express a PARN fusion protein with the widely used N-terminal pre-sequence from Neurospora crassa ATP9 and C-terminal FLAG tag as described in ‘Materials and Methods’ section. To control for any effects due to targeting of a soluble exogenous protein to mitochondria, a similar design was used to generate a C-terminal FLAG tagged mitochondrially directed luciferase, mtLUC. Following induction of transfectants, both proteins were shown to be localized within isolated mitochondria and were resistant to exogenous protease K (Supplementary Figure S1). During induction over a time course of 6 days, total RNA was isolated and analysed by northern blot. As shown in Figure 1 (lanes 14–17), mtPARN degrades mitochondrial RNA substantially. Further, circularization assays followed by sequence analysis showed the degradation to be solely from the 3′ terminus as expected, but this did not stop at the end of the poly(A) additions, continuing into the open reading frames of most transcripts (data not shown). In addition, as is shown with the 12 S probe (mtRNR1), degradation is restricted neither to the poly(A) sequences nor to polyadenylated mt-mRNAs.
Figure 1.
(A) Induction of mtPARN causes a variable effect on the steady-state level of mitochondrial transcripts. Stable HEK293T transfectants expressing mtLUC (lanes 1–-6), mtPARN (lanes 13–18) or the C-terminal truncated mtPARN (mtPARNΔC, lanes 7–12) were induced the number of days indicated and isolated RNA (10 µg) was subjected to northern blot analysis with the indicated probes as detailed in ‘Materials and Methods’ section. Probe from transcript encoding β-actin (ACTB) was used a loading control. (B) Northern blots were performed with the indicated probes on at least three independent RNA isolations following induction of mtPARNΔC. Quantification of steady-state mt-RNA was calculated by PhosphorImager analysis as a percentage of uninduced control. Probes were targeted against mt-rRNA or mt-mRNA encoding components of respiratory chain complexes as indicated, below. (C) Induction of mtPARN affects mitochondrial protein synthesis. Transfectants were induced for 2 days prior to in vivo mitochondrial protein synthesis with 35S methionine/cysteine for 30 min as detailed. Equal amounts of mitochondrial lysate (50 µg) were separated through a 15% denaturing SDS-PAGE, the gel dried and exposed to PhosphorImager prior to visualization. Individual polypeptides were designated by their mobility (20).
As partially degraded products were not observed to be readenylated (data not shown), it was likely that degradation in this context was processive. To try and reduce processivity without losing activity, we generated a C-terminal truncated construct, exploiting the suggestion that the C-terminal cap-binding domain may interact with the R3H domain to increase processivity even in the absence of the 5′ cap structure (21). This protein, mtPARNΔC, targeted successfully to mitochondria (Supplementary Figure S1) and reduced the size of mt-mRNA products as shown in Figure 1 (lanes 8–11). Compared to mtPARN, little variation in steady-state levels of mt-mRNAs were noted between Day 1 and 6 in the time course, consistent with the relatively short turnover of mt-mRNAs (16). Following mtPARNΔC induction, sequence data of clones from numerous transcripts confirmed that all products retained the correct 5′ terminus irrespective of what species was analysed, but lost the majority of polyadenylated 3′ extensions (Supplementary Table S1). As shown in Figure 1B, loss of the poly(A) extensions had a variable effect on the stability of mt-mRNA species, ranging from an average increase in steady-state level of 368±17% for MTND2 during the 6-day induction, to a decrease to 41±3% starting levels for MTCO2. MTND3 showed only a minor variation of 114±12%. Levels of both mt-rRNA species were also very minimally affected (MTRNR1 115±2%; MTRNR2 115±5%).
Translation of truncated mt-mRNA
Polyadenylation of mt-mRNA, therefore, is not an absolute requirement for stability in human mitochondria. Is it possible that polyadenylation is necessary to promote transcript translation? Full-length mtPARN severely truncated most mt-mRNA species, degrading well beyond the poly(A) termini and in to the open reading frame (Figure 1). Further, both 12 S (MTRNR1) and 16S mt-rRNA (MTRNR2, data not shown) are also degraded. Consequently, no mitochondrial protein synthesis occurs in these cells (Figure 1C, lane 4). Protein synthesis in mitochondria containing mtPARNΔC (Figure 1C, lane 3) is more informative but as polyadenylation immediately abuts the termination codon in nine ORFs, the exonucleolytic activity of mtPARNΔC still extended into the ORF in most cases. For example, all 21 MTND2 clones rescued post mtPARNΔC expression had lost their termination codon (Supplementary Table S1), although the ORF remained almost intact. Interestingly, whilst translation is clearly affected presumably due to mitoribosomal stalling, it is not lost completely. Sequences suggest that the ND2 gene product that is observed in Figure 1C will be minimally truncated and we speculate that the decrease in translation is due to a problem with recycling as the mitochondrial ribosome will be left without a codon to translate in the A site. There are a limited number of mRNA species that have 3′ untranslated regions (UTRs), amongst these are MTND5 and the upstream ORFs (encoding ATP8 and ND4) of the two bicistronic units RNA14 and RNA7. In these three cases, the coding sequences were not degraded and so translation was relatively spared. MTCO1 also has a 3′ UTR and sequence analysis revealed 96% of MTCO1 transcripts to have retained both the stop codon and at least part of the 72-nt 3′ UTR. Translation of MTCO1 however, was markedly affected but this may reflect the loss of potential interactions with a translational activator as discussed below.
The cytosolic poly(A) binding protein, PABP1, can be targeted and imported into human mitochondria
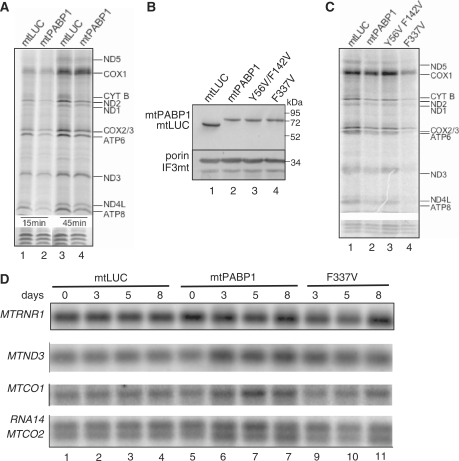
As the loss of the termination codon from most transcripts made it impossible to assess the role of polyadenylation in translation, a second approach was employed. Difficulties have also been previously reported where siRNA-mediated depletion of the mitochondrial poly(A) polymerase was unable to completely remove polyadenylated extensions (8,10). As an alternate approach, we reasoned that association of a targeted poly(A) binding protein to mt-mRNA would compete for any interaction that may be required for the poly(A) extensions to mediate mitochondrial translation. Therefore, to determine the effect of coating the polyadenylated termini of human mt-mRNA with an RNA-binding protein, the major human cytosolic poly(A) binding protein, PABPC1 (uniprot P11940), was engineered to facilitate mitochondrial targeting and import as described in the ‘Materials and Methods’ section (mtPABP1). Following transfection, clones were identified that expressed mtPABP1 and mtLUC in response to tetracycline induction. To confirm mitochondrial localization, mitochondria were isolated from induced cells and subjected to protease K shaving. As shown in Figure 4B, mtPABP1 located to the mitochondrial fraction and was resistant to the addition of protease.
Figure 4.
Occlusion of the poly(A) tail inhibits mitochondrial translation. (A) In vivo mitochondrial translation assays were performed for 15 min (lanes 1 and 2) and 45 min (lanes 3 and 4) exposure to 35S methionine/cysteine after expression of mtPABP1 (lanes 2 and 4) or mtLUC (lanes 1 and 3) for 5 days as described in ‘Materials and Methods’ section. Cell lysates (50 µg) were separated through a 15% denaturing PAG, visualized and quantified using ImageQuant software following exposure to a PhosphorImager cassette. Individual polypeptides were designated on the basis of their mobility (20). To confirm equal loading, a small section of the gel is shown after exposure following Coomassie staining. (B) mtPABP1 and variants are expressed and imported into mitochondria with equal efficiency. Transfectants were induced for 3 days, mitochondria isolated and proteinase K treated as described in ‘Materials and Methods’ section. Resulting mitochondrial lysates (10 µg) were separated by 12% denaturing SDS-PAGE. Western blots were performed with anti-FLAG antibody to detect mtPABP1 and variants. Equal loading was confirmed using two antibodies specific to mitochondrial proteins, the outer membrane protein VDAC (porin) and the soluble matrix marker translation initiation factor 3 (IF3mt). Molecular weight size markers are indicated. (C) Similar in vivo labelling experiments were performed (10-min pulse) on the indicated transfectants after 3 days induction. Lysate (25 µg) was separated and visualized as described in (A). Lane 3, variant mtPAPB1(Y56V/F142V); lane 4, variant mtPABP1(F337V). (D) Induction of mtPAPB1 and variant mtPABP1(F337V) do not increase the turnover of mt-mRNA. RNA was isolated from the indicated transfectants after 0-, 3-, 5- or 8-day induction and aliquots (10 µg) subjected to northern blot analysis as detailed in ‘Materials and Methods’ section. The indicated mt-mRNA probes were used. A probe to the 12 S mt-rRNA (MTRNR1) was used as loading control.
Expression of mtPABP1 causes a severe mitochondrial phenotype
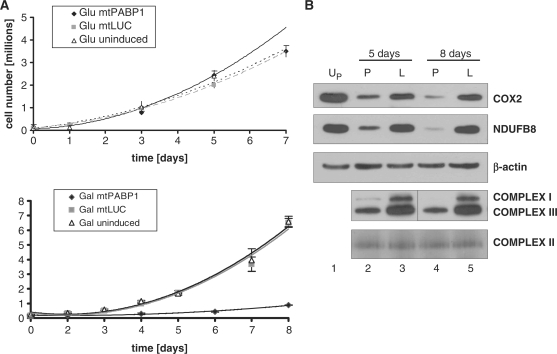
It was immediately noticeable that on induction of mPABP1, growth media became highly acidic, suggesting the cells were acidotic but on media replacement every 2 days there was only limited evidence of cell death. To measure any growth defect likely to be due to mitochondrial OXPHOS dysfunction, growth curves were produced either with standard concentrations of glucose as a carbon source, or in media substituting glucose for galactose, which forces cells to depend more heavily on oxidative phosphorylation for ATP production. Initially, all cells were grown in glucose and induced for 5 days in tetracycline. Cells were then re-plated and grown in either glucose (∼1 × 105 cells/well) or galactose (2 × 105 cells/well) media as detailed in ‘Materials and Methods’ section. As shown in Figure 2A (upper panel), growth in glucose was similar, irrespective of induction of either construct, providing that the growth media was changed after every 2 days. In galactose, however, growth was severely compromised but only in cells expressing mtPABP1, suggestive of mtPABP1 expression causing a severe defect in oxidative metabolism (Figure 2A, lower panel).
Figure 2.
Expression of mtPABP1 causes a severe mitochondrial OXPHOS defect. (A) HEK293T transfectants expressing mtPABP1 or mtLUC were induced over 5 days in standard growth media and plated into 10 wells at ∼1 × 105 (glucose, upper panel) or 2 × 105 (galactose, bottom panel) cells/well. Ten counts were performed on duplicate wells at the indicated time points and are shown as the average ± SEM. Uninduced mtPABP1 transfectants (Glu uninduced, upper panel; Gal uninduced, lower panel) were seeded and counted in a similar fashion. (B) Samples were prepared from transfectants expressing mtLUC (L, lanes 3 and 5) or mtPABP1 (P, lanes 2 and 4) for the indicated times and subjected to western blotting following SDS- (top three panels) or blue native (bottom two panels) gel electrophoresis as detailed in ‘Materials and Methods’ section. A sample from the uninduced mtPABP1 (UP, lane 1) is also shown. Blots of the denatured proteins were probed with antibodies to complex IV (COX2) and complex I (NDUFB8). The native gel is probed with antibodies to complexes I (NDUFA9) and III (Core 2). Loading controls are performed with antibodies to β-actin and complex II (SDH 70 kDa).
Steady-state levels of respiratory complexes are depleted on expression of mtPABP1
To determine why expression of mtPABP1 causes a respiratory defect, cells were induced for varying periods of time. Western blots of whole cell lysates were then probed with antibodies specific to several members of the respiratory chain. Figure 2B shows a substantial decrease in subunits of complexes I (NDUFB8) and IV (COX2) after the 5- and 8-day induction of mtPABP1. The loss of nuclear encoded complex I subunit (NDUFB8) is likely to reflect the instability of the complex due to the loss of mtDNA encoded components as evidenced by lack of intact complex I detected by blue native gel electrophoresis. A depletion of complex III was also noted, although, SDHA, the 70-kDa FAD containing subunit of the entirely nuclear encoded complex II, did not display any significant changes in steady state level. Whole-cell respirometry was also performed with cells expressing mtPABP1 or mtLUC for 8 days as detailed in ‘Materials and Methods’ section and (19) (Figure S2A). Resting rates of oxygen consumption show a decrease for the mtPABP1 expressors, with the consumption being less coupled to ATP production than the control, consistent with proton slip or electron/proton leak, potentially through partially assembled OXPHOS complexes. Rotenone insensitive respiration is also increased in the mtPABP1 expressor, potentially due to increased flux through complex II, by-passing the residual complex I.
mtPABP1 binds to mitochondrial mRNA
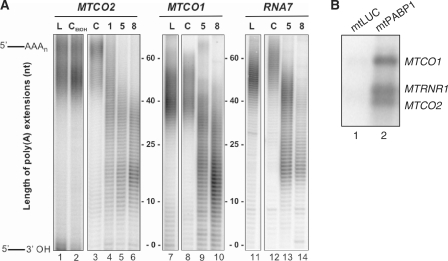
These data are likely to be due to mtPABP1 binding to the polyadenylated extensions of mt-mRNA but this had not been formally shown. To confirm binding to the poly(A) termini, total RNA was extracted from cells expressing mtPABP1 at various time points and subjected to mitochondrial poly(A) tail analysis (7). Three representative mt-mRNAs are shown for the uninduced cells where the majority of poly(A) extensions are between 40 and 60 nt (Figure 3A, lanes 3, 8 and 12). After a 2-day induction of mtPABP1, this tail became shortened, with two subpopulations predominating, one of 30–40 nt and an oligoadenylated population of 10–20 nt (see MTCO1, 2 and RNA7 Figure 3A). This alteration in profile is consistent with mtPABP1 either displacing any endogenous binding protein, with the unprotected extension being trimmed or that mtPABP1 binding prevents further extension by the relevant poly(A) polymerase. The two observed lengths of poly(A) tails may reflect the footprint of the binding factor. In vitro studies revealed that cytosolic PABPs from various organisms form a repeating unit covering 27 A residues (22), but the minimal binding site requires 12 adenosines (23,24).
Figure 3.
mtPABP1 binds mitochondrial mRNA. (A) RNA was isolated from cells expressing mtPABP1 over an 8-day period prior to the analysis of poly(A) tail extensions as described (7). 32P-end-labelled products corresponding to the 3′ termini of MTCO2 (lanes 1–6), MTCO1 (lanes 7–10) and MTND4 (lanes 11–14) were separated through an 8% denaturing PAG and visualized by PhosphorImager and ImageQuant analysis. Zero extension is taken as the position of migration predicted on 3′ processing from the polycistronic transcript prior to any addition. Products were generated in the absence of mtPABP1 expression (UP, lanes 3, 8 and 12) or after 1 (lane 4), 5 (lanes 5, 9 and 13) or 8 (lanes 6, 10 and 14) days induction. MPAT assays were also performed on RNA isolated from cells expressing mtLUC for 8 days (L, lanes 1, 7 and 11). Lane 2 demonstrates MPAT on RNA isolated from uninduced mtPABP1 transfectants grown in 1 µg/ml ethanol for 8 days (UP ETOH). (B) Immunoprecipitated mtPABP1 associates with RNA. Immunoprecipitation of FLAG tagged mtLUC and mtPABP1 was performed as described in ‘Materials and Methods’ section. Equal volumes of RNA extracted from immunoprecipitated samples were analysed by northern blot with the indicated probes as detailed in ‘Materials and Methods’ section.
To confirm more directly that mtPABP1 was able to bind mt-mRNA, the protein was immunoprecipitated from mitochondrial fractions prepared from cells after a 3-day induction as detailed in ‘Materials and Methods’ section. Immunoprecipitated mtPABP1-FLAG was then denatured and associated nucleic acids extracted. Bound mt-mRNA was quantified either directly, by northern blot (Figure 3B) or reverse-transcribed and cDNA subjected to qPCR (Table S2). A substantial enrichment was noted for all mitochondrial transcripts analysed, confirming that mtPABP1 is able to interact with mt-mRNA. An enrichment of 12S mt-rRNA, however, was also noted. Western blotting of the eluates from the immunoprecipitations showed evidence of large and small mitoribosomal subunit proteins (Supplementary Figure S2B) from mtPABP1-FLAG, alone. Taken together, these data are consistent with mtPABP1 associating with polyadenylated mRNA when loaded onto mitoribosomes, although non-specific binding cannot be ruled out (see below).
Binding of mtPABP1 to the poly(A) tail causes inhibition of translation
To assess whether binding of mtPABP1 could affect mitochondrial protein synthesis, in vivo translation was performed with cells expressing mtPABP1 or mtLUC (Figure 4A) in the presence of radiolabelled 35S methionine/cysteine for 15 min (lanes 1, 2) or 45 min (lanes 3, 4) pulses. Protein synthesis was compromised by expression of mtPABP1. Most mitochondrial transcripts contain no 3′ untranslated region and often require polyadenylation to generate a UAA translation termination codon. Therefore, binding of mtPABP1 to the poly(A) tail could occlude the stop codon of such transcripts. ND5 and COX1 are encoded by transcripts containing 3′ UTRs of 568 and 72 nt, respectively. Additionally, the bi-cistronic units encoding ND4L/4 (RNA7) and ATP8/6 (RNA14) ensure that the ND4L and ATP8 encoding open reading frames would also be extended by relatively long 3′ heteropolymeric sequences prior to poly(A) extension. Synthesis of all these proteins was clearly decreased (Figure 4A), arguing against a facile explanation for the phenotype.
The observed decline in mitochondrial translation is likely due to a dominant negative action of mtPABP1 through disruption of essential protein–protein interactions in the poly(A) extension. Immunoprecipitation with mtPABP1-FLAG clearly pulled down a level of mitoribosome (Supplementary Figure S2), possibly explaining the increase in immunoprecipitation of the 12 S mt-rRNA (Figure 3B). However, all these data do not completely negate the possibility of non-specific binding of mt-RNA by mtPABP1 causing the inhibition of translation. To address this issue, mutants of mtPABP1 were constructed with varying affinities for RNA. PABPC1 is a complex protein containing four RNA-binding domains, RRM1-4 (25). Previous mutagenic studies of yeast PAB1p had identified mutants that showed markedly altered binding properties. One such mutation, F366V (second aromatic residue in RNP1 of RRM4) displayed only a 4-fold reduction in poly(A) specific affinity (KD 50 nM) but with an immeasurably weak affinity for mixed RNA sequences. A second, harbouring substitutions in positions Y83V (RRM1) and F170V (RRM2), diminished poly(A) affinity to the micromolar range (25). Mutants containing the equivalent critical residues in mtPABP1; F337V (second aromatic residue in RNP1 of RRM4) and Y56V/F142V (second residues of RNP1 of RRM1 and RRM2, respectively) were generated as described in ‘Materials and Methods’ section. As shown in Figur 4B, induced expression of the constructs resulted in comparable amounts of the chimeric proteins present in protease K shaved mitochondria. Further, although a similar increase in acidity was noted in culture media after a 5-day induction of mtPABP1 and mtPABP1(F337V), no such acidosis was noted with mtPABP1(Y56V/F142V), consistent with OXPHOS defects in cells expressing PABP1 that bound to mt-mRNA poly(A) tails (data not shown).
To confirm that poly(A) binding was essential for the inhibition of protein synthesis, in vivo translation was performed with cells expressing the mutated forms of mtPABP1. As shown in Figure 4C, expression of mtPABP1(Y56V/F142V), equivalent to the previously characterized mutant PAB1p with more than 100-fold decrease in poly(A) affinity (25), caused only a marginal translation defect (lane 3). Conversely, mtPABP1(F337V) with a strong poly(A) affinity but very limited non-specific binding (25), consistently caused an even more profound translation inhibition than mtPABP1 (Figure 4C, compare lanes 4 and 2). This marked increase in inhibition over wild-type mtPABP1 was possibly due to the absence of any non-specific RNA binding, which could have partially titrated the wild-type protein in the mitochondrial matrix. These observations strongly support the hypothesis that binding of foreign proteins to the poly(A) extensions of mt-mRNA can substantially inhibit translation.
Binding of mtPABP1 does not cause destabilization of mt-mRNA
A substantial decrease in protein synthesis occurs on induction of mtPABP1 or the mutant mtPABP1(F337V). Although this was likely to be due to poly(A) interaction it was still possible that induction led to a decrease in mt-mRNA stability with a consequent decrease in translation. To determine whether occupation of the 3′ termini by mtPABP1 may affect the stability of mt-mRNA, total cytoplasmic RNA was isolated from cells following 3, 5 or 8 days of induction and was assessed by northern blot analysis. As shown in Figure 4D, expression of mutant or wild type mtPABP1 did not cause any decrease in the stability of mitochondrial encoded transcripts. Indeed, an increased steady-state level was noted for some species on induction of mtPABP1 (MTND3 171±10%; MTCO1 148±7%). The steady-state level of 12 S rRNA (MTRNR1) did not display any significant changes with the expression of either mtPABP1 form.
DISCUSSION
At steady state, human mt-mRNAs are virtually all oligo- or polyadenylated. For seven of the 11 protein coding transcripts, one function is clear; oligoadenylation at the 3′ terminus is necessary to generate a complete UAA termination codon that is recognized by the single mitochondrial release factor, mtRF1a. Has the mitochondrial poly(A) polymerase (mtPAP) been retained solely to provide this critical function, or is polyadenylation necessary for other functions? The standard approach to assess poly(A) function is to prevent its synthesis by depleting mtPAP with siRNA, an experiment that has been performed by two laboratories which, although invariably producing shortened poly(A) extensions unfortunately resulted in conflicting conclusions, as mentioned in the introduction (8,9). A second approach is to interfere with the turnover of the poly(A) tail. To date, it is not clear which enzymes are responsible for this important role. The core yeast mitochondrial RNA degradosome consists of a helicase, Suv3p and a ribonuclease, Dss1p (26). Although a human Suv3p orthologue has been identified (27), no Dss1p orthologue is present in the human genome. The human polynucleotidyl phosphorylase (hPNPase) has been suggested as a candidate. A report by Slomovic and Schuster showed that downregulation of hPNPase can cause a profound but unpredictable modulation of poly(A) tail length of human mitochondrial mRNA (10). No direct measurement of mt-mRNA stability was performed, but mitochondrial translation was unaffected. These authors showed that MTCO1 transcripts were often correctly processed at their 3′ termini but the majority were not adenylated. Although there was a 3′ addition of ribonucleotides, surprisingly these were commonly found to be uridylates, but no evidence of translation inhibition was noted for MTCO1. Polyuridylation has also been found on incorrectly processed intermediates and was suggested to tag incorrect transcripts for degradation (28). This observation prevents us from being able to eliminate this 3′ extension from playing an essential role in translation. This intriguing data is further complicated by the report that the substantial majority of hPNPase is not actually located in the mitochondrial matrix but is in the intermembrane space (29), effectively separated from mitochondrial RNA, raising the question of whether hPNPase is indeed involved in poly(A) turnover in human mitochondria.
The approach taken in this report was to manipulate the mt-mRNA poly(A) tail by targeting cytosolic poly(A) modifying enzymes to the mitochondrion. Deadenylation of mt-mRNAs by mtPARNΔC led to a decrease in steady-state level of MTCO1, MTCO2 and RNA14, similar to the previous report by Nagaike et al. (9) who siRNA-depleted mtPAP. However, in contradistinction we show that certain species are increased in abundance. For example, MTND2 showed an almost 4-fold increase, with MTND1 also markedly increased. These data argue against a simple explanation that polyadenylation of mt-mRNA always promotes stability. Indeed, our data is similar to the variability in stability found by Slomovic and Schuster (10) on depletion of human PNPase. Further, we show that this variability is not linked to translatability, as mitochondrial translation of all species was decreased.
Our data with mtPABP1 shows that the poly(A) tail is likely to be necessary for optimizing translation efficiency. Poly(A) specific binding of the mutant F337V also resulted in decreased translation rates of all mitochondrial proteins, confirming the decrease was linked to poly(A) binding and not due to non-specific interactions. We believe this is due to mtPABP1 competing for binding at poly(A) region, indicating that poly(A) normally interacts with endogenous components that promote translation. Proteomics analysis of proteins bound to mt-mRNA revealed some interesting candidates that may interact with poly(A) in vivo. One candidate, SLIRP, was found in all analyses (MW, RNL, ZMACL unpublished observation). This small RNA binding protein has one predicted RRM domain and has recently been shown to be essential for RNA stability in mitochondria (30). Could this protein be the elusive mitochondrial poly(A) binding protein ? SLIRP and mt-mRNA were shown to be mutually dependent. As the poly(A) extension is not essential for mt-mRNA stability, it is unlikely that the loss of a poly(A) specific binding protein would lead to dramatic loss of stability. A previous study reported tight binding of SLIRP to RNA stem–loop structures, but it will be interesting to determine whether the protein is also able to recognize poly(A) sequences. As yet, we do not know which aspect of mitochondrial translation is affected by mtPABP1 binding, initiation, elongation, termination or recycling. Preliminary analysis of RNA profiles in isokinetic density gradients suggest that on induction of mtPABP1 the RNA occupancy of mitoribosomes is decreased, whilst association with the small subunit is increased, indicating that termination and recycling, at least, are not affected.
Finally, although mtPARN translation studies were complicated by the loss of termination codons in most transcripts, translational inhibition of MTCO1 was difficult to interpret, as sequence analysis revealed that almost all transcripts retained a complete open reading frame. Translation and membrane insertion of the yeast COX1p orthologue requires the assistance of translational activators [MSS51p (31), PET309p (32)] which interact with UTRs of the COX1 transcript. Recently, a human translational activator of MTCO1, TACO1, was identified (33). Although it has yet to be formally shown, it is possible that this protein interacts within this 3′ UTR to promote translation in an unspecified manner. Consequently, the loss of this part of the MTCO1 transcript may result in the loss of the TACO1 interaction site and therefore the loss of translation. A second protein, LRPPRC, has been suggested to be the human orthologue of another yeast translational activator, PET309 (34). This human mitochondrial RNA binding protein has also been implicated in MTCO1 association and a similar problem may occur on loss of the 3′ UTR.
In summary, our data strongly suggests that in addition to being required for completing the termination codon of 7 ORFs, it is also required for the optimizing mitochondrial protein synthesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Wellcome Trust (grant number 074454/Z/04/Z); Biotechnology and Biological Sciences Research Council (grant number BB/F011520/1); and EU_FP6 (grant number MCEST-CT-FP6-503684). Funding for open access charge: Biotechnology and Biological Sciences Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Joanna Rorbach for help with tissue culture.
REFERENCES
- 1.Anderson S, Bankier AT, Barrell BG, De Bruijn M.HL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 2.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 3.Gagliardi D, Stepien PP, Temperley RJ, Lightowlers RN, Chrzanowska-Lightowlers ZM. Messenger RNA stability in mitochondria: different means to an end. Trends Genet. 2004;20:260–267. doi: 10.1016/j.tig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Schuster G, Stern D. RNA polyadenylation and decay in mitochondria and chloroplasts. Prog. Mol. Biol. Transl. Sci. 2009;85:393–422. doi: 10.1016/S0079-6603(08)00810-6. [DOI] [PubMed] [Google Scholar]
- 5.Osinga KA, De Vries E, Van der Horst G, Tabak HF. Processing of yeast mitochondrial messenger RNAs at a conserved dodecamer sequence. EMBO J. 1984;3:829–834. doi: 10.1002/j.1460-2075.1984.tb01892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008;27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Temperley RJ, Seneca SH, Tonska K, Bartnik E, Bindoff LA, Lightowlers RN, Chrzanowska-Lightowlers ZM. Investigation of a pathogenic mtDNA microdeletion reveals a translation-dependent deadenylation decay pathway in human mitochondria. Hum. Mol. Genet. 2003;12:2341–2348. doi: 10.1093/hmg/ddg238. [DOI] [PubMed] [Google Scholar]
- 8.Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 2004;32:6001–6014. doi: 10.1093/nar/gkh923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J. Biol. Chem. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- 10.Slomovic S, Schuster G. Stable PNPase RNAi silencing: its effect on the processing and adenylation of human mitochondrial RNA. RNA. 2008;14:310–323. doi: 10.1261/rna.697308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakagawa J, Waldner H, Meyer-Monard S, Hofsteenge J, Jeno P, Moroni C. AUH, a gene encoding an AU-specific RNA binding protein with intrinsic enoyl-CoA hydratase activity. Proc. Natl Acad. Sci. USA. 1995;92:2051–2055. doi: 10.1073/pnas.92.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ponamarev MV, She YM, Zhang L, Robinson BH. Proteomics of bovine mitochondrial RNA-binding proteins: HES1/KNP-I is a new mitochondrial resident protein. J. Proteome Res. 2005;4:43–52. doi: 10.1021/pr049872g. [DOI] [PubMed] [Google Scholar]
- 14.Preiss T, Chrzanowska-Lightowlers ZM, Lightowlers RN. Glutamate dehydrogenase: an organelle-specific mRNA-binding protein. Trends Biochem. Sci. 1997;22:290. doi: 10.1016/s0968-0004(97)82219-0. [DOI] [PubMed] [Google Scholar]
- 15.Soleimanpour-Lichaei HR, Kuhl I, Gaisne M, Passos JF, Wydro M, Rorbach J, Temperley R, Bonnefoy N, Tate W, Lightowlers R, et al. mtRF1a is a human mitochondrial translation release factor decoding the major termination codons UAA and UAG. Mol. Cell. 2007;27:745–757. doi: 10.1016/j.molcel.2007.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chrzanowska-Lightowlers ZM, Preiss T, Lightowlers RN. Inhibition of mitochondrial protein synthesis promotes increased stability of nuclear-encoded respiratory gene transcripts. J. Biol. Chem. 1994;269:27322–27328. [PubMed] [Google Scholar]
- 17.Nijtmans LG, Henderson NS, Holt IJ. Blue Native electrophoresis to study mitochondrial and other protein complexes. Methods. 2002;26:327–334. doi: 10.1016/S1046-2023(02)00038-5. [DOI] [PubMed] [Google Scholar]
- 18.Yokobori S, Paabo S. Transfer RNA editing in land snail mitochondria. Proc. Natl Acad. Sci. USA. 1995;92:10432–10435. doi: 10.1073/pnas.92.22.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutter E, Renner K, Pfister G, Stockl P, Jansen-Durr P, Gnaiger E. Senescence-associated changes in respiration and oxidative phosphorylation in primary human fibroblasts. Biochem. J. 2004;380:919–928. doi: 10.1042/BJ20040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chomyn A. In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol. 1996;264:197–211. doi: 10.1016/s0076-6879(96)64020-8. [DOI] [PubMed] [Google Scholar]
- 21.Wu M, Reuter M, Lilie H, Liu Y, Wahle E, Song H. Structural insight into poly(A) binding and catalytic mechanism of human PARN. EMBO J. 2005;24:4082–4093. doi: 10.1038/sj.emboj.7600869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baer BW, Kornberg RD. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J. Cell. Biol. 1983;96:717–721. doi: 10.1083/jcb.96.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sachs AB, Davis RW, Kornberg RD. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn U, Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J. Mol. Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- 25.Deardorff JA, Sachs AB. Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast poly(A)-binding protein. J. Mol. Biol. 1997;269:67–81. doi: 10.1006/jmbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- 26.Dziembowski A, Piwowarski J, Hoser R, Minczuk M, Dmochowska A, Siep M, van der Spek H, Grivell L, Stepien PP. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J. Biol. Chem. 2003;278:1603–1611. doi: 10.1074/jbc.M208287200. [DOI] [PubMed] [Google Scholar]
- 27.Minczuk M, Piwowarski J, Papworth MA, Awiszus K, Schalinski S, Dziembowski A, Dmochowska A, Bartnik E, Tokatlidis K, Stepien PP, et al. Localisation of the human hSuv3p helicase in the mitochondrial matrix and its preferential unwinding of dsDNA. Nucleic Acids Res. 2002;30:5074–5086. doi: 10.1093/nar/gkf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szczesny RJ, Borowski LS, Brzezniak LK, Dmochowska A, Gewartowski K, Bartnik E, Stepien PP. Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res. 2010;38:279–298. doi: 10.1093/nar/gkp903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen HW, Rainey RN, Balatoni CE, Dawson DW, Troke JJ, Wasiak S, Hong JS, McBride HM, Koehler CM, Teitell MA, et al. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol. Cell. Biol. 2006;26:8475–8487. doi: 10.1128/MCB.01002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baughman JM, Nilsson R, Gohil VM, Arlow DH, Gauhar Z, Mootha VK. A computational screen for regulators of oxidative phosphorylation implicates SLIRP in mitochondrial RNA homeostasis. PLoS Genet. 2009;5:e1000590. doi: 10.1371/journal.pgen.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez-Martinez X, Broadley SA, FoxT D. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003;22:5951–5961. doi: 10.1093/emboj/cdg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manthey GM, McEwen JE. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 1995;14:4031–4043. doi: 10.1002/j.1460-2075.1995.tb00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weraarpachai W, Antonicka H, Sasarman F, Seeger J, Schrank B, Kolesar JE, Lochmuller H, Chevrette M, Kaufman BA, Horvath R, et al. Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat. Genet. 2009;41:833–837. doi: 10.1038/ng.390. [DOI] [PubMed] [Google Scholar]
- 34.Xu F, Morin C, Mitchell G, Ackerley C, Robinson BH. The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem. J. 2004;382:331–336. doi: 10.1042/BJ20040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.