Abstract
A systematic search was performed for DNA-binding sequences of YgiP, an uncharacterized transcription factor of Escherichia coli, by using the Genomic SELEX. A total of 688 YgiP-binding loci were identified after genome-wide profiling of SELEX fragments with a high-density microarray (SELEX-chip). Gel shift and DNase-I footprinting assays indicated that YgiP binds to multiple sites along DNA probes with a consensus GTTNATT sequence. Atomic force microscope observation indicated that at low concentrations, YgiP associates at various sites on DNA probes, but at high concentrations, YgiP covers the entire DNA surface supposedly through protein–protein contact. The intracellular concentration of YgiP is very low in growing E. coli cells under aerobic conditions, but increases more than 100-fold to the level as high as the major nucleoid proteins under anaerobic conditions. An E. coli mutant lacking ygiP showed retarded growth under anaerobic conditions. High abundance and large number of binding sites together indicate that YgiP is a nucleoid-associated protein with both architectural and regulatory roles as the nucleoid proteins Fis and IHF. We then propose that YgiP is a novel nucleoid protein of E. coli under anaerobiosis and propose to rename it Dan (DNA-binding protein under anaerobic conditions).
INTRODUCTION
The complete sequence of Escherichia coli genome has revealed the presence of 4453 protein-coding sequences shared between two laboratory strains, MG1655 and W3110, of K-12 lineage (1). The function of each gene product has been experimentally determined or computationally predicted for about half of the genes, but a large number of genes are still left uncharacterized (1) even though E. coli has long been used as a model organism in the modern molecular biology since its isolation (2). One major research subject of the post-genome sequencing era is the identification of physiological functions for all these uncharacterized genes. Most of these genes are, however, not expressed in laboratory culture conditions and thus supposed to be needed for the survival of E. coli under stressful conditions in nature. The expression of these uncharacterized stress-response genes must be under the control of stress-response regulatory systems. This line of research encounters with the difficulty that arises from the lack of knowledge about factors and conditions required for expression of each of these silent genes under laboratory culture conditions.
Transcription of the E. coli genome is considered to be under the control of approximately 300 species of transcription factors (3–5), which altogether control the distribution of about 2000 molecules of RNA polymerase among 4453 genes within the genome (3,5,6). The regulatory roles, however, remain unidentified for about 100 species of the transcription factor (5) presumably because those putative factors are needed for regulation of the genes, which are expressed in response to as-yet unidentified environmental stresses. Ordinary genetic approach is not useful without knowledge of factors and conditions for expression of their regulatory functions. To identify the regulatory roles of hitherto uncharacterized putative transcription factors, we have performed a systematic search for the identification of DNA sequences recognized by each of 300 species of DNA-binding transcription factors from E. coli by using the newly developed Genomic SELEX (systematic evolution of ligands by exponential enrichment) system (7). Since the binding sites of prokaryotic transcription factors are generally located within or near the promoters, we can predict the target promoters, genes and operons from the binding site maps of test factors as have been successfully employed for Cra (7), RstA (8), PdhR (9), RutR (10), TyrR (11), NemR (12), AllR (13), CitB (14), LeuO (15) and AscG (16). Once the consensus recognition sequence is identified after genomic SELEX screening, the in silico search for other targets becomes possible using the information of whole genome sequence.
The LysR family of DNA-binding regulators exists in diverse bacterial genera, archea and algal chloroplasts, and participate in diverse cellular activities, including nitrogen fixation, oxidative stress response and bacterial virulence (17). Escherichia coli contains a total of 54 families of transcription factor, of which the LysR family is the most abundant including 45 members (4,5). LysR family proteins contain an N-terminal helix–turn–helix motif, but the consensus sequence of DNA binding is different between LysR family members, indicating that each controls a different set of genes or promoters with a unique recognition sequence. In this study, a systematic search was performed for identification of the target genes under the control of YgiP, an uncharacterized LysR-family transcription factor, by using this newly developed genomic SELEX system. In prokaryotes, the genes encoding transcription regulatory proteins are generally located on the genome adjoining to the target genes for regulation. Relying on this rule, Oshima and Biville (18) predicted that YgiP is involved in regulation of the adjacent ttdA-ttdB genes encoding tartrate dehydrogenase and propose to name YgiP as TtdR. After cloning-sequencing (SELEX-clos) and tilling array analysis (SELEX-chip) of SELEX fragments, however, we have obtained a large number of YgiP-binding sequences from hundreds of loci within the E. coli genome, including the ttdR-ttdA spacer sequence. After gel-shift and DNase-footprinting assays, YgiP was found to be a DNA-binding protein with affinity to AT-rich sequences. Both the number of binding sites on the genome and the sequence preference are similar to the known nucleoid proteins such as Fis and IHF (5,19,20). By using the quantitative immuno-blotting (21), we found the intracellular concentration of YgiP was very low in growing E. coli cells under aerobic conditions; however, under hypoxic or anaerobic culture conditions, the YgiP level increased more than 100-fold, reaching to the level as high as those of major nucleoid proteins HU and IHF. A mutant E. coli lacking ygiP showed retarded growth under anaerobic conditions. The binding mode of YgiP to DNA was investigated by atomic force microscory (AFM). These observations altogether indicate that YgiP has dual functions, playing the architectural role for genome folding and the global regulatory role in transcription of a number of genes, as in the case of the nucleoid proteins Fis and IHF. We then conclude that YgiP is a hitherto unidentified nucleoid protein in E. coli growing under anaerobic conditions, and propose to rename YgiP (or TtdR) to Dan (DNA-binding protein under anaerobic conditions), which belongs to the same family of growth phase-specific nucleoid proteins as Fis in exponential growth phase (22,23) and Dps in stationary phase (24).
MATERIALS AND METHODS
Bacterial strains and culture conditions
The E. coli strains used in this work were KP7600 (W3110 typeA lacIq lacZΔM15 galK2 galK22) and JD24074 (KP7600 dan) which was constructed by a transposon insertion method (a gift from T. Miki). Cells were grown at 37°C in Luria-Bertani (LB) or M9–0.4% glucose medium. Overnight culture in LB medium was diluted 1000-fold into fresh LB or M9 medium. For the aerobic culture, the incubation was carried out at 37°C with a constant sharking to an appropriate cell density. The cell density was determined by measuring the absorbance at 600 nm.
For hypoxic culture, bacteria were suspended in 15-ml centrifuge tubes with silicone stoppers to make the initial cell density of 0.1 OD600 and anoxia was achieved to expel any remaining air by chemical duty pump (Millipore) for 60 min. Cell culture was maintained at 37°C without shaking. On the other hand, anaerobic culture was carried out using COY chamber (COY Laboratory Products, Inc.) under the constant circulation of nitrogen (95%)-hydrogen (5%) mixed gas equipped with a palladium catalyst.
Purification of Dan protein
For construction of plasmid pDan for Dan expression, a DNA fragment corresponding to the Dan-coding region was amplified by PCR using E. coli W3110 genome DNA as a template and a pair of primers, which were designed so as to hybridize upstream or downstream of the Dan-coding sequence. After digestion with NdeI and NotI, PCP products were cloned into pET21a(+) (Novagen) between NdeI and NotI sites. The plasmid construct was confirmed by DNA sequencing. For protein expression, the pDan plasmid was transformed into E. coli BL21 (DE3). Expression of Dan was induced by adding IPTG (isopropyl-β-d-1-thiogalactopyranoside). Purification of over-expressed His-tagged Dan was performed under the standard purification procedure (7–16).
Genomic SELEX
The genomic SELEX screening was performed as described previously (7–16). In brief, 5 pmol of a mixture of genome DNA fragments of 150–300 bp in length and 10 pmol His-tagged Dan were mixed in a binding buffer (10 mM Tris–HCl, pH 8.0 at 4°C, 3 mM Mg-acetate, 150 mM NaCl and 0.1 mM EDTA) and incubated for 30 min at 37°C. The mixture was applied onto Ni-NTA column and after washing unbound DNA with the binding buffer containing 10 mM imidazole, DNA-Dan complexes were eluted with an elution buffer containing 200 mM imidazole.
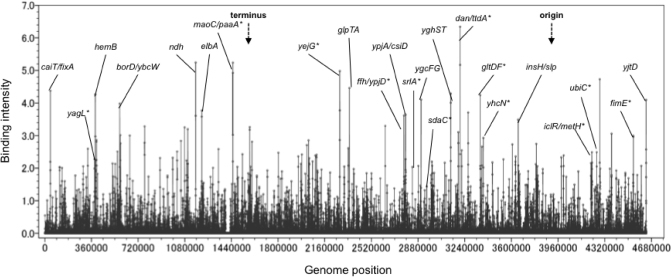
The sequences of DNA fragments obtained by the genomic SELEX screening were identified by both SELEX-chip and SELEX-clos procedures. For SELEX-chip analysis, DNA fragments were subjected to the Microarray assay using a tilling array consisting of 22 000 60-bp-long oligonucleotide probes aligned at 160 bp intervals along the E. coli genome (25). Dan-bound SELEX DNA fragments and the original mixture of genome DNA fragments used in the genomic SELEX screening were differentially labeled with Cy5 and Cy3, respectively. The two labeled DNA samples were combined and hybridized to the tilling microarray (Oxford Gene Technology, Oxford, UK). The Cy5/Cy3 intensity ratio was measured for each spot, and plotted against the corresponding position on the E. coli KP7600 chromosome, creating the genome-wide profiles of Dan binding. Peaks that exhibited a Cy5/Cy3 ratio of >3.5 with two or more consecutive probes were selected as Dan-binding sites (for the typical pattern see Figure 1).
Figure 1.
Distribution of Dan-binding sequences on the E. coli genome. Mixtures of Dan-binding DNA fragments isolated by the genomic SELEX were subjected to the DNA microarray analysis. Binding signals (y-axis) are plotted against the location along the E. coli genome (x-axis). The genes showing high-level peaks above the cutoff level of 3.5 are indicated. The genes with asterisk represent those identified by SELEX-clos (see Supplementary Table 1).
In SELEX-clos (cloning and sequencing) procedure, the same SELEX DNA fragments as used for SELEX-chip were cloned into pT7 into pT7 Blue-T vector (Novagen) and transformed into E. coli DH5α for amplification. The DNA fragments were regenerated by polymerase chain reaction (PCR) using amplified plasmids as template and fluorescent-labeled T7-primer (5′-TAATACGACTCACTATAGGG-3′) for sequencing with ABI DNA sequencer 3130×.
Gel mobility shift assay
Gel shift assay was performed as described previously (8,9). In brief, probes were generated by PCR amplification of Dan-binding sequences in SELEX using a pair of primers, 5′ fluorescein isothiocyanate (FITC)-labeled T7-F primer (5′-TAATACGACTACTATAGGG-3′) and T7-R primer (5′-GGTTTTCCCAGTCACACGACG-3′), the genomic SELEX plasmids containing the respective Dan recognition sequences as templates, and Ex Taq DNA polymerase. PCR products with FITC at their termini were purified by PAGE. For gel shift assays, 0.5 pmol each of the FITC-labeled probes was incubated at 37°C for 30 min with various amounts of Dan in 15 µl of gel shift buffer consisting of 10 mM Tris–HCl, pH 7.8 at 4°C, 150 mM NaCl, 3 mM Mg acetate. After the addition of DNA dye solution, the mixture was directly subjected to 6% PAGE. Fluorescent-labeled DNA in gels was detected using Pharos FX plus system (Bio-Rad). The setting of excitation and emission wavelength was 488 and 532 nm, respectively.
DNase-I footprinting assay
DNase-I footprinting assay was carried out using FITC-labeled DNA fragments as described previously (8,9). Each 2.0 pmol of FITC-labeled probes was incubated at 37°C for 30 min with various amounts of Dan in DNase-I footprinting buffer consisting of 25 μl of 10 mM Tris–HCl (pH 7.8), 50 mM NaCl, 3 mM magnesium acetate, 5 mM CaCl2 and 25 mg/ml BSA. After incubation for 30 min, DNA digestion was initiated by the addition of 0.05 U of DNase I (TAKARA). After digestion for 40 s at 25°C, the reaction was terminated by the addition of 25 μl of phenol. Digested products were precipitated with ethanol, dissolved in formamide-dye solution and analyzed by electrophoresis on a 6% polyacrylamide gel containing 7 M urea.
AFM observation
For observation of Dan–DNA complexes by AFM, plasmid pGRdan was constructed, which carried the ttdA-dan spacer sequence within the promoter assay vector pGR (26). Mixtures of 200 ng pGRdan DNA and 5–800 ng of Dan protein in a binding buffer [50 mM HEPES-KOH (pH 7.6), 1 mM EDTA, 500 mM NaCl, 1 mM DTT] were incubated for 30 min at 37°C. After cross-linking DNA–protein by treatment with 0.1% glutaraldehyde for 1 h at 4°C, the samples were diluted to make the final DNA concentration of ∼5 ng/μl with a dilution buffer [5 mM HEPES-KOH (pH 7.6), 2 mM MgCl2] and directly spotted onto a freshly cleaved mica substrate pretreated with 10 mM spermidine. After 10 min, the mica was gently washed with distilled water and dried under nitrogen gas. Imaging was performed in Tapping ModeTM with a MultimodeTM AFM (Veeco, Santa Barbara, CA, USA) operation with a Nanoscope IIIaTM controller. For scanning, Olympus silicon cantilevers (OMCL-AC160TS-W2) with spring constants between 36 and 75 N/m were used. The scan frequency was typically 1.5 Hz per line and the modulation amplitude was a few nanometers. Analysis of the DNA–Dan complexes was performed using the Nanoscope software. Two-dimensional images in brightness and contrast were optimized for the purpose of clarity. Three-dimensional images were created with the Nanoscope software and exported in TIFF format.
Quantitative immunoblot analysis
A quantitative western blot analysis was carried out by standard method as described previously (21). In brief, E. coli cells grown in 10 ml of either LB or M9-0.4% glucose medium under both aerobic and anaerobic culture conditions were harvested by centrifugation and resuspended in 0.3 ml lysis buffer (50 mM Tris–HCl, pH 7.5, 50 mM NaCl, 5% glycerol and 1 mM dithiothreitol), and then lysozyme was added to a final concentration of 20 µg/ml. Total proteins were subjected to 12% SDS-PAGE and blotted on to a plyvinylidene difluoride (PVDF) membranes using semi-dry transfer apparatus. Membranes were first immuno-detected with anti-Dan and then developed with an enhanced chemiluminescence kit (Amersham-Pharmacia Biotech). The image was analyzed with a LAS-1000 Plus Lumino-Image Analyzer and Image Gauge (Fuji Film).
Indirect immuno-fluorescence microscopy
The procedure for indirect immuno-fluorescence microscopy was essentially the same as previously described (27,28). In brief, cells were fixed by treatment with 80% methanol for 1 h at 25°C, and deposited on to a poly l-lysine-coated slide. After air drying, cells were treated with 2 mg/ml lysozyme solution at room temperature for 5 min. The slide was treated 99% methanol for 1 min, and then with acetone for 1 min until complete dryness. After washing with PBS containing 0.05% Tween 20) and 2% bovine serum albumin (BSA) for 15 min for blocking, the sample was treated with 200–500-fold diluted anti-Dan antibodies for 1 h at room temperature. After washing with Tween-containing PBS, the sample was treated at room temperature for 1 h with 500-fold diluted goat anti-rabbit IgG antibodies conjugated with a fluorescence compound Cy3. After washing with Tween-containing PBS, the sample was treated with 10 µl of DAPI (4′,6-diamidino-2-phenylindole dihydrochloride, 10 µg/ml) for 1 min for staining DNA, and covered with one drop of mountain medium (1 mg/ml p-phenylenediamine and 90% glycerol in PBS, pH 9.0). The immuno-stained samples were observed with a fluorescence and phase-contrast microscope (100×objective lens; Olympus, Japan) equipped with a chilled, high sensitive digital CCD camera, Regita EXi (NIPPON ROPER, Japan), connected to a computer. Different filter cassettes were used for observation of Cy3 and DAPI fluorescence. The images were transferred directly to a Windows XP and processed using Image-Pro Plus version 6.0 software. The specificity of the observed immuno-stained fluorescent signal was verified by the absence of the signal in corresponding immuno-stained samples prepared using the respective dan null mutant cells.
RESULTS
Genomic SELEX search for Dan-binding sequences
For the identification of DNA sequences that are recognized by E. coli Dan, we employed the ‘Genomic SELEX’ system (7), in which a mixture of genome DNA fragments was used instead of mixtures of oligonucleotides with all possible sequences as used in the original SELEX (systematic evolution of ligands by exponential enrichment) (29–31). The ‘Genomic SELEX’ has since been successfully employed for identification of the sets of genes under the control of some known and unknown transcription factors (7–16). From a mixture of DNA fragments of E. coli K12 W3350 and a two-fold molar excess of the purified His-tagged Dan protein, Dan–DNA complexes were affinity purified using Ni-NTA resin. In the early stage of this genomic SELEX cycle, the Dan-bound DNA fragments formed smear bands on PAGE as did the original genome fragment mixture. After four and five SELEX cycles, however, the width of gel bands decreased, indicating certain enrichment of specific DNA fragments with Dan-binding activity. For sequence identification of Dan-bound DNA fragments, we first employed the cloning-sequencing method (SELEX-clos), in which the DNA fragment mixtures were recovered from the gels, PCR-amplified and subjected to the ordinary procedure of cloning and sequencing (7).
After four cycles of the ‘Genomic SELEX’ screening, a total of 333 independent SELEX clones were isolated by the SELEX-clos method, which were classified into two different groups based on the location on the E. coli genome relative to the genes (Table 1). Group A included 149 independent clones, each carrying a unique sequence from 32 different spacer regions between two neighboring genes on the E. coli genome (Supplementary Table 1A). In prokaryotes, the recognition DNA sequences by transcription regulatory factors are generally located within or near the promoters of target genes. If Dan is such a specific DNA-binding transcription factor, the regulation targets may be the genes, which are located downstream of these Dan-binding sites (shown by arrows in Supplementary Table 1A).
Table 1.
Dan-bound DNA fragmentsa
| Group | No. of clones | No. of loci | Total no.of clones | Group | No. of clones | No. of loci | Total no.of clones |
|---|---|---|---|---|---|---|---|
| A | 69 | 1 | 69 | B | 19 | 1 | 19 |
| 22 | 1 | 22 | 15 | 1 | 15 | ||
| 10 | 1 | 10 | 10 | 1 | 10 | ||
| 4 | 4 | 16 | 4 | 3 | 12 | ||
| 2 | 7 | 14 | 3 | 4 | 12 | ||
| 1 | 18 | 18 | 2 | 34 | 68 | ||
| 1 | 48 | 48 | |||||
| 32 | 149 | 92 | 184 |
aUsing the genomic SELEX method, a total of 333 DNA fragments have been isolated as complexes with the purified Dan protein.
A total of 149 group A clones contained the sequences from spacer regions between the neighboring genes, while a total of 184 group B clones carried the sequences within the coding frames.
The location of all the SELEX fragments on the genome is described in Table 2 and Supplementary Tables 1 and 2.
Of the total 149 group-A library, 69 clones contained two different but partially overlapping 297-bp-long segments of the spacer region of divergently transcribed dan (renamed from ygiP and ttdR) and ttdA (between nucleotides 3 204 211 and 3 204 508 of the E. coli MG1655 genome) (Supplementary Table 1A). The two different 297-bp-long segments overlapped in 63-bp sequence, indicating that the Dan-binding sequence is located within this narrow region of 63 bp in length. The number of SELEX isolates by a test transcription factor correlates with its affinity to the target sequence (7–16), we predicted that Dan has a strong affinity to this dan-ttdA spacer region. This sequence with strong affinity to Dan is located upstream of its own gene and upstream of the divergently transcribed ttdAB genes encoding L-tartrate dehydratase. One possible function of Dan is the autogenous control of its own expression, but Dan may also play a regulatory role in expression of the neighboring and divergently transcribed ttdA-ttdB-ygjE operon as predicted based on the gene organization (18). A total of 22 clones from group-A library carried sequences from the ffh-ypjD spacer; 10 clones from the maoC-paaA spacer; four clones each from the yfaZ-yfaO, ygdH-sdaC, fimB-fimE and cadB-cadC spacers; two clones each from seven different spacers; and one clone each from other 18 spacers (Supplementary Table 1A).
On the other hand, group B included 184 independent clones (Supplementary Table 1B), which were derived from a total of 95 protein-coding regions, including 19 clones from nrfE; 15 clones from yhfS; 10 clones from ebgA; 4 clones each from yagM, mltB, yegB, and yiiG; three clones each from kdpB, yegB, yfcN and yjbI; and two clones each from 35 different loci (Supplementary Table 1B). After the genome-wide screening by the Genomic SELEX, we realized that the DNA-binding sites are located within protein-coding sequences for a specific set of transcription factors (32). The ratio between group-A and group-B libraries is different between more than 50 species of transcription factor so far examined by the Genomic SELEX (Ishihama, A., unpublished data), implying that transcription factors associated within protein-coding sequences play an as yet unidentified regulatory role(s).
The high level of heterogeneity in the SELEX DNA library indicates that Dan binds to a larger number of sites on the genome. With use of the SELEX-clos method, we must have identified only a fraction of DNA fragments with high affinity to Dan. In order to identify the whole set of potential Dan-binding sites, we then subjected the DNA fragment mixtures to a tilling array analysis (SELEX-chip). The array herein employed consists of 22 000 60-mer-oligonucleotides that correspond to evenly spaced sections of the E. coli genome at ∼160-bp intervals (25). Dan-binding SELEX fragments isolated after five and six cycles of the Genomic SELEX were labeled with Cy5 while the original DNA fragment library used for SELEX screening were labeled with Cy3. The fluorescent-labeled DNA mixture was subjected to hybridization with the tilling array (an Oxford Gene Technology product). After washing and scanning, Cy5/Cy3 signal intensity ratio was calculated for each of 22 000 probes.
A total of 688 peaks with Dan-binding activity could be identified when the cutoff level of Cy5/Cy3 ratio was set at 3.5 (Figure 1). A total of 333 independent clones were isolated from 124 different locations by the SELEX-clos analysis, while a total of 688 sites were identified by the SELEX-chip analysis, indicating that the screening was not saturated for the SELEX-clos analysis. Most of the Dan-binding sites identified by the SELEX-clos analysis (see Supplementary Table 1) were included in the data collection of SELEX-chip analysis (see Supplementary Table 2. Note that the genes identified by SELEX-clos are indicated by asterisk in Figure 1). In agreement with the results of SELEX-clos analysis, the signals were detected on both coding and non-coding sections of the E. coli genome (Table 2 ). Of a total of 688 Dan-binding sites, 504 were located within intergenic non-coding regions (group-A) (Table 2; for details see Supplementary Table 2A), while 184 were within coding regions (group-B) (Table 2; for details see Supplementary Table 2B). Even though the combination of total non-coding sequences is <10% of the whole E. coli genome, ∼80% of the Dan-binding sites are located within the non-coding sequences (Table 2). This supports the prediction that Dan plays a regulatory role of the genome function.
Table 2.
Location of Dan-binding DNA fragments identified by SELEX-chipa
| Classification | Group-A | Group-B |
|---|---|---|
| Gene for metabolism | 78 | 25 |
| Genetic information genes | 77 | 19 |
| Information processing genes | 76 | 13 |
| Membrane-related Genes | 25 | 3 |
| Phage-derived genes | 21 | 13 |
| Genes for other functions | 43 | 20 |
| Conserved protein genes | 59 | 23 |
| Genes with unknown function | 125 | 68 |
| 504 | 184 |
aDNA fragment mixtures obtained by the Genomic SELEX were subjected to the SELEX-chip analysis.
When Dan-binding sites were picked up at the Cy5/Cy3 ratio above 3.5, a total of 688 peaks were identified (see Figure 1), which could be classified into two groups: A, Dan-binding sites within spacer regions between two genes; A, Dan-binding sites within spacer regions; B, Dan-binding sites are located within open reading frames. These genes are categorized based on the functions of gene products (for details see Supplementary Tables 3 and 4).
DNA-binding activity of Dan: Gel mobility shift assay
One unique feature of the SELEX-clos and SELEX-chip analyses with Dan is the large number of its binding sites within the entire E. coli genome. With respect to the number of DNA-binding sites, Dan is different from the well-characterized global regulators so far examined by the genomic SELEX, including CRP, Cra, RstA, PdhR, RutR, TyrR, NemR, AllR and CitB (7–10,12,13). Instead Dan is rather similar to those of major nucleoid proteins such as HU, IHF, H-NS and Fis (5). The number of DNA-binding sites by the nucleoid proteins with regulatory roles such as Fis and IHF ranges several hundreds as detected by Genomic SELEX screening (Kori,A. and Ishihama,A., unpublished data).
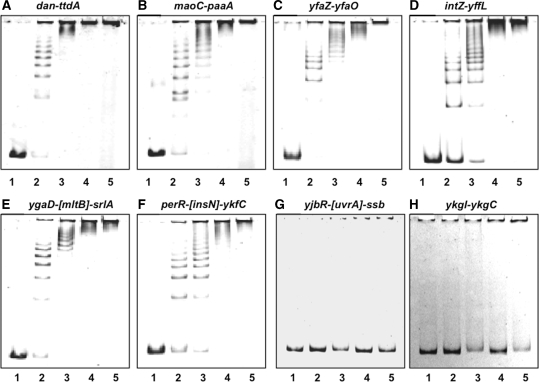
For the confirmation of Dan binding to the DNA sequences identified by the Genomic SELEX screening, we performed the gel mobility shift assay for five representative DNA probes from SELEX-clos and SELEX-chip library: dan-ttdA (Figure 2A), maoC-paaA (Figure 2B), yfaZ-yfaO (Figure 2C) and intZ-yffL (Figure 2D) from group A, and ygaD-[mltB]-srlA (Figure 2E) and perR-[insN]-ykfC (Figure 2F) from group B. In parallel, we analyzed several DNA probes from non-peak fractions of the SELEX-chip pattern, including ykgI-ykgC (Figure 2G) and yjbR-[uvrA]-ssb (Figure 2H). The 179-bp-long dan-ttdA probe with high-affinity to Dan formed a ladder consisting of at least eight bands on PAGE (Figure 2A), indicating that more than eight Dan molecules are able to bind to this 179-bp-long DNA segment (one Dan molecule per 20-bp-long DNA on average) in a sequential manner. Similar pattern of Dan binding was observed in all other SELEX DNA fragments tested, including all four group-A probes, dan-ttdA (Figure 2A), maoC-paaA (Figure 2B), yfaZ-yfaO (Figure 2C), intZ-yffL (Figure 2D) and two group-B probes, ygaD-[mltB]-srlA (Figure 2E) and perR-[insN]-ykfC (Figure 2F). However, gel retardation was not observed with probes with non-peak probes, yjbR–[uvrA]-ssb (Figure 2G) and ykgI-ykgC (Figure 2H), selected from the SELEX-chip pattern (Figure 1). The gel shift assay indicates that a certain level of the sequence selectivity of DNA binding by Dan. The gel mobility shift pattern of multi-step-complex formation is essentially the same with those of the major nucleoid proteins, HU and IHF (19), indicating that protein oligomerization takes place on DNA-bound Dan molecules (see below for AFM images).
Figure 2.
Gel mobility shift assay. Fluorescent-labeled DNA probes, each containing a SELEX segment from the dan-ttdA (A), maoC-paaA (B), yfaZ-yfaO (C), intZ-yffL (D), ygaD-[mltB]-srlA (E) and perR-insN (F) regions, or an unrelated reference fragment from yjbR-[uvrA]-ssb (G) and ykgI-ykgC spacer (H) were incubated at 37°C for 30 min with the indicated amounts (lane 1, 0; lane 2, 5 pmol; lane 3, 10 pmol; lane 4, 20 pmol; and lane 5, 30 pmol) of Dan. The reaction mixtures were directly subjected to PAGE.
The apparent Kd value of Dan ranges 10–100 nM depending on the probe DNA as estimated from the level of conversion of free DNA to Dan–DNA complexes. This value is slightly higher than those of Fis and IHF (19). The order of Dan-binding affinity among the probes examined was: dan-ttdA, yfaZ-yfaO > ygaD-[mltB]-srlA, intZ-yffL > maoC-paaA, perR-[insN]-ykfC (Figure 2).
DNA-binding specificity of Dan: DNase-I footprinting assay
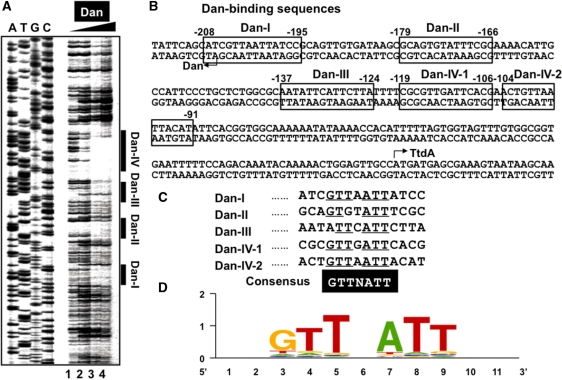
The sequence specificity of DNA binding by Dan was then analyzed by DNase-I footprinting using the dan-ttdA spacer probe, which was the most abundant segment isolated by the Genomic SELEX screening (see Supplementary Table 1A) and formed at least eight Dan–DNA complex bands by gel shift assay (Figure 2A). After brief treatment with DNase I, at least four protected regions were identified (Figure 3A), of which three (Dan-I, Dan-II and Dan-III) covered 14-bp-long sequences and one (Dan-IV) covered 29-bp sequence, suggesting that two molecules of Dan are associated with Dan-IV site, designated as Dan-IV-1 and Dan-IV-2 (Figure 3B). Besides the protection against DNase-I digestion, a hypersensitive site was also detected near the center within the protected regions as in the case of Dan-III region (Figure 3A). All the protected regions by Dan against DNase-I digestion included GTTNATT consensus sequence (Figure 3C). Similar GTTNATT-like sequences were identified in all other Dan-binding sites identified by both SELEX-clos and SELEX-chip (data not shown). After search of GTTNATT consensus sequence in the entire E. coli genome using the coliBASE database program (http://xbase.bham.uk/), a total of 1869 Dan-binding sites on top strand and 1860 sites on bottom strand were predicted to exist in the E. coli MG1655 genome. Sequence logo analysis of all these sequences indicates a high-level conservation of this 6-bp consensus GTTNATT sequence (Figure 3D).
Figure 3.
Identification of the Dan box sequence. (A) Fluorescent-labeled SELEX segment (1.0 pmol) from the dan-ttdA spacer was incubated in the absence or presence of increasing concentrations of purified Dan (10, 20 and 40 pmol from left to right) and then subjected to DNase-I foot-printing assays. Lanes A, T, G and C represent the respective sequence ladders. (B) Four Dan-binding sites were identified on the dan-ttdA spacer sequence. (C) The Dan-box sequence GTTAAT was predicted after sequence comparison of four Dan-binding sites. Similar sequences were identified among a total of 688 Dan-binding fragments selected by Genomic SELEX. Using this Dan-box sequence, a total of about 1860 sites were predicted to be present in the entire E. coli genome. (D) Logo representation of Dan-binding site derived from sequences in in silico analysis. Logos were generated using weblogo (http://weblogo.berkeley.edu/).
AFM imaging of Dan–DNA complexes
The binding mode of major E. coli nucleoid proteins has been studied in vitro by observing their DNA complexes immobilized on mica using atomic force microscopy (AFM) (33–38). The native nucleoid conformation of E. coli was also investigated successfully with AFM (39,40). AFM analysis provides some distinct advantages over bulk biochemical assays, particularly when studying DNA–protein interactions. For analysis of structural characteristics of Dan–DNA interaction, we also employed AFM to directly visualize Dan–DNA complexes. The mode of Dan binding to DNA was studied using a circular pGRdan plasmid (10-bp dan-promoter assay vector) as a probe, which contains the dan-ttdA spacer region with Dan-binding activity in the promoter assay vector pGRP (26). By gel mobility shift assay, this sequence formed multiple bands of Dan–DNA complexes at moderate protein concentrations below the aggregation thresholds (Figure 2A).
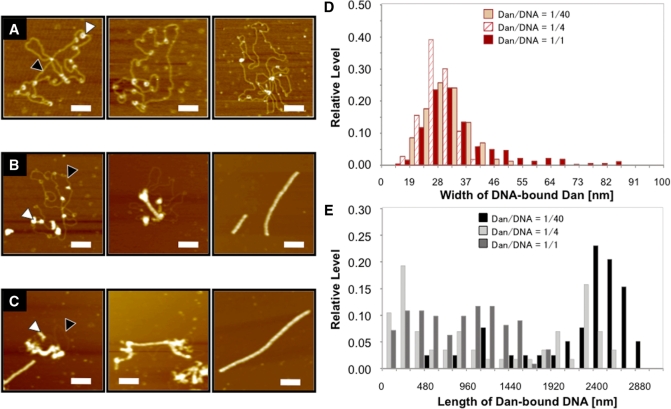
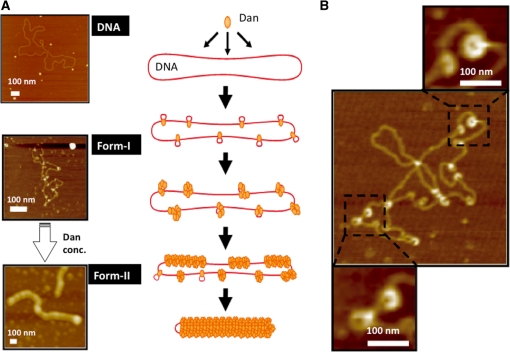
Dan–DNA complexes were observed with AFM in the presence of various concentrations of Dan (Figure 4A–C; also see Supplementary Figure 1). At low Dan concentrations below the Dan/DNA molar ratio of 1/40, Dan bound to various positions along the entire plasmid DNA (Figure 4A). At high Dan concentrations >1/10, the Dan–DNA complexes were converted into rod-like Dan–DNA complexes fully covered with Dan molecules (Figure 4C). At intermediate Dan concentrations, both forms of Dan–DNA complexes, i.e. plasmid DNA with Dan associated at various positions (form-I) and DNA fully covered with Dan, were observed (form-II) (Figure 4B and Supplementary Figure 1; for the model Figure 8).
Figure 4.
AFM images of Dan–DNA complexes. (A–C) Dan-pGRdan DNA complexes were formed at input Dan/DNA weight ratios of 1/40 (A), 1/4 (B) and 1/1 (C), respectively. AFM images were taken by the standard procedure as described in ‘Materials and Methods’ section. Black arrowheads show naked DNA region, while white arrowheads highlight DNA-bound Dan molecules. All images represent 500 × 500 nm2 area and scale bars indicate 100 nm. (D) Width of DNA-bound Dan dots was measured for a number of Dan dots observed in each AFM image. (E) DNA contour length was measured for a number of Dan–DNA complexes in each AFM image.
Figure 8.
Model of Dan-induced DNA compaction. (A) On the bases of AFM image analysis of Dan-pGRdan DNA complexes formed at increasing concentrations of Dan (see Figure 1), we propose a model of the conformational change of plasmid DNA. At low Dan concentrations, Dan binds various positions on pGRdan in non-specific manner (Form I). Each Dan may play a role of seeding for further assembly of additional Dan molecules. At high Dan concentration, the increased number of Dan molecules bound to DNA strand, leading to extension of Dan oligomers along the DNA strand and ultimately covering the entire DNA surface (Form 2). Typical AFM images of form-I and form-II Dan-DNA complexes are shown on left (500 × 500 nm2 area; scale bars, 100 nm). DNA strand is represented by a red line while Dan is represented by an orange rod. (B) A model of local conformation of DNA-bound Dan dots. Shortening of the contour length of pGRdan DNA might be due to local folding of DNA at the site of Dan binding (for details see text).
A number of AFM images were subjected to a statistical analysis to identify the width, height and length of Dan–DNA complexes. The histograms of width and length are shown in Figure 4D and E, respectively, while the histogram of height is shown in Supplementary Figure 2. The analysis of AFM images clearly indicates the drastic conformational transitions of Dan–DNA complexes with the increase in the amount of associated Dan molecules.
Width distribution
The width of Dan–DNA complexes at the site of Dan binding slightly increased upon increase in Dan concentration (Figure 4D and Supplementary Figure 2). At low Dan concentrations below the Dan-to-DNA molar ratio of 1/40, the width of single Dan molecule on DNA was estimated to be 27–32 nm. The width of peak position increased 1.5–2-fold reaching to around 45 nm concomitant with the increase in Dan concentration. At the Dan/DNA input ratio above 1, Dan–DNA complexes with the width larger than 50 nm were observed. This finding implies that Dan induces local DNA folding, giving the larger image of Dan–DNA dots at the site of Dan binding (for a model Figure 8).
Length distribution
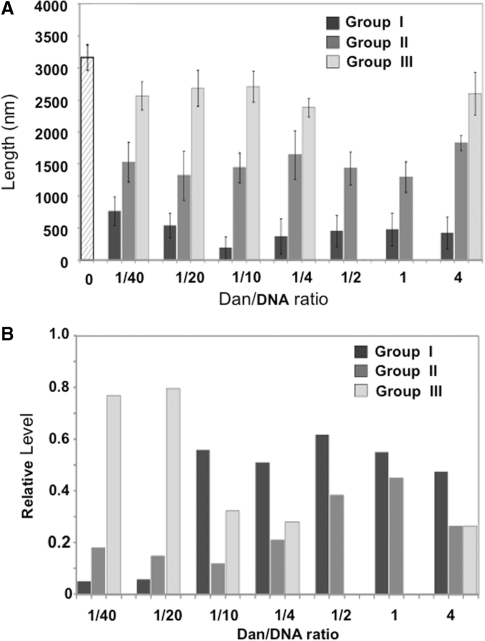
The contour length of Dan–DNA complexes gradually decreased ∼2-fold from 2.5–2.7 µm to 0.8–1.2 µm concomitant with the increase in Dan concentration (Figure 4E), supporting the notion that Dan binding induces DNA compaction. Since the length fluctuation increased upon increase in Dan concentration, the images were classified into three groups, group-I (<0.96 μm), group-II (between 0.96 and 2.08 μm) and group-III (2.08–3.20 μm), with respect to the contour length (Figures 4E and 5). Under the AFM imaging condition employed, the average length of naked plasmid DNA was estimated to be 3.265 ± 0.201 μm. At low Dan concentrations, the proportion of group-III images dominated ∼80% (close to naked DNA) (Figure 5). Under the same conditions, a single molecule of Dan binds, on average, at various positions along each probe DNA (Figure 4A). The slight decrease in DNA length after binding Dan may be explained by local folding of DNA at the site of Dan binding. At intermediate Dan concentrations, a number of Dan molecules bind to the probe DNA and as a result, the proportion of shortest group-I complexes became maximum (Figure 5) in agreement with the interpretation that local folding takes place at each site of Dan binding. Accordingly, the longest group-III complexes disappeared upon further increase in Dan concentration, and instead the proportion of intermediate-sized group-II increased linearly (Figure 5). At the highest Dan concentration used, group-III complexes appeared again, presumably due to tail-to-head association of rod-like complexes fully covered with Dan proteins.
Figure 5.
Contour length of Dan–DNA complexes. (A) Dan–pGRdan DNA complexes were formed at various input Dan/DNA ratios (1/40, 1/20, 1/10, 1/4, 1/2, 1/1 and 1/0.25). Contour length of AFM images was measured for a number of complexes from each AFM image. The Dan–pGRdan complexes were classified into three groups based on the length: group-I, shorter than 960 nm; group-II, 960–2080 nm; and group-III, 2080–3200 nm. Data represent the mean value obtained from more than 100 samples (N > 100). (B) The relative amounts of three groups of Dan–DNA complex were plotted against the input Dan/DNA ratios.
Height distribution
The height of naked DNA was around 0.5–0.6 nm, but upon binding of Dan, increased almost 10-fold to around 5 nm (Supplementary Figure 3). Thereafter, the height stayed rather constant around 5 nm up to the Dan concentrations analyzed, i.e. Dan/DNA ratio of 1/0.25 (Supplementary Figure 3F).
Role of Dan in transition of the nucleoid architecture
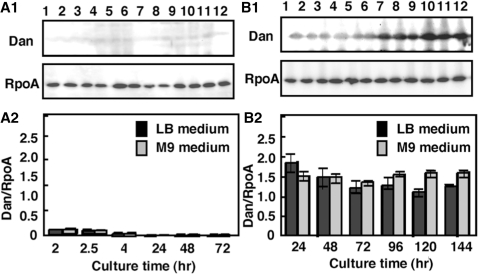
The protein composition of nucleoid has been studied in details for E. coli grown under laboratory culture conditions (20,41), but Dan has never been identified as a major nucleoid component in steady-state growing cells under the ordinary laboratory culture conditions. We then measured the intracellular concentration of Dan under various culture conditions by using the quantitative western blot analysis (5,21). When E. coli cells were grown under aerobic conditions in either rich (LB) or poor (M9–0.4% glucose) medium, the level of Dan was <10% of the level of RNA polymerase α subunit (Figure 6A). Under the same culture conditions, the level of α subunit is ∼5000 molecules per genome equivalent of DNA (3,41), while the major nucleoid proteins (HU, IHF and H-NS) in exponentially growing E. coli cells range from 5000 to 50 000 molecules per genome (5,20,41). Thus, Dan has never been identified as a structural component of the E. coli nucleoid because the intracellular level of Dan is <1% of the major nucleoid proteins.
Figure 6.
Intracellular concentration of the Dan protein. (A1) Wild-type E. coli KP7600 was grown in both LB and M9–0.4% glucose media at 37°C for various times under aerobic conditions. Cell lysates were prepared at various growth phases (lanes 1 and 7, log-phase at 0.3 OD600; lanes 2 and 8, log-phase at 0.6 OD600; lanes 3 and 9, late log-phase, 1.0 OD600; lanes 4 and 10, 24-h stationary-phase; lane 5 and 11, 48-h stationary-phase; lanes 6 and 12, 72-h stationary-phase) and subjected to quantitative western blots analysis against anti-Dan and anti-RpoA antibodies. (A2) The intensity of immuno-blot bands was measured with a LAS-1000 Plus Lumino-Image analyzer and IMAGE GAUGE (Fuji Film). (B1) Wild-type E. coli KP7600 was grown in both LB and M9–0.4% glucose media at 37°C for various times under hypoxic culture conditions. Cell lysates were prepared at various growth phases (lanes 1 and 7, log-phase at 0.3 OD600; lanes 2 and 8, log-phase at 0.6 OD600; lanes 3 and 9, late log-phase, 1.0 OD600; lanes 4 and 10, 24-h stationary-phase; lane 5 and 11, 48-h stationary-phase; lanes 6 and 12, 72-h stationary-phase) and subjected to western blots analysis against anti-Dan and anti-RpoA antibodies. (B2) The intensity of immuno-blot bands was measured with a LAS-1000 Plus Lumino-Image analyzer and IMAGE GAUGE (Fuji Film).
Next, we measured the Dan level in E. coli cells grown under various stressful conditions. Under a hypoxic culture condition, the level of Dan was 1.5- to 2.0-fold higher than the RpoA level in both rich and poor media and from exponential growth to stationary phase (Figure 6B). On the basis of RpoA level (∼5000 molecules per genome equivalent DNA), the level of Dan was estimated to range 7000–9000 molecules per genome. In late stationary phase, the Dan level was significantly higher in the poor medium than in the rich medium (Figure 5B, 96–144 h culture). This finding suggests that Dan plays a role in either maintenance of the nucleoid architecture and/or expression of the nucleoid function under the anaerobic conditions.
We also examined the Dan level at both log and stationary phase and in not only standard LB medium but also 20% LB, 20% LB containing 0.2% glucose or 0.2% arabinose, and DMEM medium (data not shown). The intracellular level of Dan was as close as that in aerobic culture in LB and M9–0.2% glucose medium (Figure 5A). Little induction of Dan was also observed at both low (4.5) and high (9.0) pH in M9-0.2% glucose, and at high osmolarity (3% NaCl in LB, and 20% polyethylene glycol-6000 in LB) (data not shown). None of these culture conditions induced the synthesis of Dan protein, supporting the notion that Dan is specifically induced under anaerobic conditions.
Intracellular localization of Dan
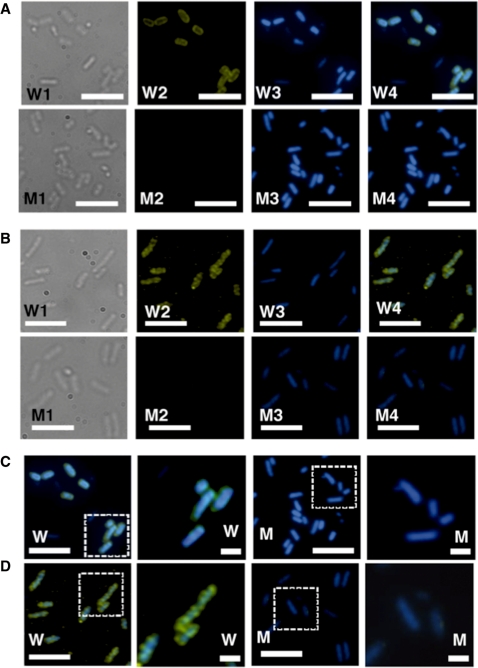
Previously we analyzed the intracellular localization of nucleoid proteins by indirect immuno-fluorescent labeling, and classified them into two groups: one group is proteins including HU, IHF, H-NS and Dps uniformly distributed within the nucleoid; another group is proteins including Fis located at specific loci within the nucleoid (28). Consequently, we observed the intracellular distribution of Dan in E. coli grown under aerobic and hypoxic conditions. The intracellular level of Dan within the aerobically grown wild-type KP7600 cells is very low (Figure 6A), showing a faint staining with anti-Dan antibodies (Figure 7A, W1–W4). This faint staining was not detected in the mutant JD24074 lacking the dan gene (Figure 7A, M1–M4).
Figure 7.
Intracellular localization of the Dan protein. Wild-type E. coli KP7600 and its dan mutant JD24074 were grown in M9–0.4% glucose media for 24 h at 37°C under hypoxic conditions, and subjected to the indirect immuno-fluorescent microscopy according to the standard procedure described in ‘Materials and Methods’ section. (A) Wild-type strain KP7600 (W1–W4) and the dan mutant JD24074 (M1–M4) were grown under aerobic (A) or hypoxic (B) conditions. W1 and M1 represent; W2 and M2, Indirect immuno-blot against anti-Dan; W3 and M3, DAPI-staining; W4 and M4, merged images of DAPI and immuno-stained patterns. (C, D) The area shown by dotted square is expanded. Anti-Dan antibody was raised in rabbits using the purified Dan as an antigen. Cys3-labeled anti-rabbit IgG antibody was used as the secondary antibody. White bars indicate 5 μm.
Under the hypoxic conditions, Dan was markedly induced (Figure 6B), and formed highly stained dots (Figure 7B, W1–W4; Figure 6D, W), indicated that Dan belongs to the group-II nucleoid proteins, including Fis, Rob (CbpB), CbpA and IciA, which show irregular distribution within the nucleoid (28). The distribution pattern is mostly similar to that of Fis, a bifuctional nucleoid protein with the regulatory activity of a set of growth-related genes. In the highly induced wild-type E. coli cells under the hypoxic conditions, Dan is enriched near the interface between the nucleoid and cytoplasm (or the surface of nucleoid), suggesting that some of the over-produced Dan is enriched in the boundary between nucleoid and cytoplasm. In the dan mutant, no immuno-stained Dan dots were detected (Figure 7B, M1–M4; Figure 6D, M), indicating that Dan could not be detected by the immuno-staining method using the same anti-Dan antibody.
Considering all these results, we conclude that Dan is a nucleoid protein specific in cells growing under anaerobic conditions, and thus belongs to the family of nucleoid proteins that are produced only at specific growth phases or under specific growth conditions. As in the case of other major nucleoid proteins (3,5,36), Dan may have functional dichotomy, controlling both the structure and function of the nucleoid. In fact, Dan was proposed to regulate the neighboring ttdA-ttdB-ygiE operon (18).
Growth of Dan-defective E. coli mutant under anaerobic conditions
Dan was identified as a nucleoid protein in cells growing under hypoxic or anaerobic conditions, implying that Dan plays a role in cell survival under anaerobic conditions. We constructed an E. coli mutant defective in the dan gene and compared the cell growth between wild-type and dan-defective mutants. In M9–0.4% glucose medium, the growth rate of the mutant was about 10–20% slower than the rate of wild-type E. coli even under aerobic conditions. Under the anaerobic culture condition, both the growth rate and saturation level of the dan-defective mutant were approximately 50% lower than those of wild-type control is also reduced (Supplementary Figure 4). The mutant cells tended to aggregate, forming sediments on the bottom of culture flask. These observations altogether indicate that Dan is needed for full growth under anaerobic conditions.
DISCUSSION
The aim of this research was to identify target genes or promoters recognized by Dan, one of the uncharacterized putative transcription factors, renamed from YgiP and TtdR (18). A collection of DNA fragments isolated after the genomic SELEX screening included at least 688 different sequences. Taken together with the gel shift and DNase-footprinting patterns, we concluded that Dan is a DNA-binding protein with recognition preference of GTTNATT sequence.
DNA-binding mode of Dan
Direct observation of nucleoid protein–DNA complexes with AFM has been successfully employed for analysis of protein–DNA complexes with E. coli H-NS (33,34), HU (35), Fis (36), Dps (37) as well as HU-like protein HCc3 of dianoflagellate (42), The pattern of DNA compaction by Dan is rather similar to that of HCc3 from dianoflagellate, which lacks histones but retains permanently condensed chromosome. Both Dan and HCc3 proteins form bundle architectures for DNA compaction in protein concentration-dependent manner.
After statistical analysis of AFM images formed with increasing concentrations of Dan (Figure 4; Supplementary Figures 2 and 3), we propose a model of two forms of Dan–DNA complexes (Figure 8). At low Dan concentrations, Dan binds to various positions on plasmid DNA in non-specific manner (Form-I) and upon increase in the Dan concentration, more Dan molecules binds through protein-protein interactions, ultimately forming the Dan-saturated complexes (Form-II) (Figure 8A). The height of DNA-bound Dan is rather constant independent of the protein concentrations, but the width increased with the increase in Dan concentration. This extension in the contour width may be due to local folding of DNA at the site of Dan binding as observed with AFM (Figure 8B). The length of Dan–DNA form-I complexes is smaller than that of pGRdan plasmid (Figure 5), supporting our proposal that Dan plays a role in local folding of DNA, ultimately leading to DNA compaction. By adding excess Dan, the length of Dan–DNA rods increased again (Figure 5). One explanation of the elongation of Dan–DNA complex rods at high Dan concentrations might be condensation of Dan–DNA rods by forming tail-to-head joining.
Physiological roles of Dan
With respect to the mode of DNA binding, Dan is apparently similar to the major nucleoid proteins HU and IHF, but it has never been identified as a protein component of the isolated nucleoids from E. coli cells grown under laboratory culture conditions (5,19,41). After testing various culture conditions, we found that Dan is highly expressed in cells grown under hypoxic or anaerobic conditions.
In the E. coli nucleoid, two groups of the nucleoid protein exist, universal nucleoid proteins (UNP) that always stay in the nucleoid; and growth condition-specific nucleoid proteins (GNP) that appear only at specific growth phases or under specific growth conditions (5,41). For instance, Fis is synthesized preferentially in growing cells (19,43) and plays an essential role for maintenance of the nucleoid competent for transcription of the growth-related genes (3,5,41). Fis is one of the key global regulators of a number of genes for adaptation to external conditions such as the availability of oxygen and nutrients (22,23). On the other hand, Dps (DNA-binding protein form starved cells) is the major nucleoid protein only in starved stationary-phase cells (6,20,24) and plays roles in protecting resting bacterial cells from environmental stresses such as high levels of toxic iron. During the transition of E. coli cells from exponential growth to stationary phase, marked changes take place on the cell shape and properties, allowing physical separation of cells at different stages (44). Growth-coupled transformation of the E. coli nucleoid from exponential growth phase-specific fibrous structures to stationary phase-specific rod forms (41) depends on the association of Dps to the genome DNA. This group of nucleoid proteins may be stored on the nucleoid surface, forming dots as detected by indirect immuno-staining (28; also Figure 7).
Since Dan is highly expressed under anaerobic and starved conditions (Figure 6B), it may play a role in protection of the genome DNA from anaerobic stress. Dan contains two sets of Cys–Cys pair: one consisting of Cys170 and Cys193, and another of Cys288 and Cys296. In the case of OxyR transcription factor, the Cys–Cys pair is involved in redox sensing (45). This motif is also involved in metal chelating, suggesting a role of Dan in resistance of a metal, as does Dps against iron. Thus, the reversible disulfide bond formation may also be involved in structural and functional control of the Dan protein. A group of Gram-positive bacteria such as Bacillus subtilis form spores for protection of the genome and survival under harmful stresses. On the contrary, Gram-negative bacteria such as E. coli that do not form spores must harbor another system for survival under stressful conditions. Dan and Dps play essential roles for protection of the genome during dormant stage.
The major nucleoid proteins, HU, IHF, H-NS and Fis, in exponential growth phase E. coli under aerobic conditions are all known to have functional dichotomy (5). Likewise, Dan may play dual roles, i.e. an architectural role and a global regulator of transcription. Non-coding sequence of the E. coli genome is less than 10%, but approximately two-third of the total number of Dan-binding sites is located within the intergenic spacer regions where regulatory signals of the genome functions such as transcription are located. Along this line, Dan may be involved in the regulation of genome transcription under anaerobic conditions. Such a biased localization of binding sequences was also observed for the major nucleoid proteins with functional dichotomy (46).
Oshima and Biville (18) predicted that YgiP, here renamed to Dan, is involved in regulation of the adjacent ttdA-ttdB-ygiE operon. Interestingly, tartrate dehydratase encoded by ttdA-ttdB for uptake of antioxidant tartrate is required for anaerobic growth on glycerol as a carbon source. Our findings extend this concept and further indicate that Dan is a bifunctional nucleoid protein, playing dual roles both in structuring the nucleoid under anaerobic growth conditions and in regulation of a set of genes needed for growth under anaerobic conditions. Along this line, besides the ttdA-ttdB-ygiE operon, a set of genes must exist, which are induced in the presence of Dan altogether contribute to the E. coli survival under the anaerobic conditions such as within animals.
SUMMARY AND IMPLICATIONS
The bacterial genome is associated with a set of DNA-binding architectural proteins, altogether forming the nucleoid. The major nucleoid proteins can be classified into two groups, UNP and GNP. The GNP-group proteins are interchangeable depending on the growth conditions or growth phases, including Fis in exponentially growing cells in rich media and Dps in starved cells in stationary growth phase. Here we discovered Dan (DNA-binding protein under anaerobic conditions) as the third member of the GNP. The growth of mutants lacking Dan is retarded under anaerobic conditions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-Aid for (Scientific Research Priority Area 17076016 and Scientific Research 18310133) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Funding for open access charge: Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Takenori Miki for providing the E. coli mutant lacking the dan gene, Naoki Kobayashi and Minami Naruse for the initial SELEX screening, and Ayako Kori and Kayoko Yamada for expression and purification of Dan, and Tomohiro Shimda, Hiroshi Ogasawara and Kaneyoshi Yamamoto for discussion.
REFERENCES
- 1.Riley M, Abe T, Arnaud MB, Berlyn MK, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, Kosuge T, et al. Escherichia coli K-12: a cooperatively developed annotation snapshot – 2005. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann B. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt FC, et al., editors. Escherichia coli and Salmnella. Washington: ASM Press; 1996. pp. 2460–2488. [Google Scholar]
- 3.Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Ann. Rev. Microbiol. 2000;54:499–518. doi: 10.1146/annurev.micro.54.1.499. [DOI] [PubMed] [Google Scholar]
- 4.Perez-Rueda E, Collado-Vides J. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 2000;28:1838–1847. doi: 10.1093/nar/28.8.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishihama A. The nucleoid: an overview. In: Neidhardt FC, et al., editors. Escherichia coli and Salmnella, 2nd version. Washington: ASM Press; 2009. [Google Scholar]
- 6.Ishihama A. Adaptation of gene expression in stationary phase bacteria. Curr. Opn. Microbiol. 1997;7:582–588. doi: 10.1016/s0959-437x(97)80003-2. [DOI] [PubMed] [Google Scholar]
- 7.Shimada T, Fujita N, Maeda M, Ishihama A. Systematic search for the Cra-binding promoters using genomic SELEX systems. Genes Cells. 2005;10:907–918. doi: 10.1111/j.1365-2443.2005.00888.x. [DOI] [PubMed] [Google Scholar]
- 8.Ogasawara H, Hasegawa A, Kanda E, Miki T, Yamamoto K, Ishihama A. Genomic SELEX search for target promoters under the control of the PhoQP-RstBA signal cascade. J. Bacteriol. 2007;187:4791–4799. doi: 10.1128/JB.00319-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogasawara H, Ishida Y, Yamada K, Yamamoto K, Ishihama A. PdhR (pyruvate dehydrogenase complex regulator) controls the respiratory electron transport system in Escherichia coli. J. Bacteriol. 2007;189:5534–5541. doi: 10.1128/JB.00229-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimada T, Hirao K, Kori A, Yamamoto K, Ishihama A. RutR is the uracil/thymine-sensing master regulator of a set of genes for synthesis and degradation of pyrimidines. Mol. Microbiol. 2007;66:744–779. doi: 10.1111/j.1365-2958.2007.05954.x. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Ogawa Y, Camkaris H, Shimada T, Ishihama A, Pitard AJ. folA, a new member of the TyrR regulon in Escherichia coli K-12. J. Bacteriol. 2007;189:6080–6084. doi: 10.1128/JB.00482-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umezawa Y, Shimada T, Kori A, Yamada K, Ishihama A. The uncharacterized transcription factor YdhM is the regulator of the nemA gene encoding N-ethylmaleimide reductase. J. Bacteriol. 2008;190:5890–5897. doi: 10.1128/JB.00459-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasegawa A, Ogasawara H, Kori A, Teramoto J, Ishihama A. The transcription regulator AllR senses both allantoin and glyoxylate and controls a set of genes for degradation and utilization of purines. Microbiology. 2008;154:3366–3378. doi: 10.1099/mic.0.2008/020016-0. [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto K, Matsumoto F, Oshima T, Fujita N, Ogasawara N, Ishihama A. Anaerobic regulation of citrate fermentation by CitAB in Escherichia coli. Biosci. Biotechnol. Biochem. 2008;72:3011–3014. doi: 10.1271/bbb.80301. [DOI] [PubMed] [Google Scholar]
- 15.Shimada T, Yamamoto K, Ishihama A. Involvement of leucine-response transcription factor LeuO in regulation of the genes for sulfa-drug efflux. J. Bacteriol. 2009;191:4562–4571. doi: 10.1128/JB.00108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishida U, Kori A, Ishihama A. Participation of regulator AscG of the β-glucoside utilization operon in regulation of the propionate catabolism operon. J. Bacteriol. 2009;191:6136–6144. doi: 10.1128/JB.00663-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schell MA. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 18.Oshima T, Biville F. Functional identification of ygiP as a positive regulator of the ttdA-ttdB-ygjE operon. Microbiology. 2006;152:2129–2135. doi: 10.1099/mic.0.28753-0. [DOI] [PubMed] [Google Scholar]
- 19.Azam TA, Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli: Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- 20.Azam TA, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jishage M, Iwata A, Ueda S, IshIhama A. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 1996;178:2507–2513. doi: 10.1128/jb.178.18.5447-5451.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsson LA, Vanet A, Vijgenboom E, Bosch L. The role of Fis in transcription activation of stable RNA operons. EMBO J. 1990;9:727–737. doi: 10.1002/j.1460-2075.1990.tb08166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross W, Thomspon JF, Newlands JT, Gourse GL. E. coli Fis protein activates rRNA transcription in vitro and in vivo. EMBO J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almiron M, Link AJ, Furlog D, Kolter D. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2624–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 25.Grainger DC, Hurd D, Harrison M, Holdstock J, Busby S.JW. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E coli chromsome. Proc. Natl Acad. Sci. USA. 2005;102:17693–17698. doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimada T, Makinoshima H, Ogawa Y, Miki T, Maeda T, Ishihama A. Classification and strength measurement of stationary-phase promoters by use of a newly developed promoter cloning vector. J. Bacteriol. 2004;186:7112–7122. doi: 10.1128/JB.186.21.7112-7122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiraga S, Ichinose C, Niki H, Yamazoe M. Cell cycle-dependent duplication and bidirectional migration of SeqA-associated DNA-protein complexes in E. coli. Mol. Cell. 1998;1:381–387. doi: 10.1016/s1097-2765(00)80038-6. [DOI] [PubMed] [Google Scholar]
- 28.Azam TA, Hiraga S, Ishihama A. Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells. 2000;5:613–626. doi: 10.1046/j.1365-2443.2000.00350.x. [DOI] [PubMed] [Google Scholar]
- 29.Ellington AD, Szostak JW. In vitro selection of DNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 30.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 31.Singer BS, Shtatland T, Brown D, Gold L. Libraries for genomic SELEX. Nucleic Acids Res. 1997;25:781–786. doi: 10.1093/nar/25.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimada T, Ishihama A, Busby SJ, Grainger DC. The Escherichia coli RutR transcription factor binds at targets within genes as well as intergenic regions. Nucleic Acids Res. 2008;36:3950–3955. doi: 10.1093/nar/gkn339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dame RT, Luijsterburg MS, Krin E, Berlin PN, Wagner R, Wuite GJ. DNA bridging: a property shared among H-NS-like proteins. J. Bacteriol. 2005;187:1845–1848. doi: 10.1128/JB.187.5.1845-1848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dame RT, Wyman C, Goosen N. H-NS mediated compaction of DNA visualized by atomic force microscopy. Nucleic Acid Res. 2000;28:3504–3510. doi: 10.1093/nar/28.18.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kar S, Choi EJ, Guo F, Dimitriadis EK, Kotova SL, Adhya S. Right-handed DNA supercoiling by an octmeric form of histone-like protein HU: modulation of cellular transcription. J. Biol. Chem. 2006;281:40144–40153. doi: 10.1074/jbc.M605576200. [DOI] [PubMed] [Google Scholar]
- 36.Maure S, Fritz J, Muskhelishvili G, Travers A. RNA polymerase and an activator form discrete subcomplexes in a transcription initiation complex. EMBO J. 2006;25:3784–3790. doi: 10.1038/sj.emboj.7601261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ceci P, Cellai S, Falvo E, Rivetti C, Rossi GL, Chianocone E. DNA condensation and self-aggregation of Escherichia coli Dps are coupled phenomena related to the properties of the N-terminus. Nucleic Acid Res. 2004;32:5935–5944. doi: 10.1093/nar/gkh915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benevides JM, Serban D, Thomas G.J., Jr Structural perturbations induced in linear and circular DNA by the architectural protein HU from Bacillus stearothermophilus. Biochemistry. 2006;45:5359–5368. doi: 10.1021/bi0523557. [DOI] [PubMed] [Google Scholar]
- 39.Kim J, Yoshimura SH, Hizume K, Ohniwa RL, Ishihama A, Takeyasu K. Fundamental structural units of the Escherichia coli nucleoid revealed by atomic force microscopy. Nucleic Acids Res. 2004;32:1982–1992. doi: 10.1093/nar/gkh512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohniwa RL, Morikawa K, Kim J, Ohta T, Ishihama A, Wada C, Takeyasu K. Dynamic state of DNA topology is essential for genome condensation in bacteria. EMBO J. 2006;25:5591–5602. doi: 10.1038/sj.emboj.7601414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishihama A. Modulation of the nucleoid, the transcription apparatus, and the translation machinery in bacteria for stationary phase survival. Genes Cells. 1999;4:135–143. doi: 10.1046/j.1365-2443.1999.00247.x. [DOI] [PubMed] [Google Scholar]
- 42.Chan YH, Wong JT. Concentration-dependent organization of DNA by the dinoflagellate histone-like protein HCc3. Nucleic Acid Res. 2007;35:2573–2583. doi: 10.1093/nar/gkm165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ball CA, Osuna R, Ferguson KC, Johnson RC. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J. Bacteriol. 1992;174:8042–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makinoshima H, Nishimura A, Ishihama A. Fractionation of Escherichia coli cell populations at different stages during growth transition to stationary phase. Mol. Microbiol. 2002;43:269–279. doi: 10.1046/j.1365-2958.2002.02746.x. [DOI] [PubMed] [Google Scholar]
- 45.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 46.Grainger DC, Hurd D, Goldberg MD, Busby S.JW. Association of nucleoid proteins with coding gand non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 2006;34:4642–4652. doi: 10.1093/nar/gkl542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.