Abstract
The multifunctional HCV core protein consists of a hydrophilic RNA interacting D1 domain and a hydrophobic D2 domain interacting with membranes and lipid droplets. The core D1 domain was found to possess nucleic acid annealing and strand transfer properties. To further understand these chaperone properties, we investigated how the D1 domain and two peptides encompassing the D1 basic clusters chaperoned the annealing of complementary canonical nucleic acids that correspond to the DNA sequences of the HIV-1 transactivation response element TAR and its complementary cTAR. The core peptides were found to augment cTAR-dTAR annealing kinetics by at least three orders of magnitude. The annealing rate was not affected by modifications of the dTAR loop but was strongly reduced by stabilization of the cTAR stem ends, suggesting that the core-directed annealing reaction is initiated through the terminal bases of cTAR and dTAR. Two kinetic pathways were identified with a fast pre-equilibrium intermediate that then slowly converts into the final extended duplex. The fast and slow pathways differed by the number of base pairs, which should be melted to nucleate the intermediates. The three peptides operate similarly, confirming that the core chaperone properties are mostly supported by its basic clusters.
INTRODUCTION
Hepatitis C virus (HCV) (1) is an important human pathogen transmitted mainly through blood contacts and affecting ∼170 million people worldwide (2). Chronic infections are frequent and a major cause of liver cirrhosis and cancer (3). HCV is an enveloped virus of the Flaviviridae family, which has a positive-sense, single-stranded RNA genome with an open-reading frame encoding a polyprotein of 3010 amino acids or so (4). The 5′- and 3′-ends of the viral RNA correspond to non translated, highly structured and conserved regions (5′ UTR and 3′ UTR) of 340 and 230 nucleotides, respectively. The 5′ UTR consists of five stem–loops (5) and includes an internal ribosome entry site (IRES) that directs translation of the HCV genome in a cap-independent manner. The 3′ UTR is composed of three distinct parts: (i) a variable part with the VSL1 and VSL2 stem–loops, (ii) a poly (U/UC) tract and (iii) a sequence X of 98 nucleotides (6,7). Deletion of the U-rich and X sequence results in a loss of virus replication and infectivity (8).
The polyprotein precursor is co- and post-translationally processed by cellular and viral proteases at the endoplasmic reticulum (ER) membrane to yield the mature structural and nonstructural proteins. The structural proteins include the core and the envelope glycoproteins E1 and E2 (9). The N-terminal end of the polyprotein, which is a major HCV antigen (10,11), is cleaved by a cellular signal peptidase, generating the core of 191 residues which binds the ER membrane (12). This protein is then processed by a signal peptide peptidase (13,14), giving a mature protein of 179 residues or so, which is targeted to the lipid droplets (LDs) (15–17). Apart from LDs, the protein can be retained at the ER membrane and localized on mitochondria (15,18). The mature protein is a dimeric, alpha-helical protein exhibiting features that are consistent with those of a membrane protein (19) . It consists of a basic N-terminal RNA binding domain D1 (aa 2–117) with three highly basic amino acid clusters (BD1-BD3) (20), and a hydrophobic C-terminal domain D2 (aa 118–179 or so) involved in targeting the protein to LDs (21). Core attachment to LDs is critical for assembly and release of infectious HCV particles (HCVcc) (22).
The core exerts multiple roles in infected cells and virus replication. The core notably interacts with a number of cellular proteins which impacts on cell proliferation and differentiation (10,23) and also on the lipid metabolism with a possible influence on hepatocellular carcinomas (24). As a major viral component tightly binding to genomic RNA sequences, the core is also thought to drive genomic RNA packaging and nucleocapsid formation (25–27). Moreover, the core has potent nucleic acid chaperoning activities (20,28), similar to retroviral nucleocapsid (NC) proteins (29–35). Through this activity, the core can resolve RNA misfolding and promote annealing of complementary sequences and strand exchanges (28). This activity appears to be mediated by the three basic clusters (BD1–BD3) of the D1 domain (20,28).
To further characterize the core RNA chaperone properties, we investigated how the D1 domain and peptides encompassing the basic clusters direct the annealing of complementary DNA sequences represented by the canonical model of the HIV-1 transactivation response (TAR)(36) element in a DNA form (dTAR) and the complementary sequence cTAR. The extremely slow annealing of these two sequences is drastically accelerated by addition of a retroviral NC protein (30,32,34) or the HCV core (28). The nucleocapsid protein NCp7 of HIV-1 activates the transient opening of the cTAR terminal base-pairs (29,37) causing a partial melting of the stem (33,38). These partly melted TAR DNAs anneal then through their termini to form an intermediate complex which is further converted into the final extended duplex (ED) (30,33,35). To determine whether the HCV core promotes cTAR/dTAR annealing through a similar molecular mechanism, we monitored the real-time annealing kinetics of fluorescently labelled cTAR with dTAR and mutants thereof in the presence of D1 Domain, and peptides E and F corresponding to the two (BD2 and BD3) and all three (BD1, BD2 and BD3) basic clusters, respectively. Results indicate that the three core peptides destabilize cTAR and dTAR secondary structures to a small extent, but efficiently promote dTAR/cTAR annealing through the dTAR and cTAR ends.
MATERIALS AND METHODS
Core peptides and oligonucleotides
Peptides E and F were synthesized by solid-phase peptide synthesis on a 433 A synthesizer (ABI, Foster City, CA, USA) as described (20). Purification by HPLC was carried out on a C8 column (Uptisphere 300 A, 5 µm; 250×10, Interchim, France) in 0.05% TFA with a linear gradient of 10–70% of acetonitrile for 30 min. The peptide purity and molecular weight (6754 for peptide E and 9444 for Peptide F) were checked by LC/MS. Absorption coefficient of 5700 M–1 cm–1 at 280 nm was used to determine their concentration.
The D1 domain consisting of amino acids 1–117 of the core protein fused to a C-terminal 6xHis tag was prepared as previously described (19).
The unlabelled and the labeled ODNs were synthesized by IBA GmbH Nucleic Acids product Supply (Gottingen, Germany). In the case of the doubly labeled ODNs, the 5′ terminus was labeled with carboxytetramethylrhodamine (TMR) or ethyl 2-[3-(ethylamino)-6-ethylimino-2,7-dimethylxanthen-9-yl]benzoate hydrochloride (Rh6G) via an amino-linker with a six carbon spacer arm, while the 3′ terminus was labeled with either 4-(4′-dimethylaminophenylazo) benzoic acid (Dabcyl) or 5(and 6)-carboxyfluorescein (Fl) using a special solid support with the dye already attached. ODNs were purified by the manufacturer by reverse-phase HPLC and polyacrylamide gel electrophoresis.
Experiments were performed in 25 mM Tris–HCl (pH 7.5), 30 mM NaCl and 0.2 mM MgCl2 at 20°C.
Fluorescence measurements
Emission spectra and kinetic traces were recorded with Fluorolog and FluroMax spectrofluorimeters (Jobin Yvon Instruments, S.A. Inc.) equipped with a temperature-controlled cell compartment. All fluorescence intensities were corrected for buffer emission and Lamp fluctuations.
Kinetic measurements were performed in pseudo first-order conditions by using concentrations of unlabelled TAR and dTAR at least 10-fold higher than the concentration of the labeled complementary sequence. Excitation and emission wavelengths were 520 and 550 nm, respectively, to monitor the Rh6G fluorescence. The corresponding wavelengths were 480 and 520 nm, respectively to monitor the Fl fluorescence. All reported concentrations correspond to those after mixing. To avoid high local concentrations during mixing, both reactant mixtures were prepared of the same volume. Peptides were added to each reactant separately, and then the reaction was initiated by mixing the peptide-coated ODNs together. The kinetics was fast enough to monitor the fluorescence intensities continuously without photo-bleaching. The apparent rate constants kobs and the amplitudes were determined from the kinetic traces by including a dead-time correction t0 to take into account the delay between the mixing of reactants and the start of the measurements. All fitting procedures were carried out with OriginTM 7.5 software based on nonlinear, least-square methods and the Levenberg-Marquardt algorithm.
The temperature dependence of the annealing kinetics was carried out with 10 nM doubly labeled cTAR derivatives and 300-nM nonlabeled dTAR derivatives at different temperatures (10°C, 20°C, 25°C, 30°C, 35°C and 40°C) in the presence of either peptide E, peptide F and Domain D1, added at a peptide to oligonucleotide ratio of 1.4:1.
Fluorescence correlation spectroscopy setup and data analysis
Fluorescence correlation spectroscopy (FCS) measurements were performed on a two-photon platform including an Olympus IX70 inverted microscope, as described previously (37,39). Two-photon excitation at 850 nm is provided by a mode-locked Tsunami Ti:sapphire laser pumped by a Millenia V solid-state laser (Spectra Physics, USA). The experiments were performed in an eight-well lab-Tek II coverglass system, using a 400-µl volume per well. The focal spot was set about 20 µm above the coverslip. Peptide E was added to the labeled cTAR sequences at different nucleotide to peptide ratios. To avoid high local concentrations during mixing, both reactants were of the same volume. Assuming that peptide E diffuses freely in a Gaussian excitation volume, the normalized autocorrelation function, G(τ), calculated from the fluorescence fluctuations was fitted according to (40):
| (1) |
where τd is the diffusion time, N is the mean number of molecules within the excitation volume and S is the ratio between the axial and lateral radii of the excitation volume. The point spread function of the set-up was determined from a z-scan on one fluorescent bead (20 nm in diameter). The measured lateral and axial resolutions were respectively 0.3 and 1 µm. Typical data recording times were 10 min. Twenty autocorrelation curves were recorded for each sample. When spikes of high fluorescence intensity, most likely associated to aggregates, were observed in the fluorescence fluctuation profiles, the corresponding autocorrelation curves were discarded. In these conditions, the average autocorrelation curve calculated from the sum of the remaining autocorrelation curves could be adequately fitted by a single population model with a diffusion time close to that of cTAR alone.
RESULTS
Investigating the interaction of cTAR DNA with the core peptides by FCS
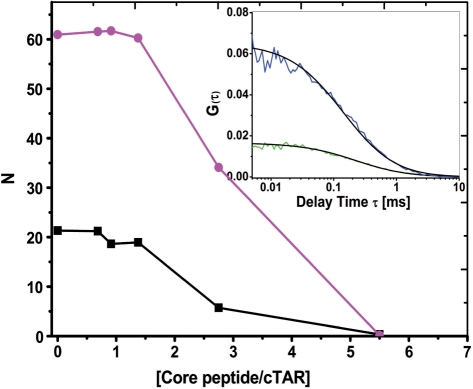
To characterize in depth the nucleic acid chaperone properties of the core protein, we first had to find experimental conditions where the core peptides did not cause DNA aggregation in a manner similar to other nucleic acid chaperones (32,34,41,42). Indeed, molecular aggregation can cause strong bias when using fluorescence-based techniques (30,32). Since nucleic acid aggregation by positively charged peptides is concentration dependent (42,43), we investigated this dependence by means of FCS. In FCS, the fluorescence intensity arising from a very small volume (about 0.2 fL) is correlated to obtain information about the processes that give rise to fluorescence fluctuations. These fluctuations are mainly governed by the diffusion of fluorescent species throughout this volume, and thus parameters such as the average number of fluorescent species within this volume and their diffusion constant can be determined. As a consequence, aggregation of the TMR-labeled 5′-cTAR molecules by the core peptides would be expected to decrease the number of fluorescent species.
Experiments were performed at two different ODN concentrations (100 and 300 nM). In the absence of peptide, the number of fluorescent TAR molecules in the excitation volume was fully consistent with the theoretical number of molecules expected from their concentration. By adding increasing concentrations of either core peptide, we found no change in the number of fluorescent species up to a peptide/ODN molar ratio of 1.4:1, indicating that no aggregation occurred under these conditions (Figure 1). In contrast, aggregation took place at higher ratios (2.8:1 and 5.6:1) as evidenced by the sharp drop in the number of fluorescent species. Changes in the diffusion constant were also observed at these higher ratios but were less prominent (data not shown), due to the dependence of the diffusion constant on the cubic root of the molecular mass of the diffusing species. As a consequence, we selected a peptide/ODN molar ratio of 1.4:1 to characterize the chaperone properties of the core peptides.
Figure 1.
Evidence by fluorescence correlation spectroscopy of core peptide-induced aggregation of TMR-5′-cTAR. The average number of fluorescent species, N, for 100 nM (black squares) or 300 nM (magenta circles) of TMR-5′-cTAR within the excitation volume was obtained by fitting the autocorrelation curves to Equation (1). The curves are given with peptide E. Data with peptide F and D1 domain were very similar and were thus not represented. (Inset) Autocorrelation curves of 100 nM (blue line) and 300 nM TMR-5′-cTAR (green line) and their fits (black line) with Equation (1) in the presence of 140 nM and 420 nM of peptide E, respectively.
Destabilization of cTAR secondary structure by the core peptides
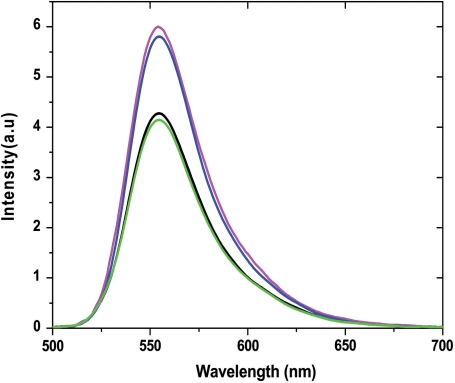
Since destabilization of nucleic acid secondary structures is a feature of the chaperone protein activity (29,32,37,41,44), we characterized the nucleic acid destabilizing activity of the core peptides, using cTAR labeled at its 5′ and 3′ ends by Rh6G and Dabcyl, respectively. In the absence of peptide, the proximity of cTAR ends induces a strong fluorescence quenching of Rh6G by the Dabcyl group (37), while the melting of the lower half of the cTAR stem increases the distance between the two dyes and thus, restores Rh6G fluorescence. Thus, the destabilizing activity of the core peptides can be evaluated from the ratio of the fluorescence intensity with or without the peptide. Addition of core peptides E and F at a peptide/cTAR molar ratio of 1.4:1 led to a 1.5-fold increased fluorescence intensity for Rh6G-5′-cTAR-3′-Dabcyl, while no change was observed with the D1 domain (Figure 2). Since full melting of cTAR would lead to a 20- to 25-fold increase of the Rh6G fluorescence, <10% of the Rh6G fluorescence was restored upon binding of the core peptides. No further increase was observed with either peptide at higher ratios (data not shown), possibly due to the fluorescence quenching related to formation of aggregates. Thus, in contrast to retroviral NC proteins (30–32,37), the core peptides can only slightly destabilize the secondary structure of cTAR, in the present experimental conditions.
Figure 2.
Destabilization of cTAR secondary structure by core peptides. Emission spectra of Rh6G-cTAR-Dabcyl (100 nM) were recorded in the absence (black) and presence of peptide E (magenta), peptide F (blue) and D1 Domain (green) at a molar ratio of 1.4 peptide per cTAR in 25 mM Tris, 30 mM NaCl and 0.2 mM MgCl2, pH 7.5. Excitation wavelength was 520 nm.
Kinetics of cTAR/dTAR annealing in the presence of peptide E
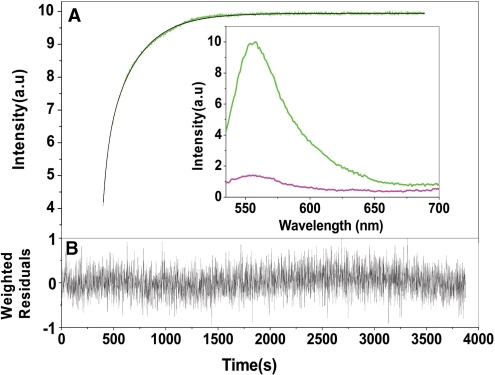
To characterize the mechanism by which the core protein can activate the annealing of the complementary cTAR and dTAR sequences, we first used the peptide E, which was easy to synthesize in large quantities and was previously shown to mimic the core chaperone properties (20,28). The real-time annealing kinetics of cTAR to dTAR was monitored by mixing Rh6G-5′-cTAR-3′-Dabcyl with an excess of non-labeled dTAR, in the presence of peptide E added at a peptide/ODN ratio of 1.4:1. Formation of the 55-bp ED strongly increases the interchromophore distance, leading to a full recovery of Rh6G emission (29). The same fluorescence plateau was obtained as in the absence of peptide, indicating that ED formation went to completion (Figure 3A). Peptide E strongly accelerated the annealing reaction, since it was complete in <30 min, while it needed more than 24 h in the absence of peptide (30). As shown by the nearly random distribution of residuals around the zero value (Figure 3B), the annealing kinetic traces could be adequately fitted using a biexponential function:
| (2) |
where, t0 being the dead time, kobs1,2 are the observed kinetic rate constants, a is the amplitude of the fast component, and I0 and If are the fluorescence intensities of the stem–loop and the ED, respectively.
Figure 3.
Kinetics of cTAR/dTAR annealing in the presence of peptide E. (A) Kinetic trace of 10 nM doubly labeled cTAR reacted with 300 nM dTAR in the presence of peptide E added at a peptide/ODN molar ratio of 1.4:1. The continuous line describes the best fit of the data according to equation (2) with kobs1 = 1.8 × 10–2 s–1, kobs2 = 2.8 × 10–3 s–1 and a = 0.7. Inset: emission spectra of the doubly labeled cTAR in the presence of peptide E added at a peptide/ODN ratio of 1.4:1 before (magenta) and after completion of the annealing reaction with dTAR (green). (B) Weighted residuals for the fit of the experimental data to Equation (2).
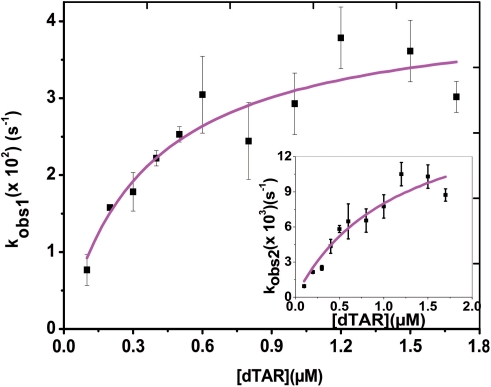
The dTAR concentration dependence indicates a saturation behavior for both kobs1 and kobs2 values (Figure 4), consistent with a two-step reaction, where a rate-limiting interconversion step is coupled to a much faster, preceding binding step, considered as a pre-equilibrium:
| (3) |
where IC corresponds to the intermediate complex. The dTAR concentration dependence of both kobs values was fitted using (45):
| (4) |
Figure 4.
Kinetic parameters of cTAR/dTAR annealing in the presence of peptide E. The fast (kobs1) and slow (kobs2) (inset) components were determined in pseudo-first-order conditions from real-time kinetics, as described in Figure 3. The solid line corresponds to the fit of the data with Equation (4) and the KM and kf values given in Table 1.
Values of 4 (±2) × 106 M–1 and 1.4 (±0.7) × 106 M–1 were obtained for KM1 and KM2, the equilibrium binding constants of the intermediate complexes associated with the fast and slow kinetic component, respectively. Moreover, values of 0.045 (±0.008) s–1 and 0.016 (±0.002) s–1 were obtained for the sum of the forward (kf1, kf2) and backward (kb1, kb2) interconversion rate constants associated to the two kinetic components. The values of kb1 and kb2, given by the intercept, were very low (<0.001 s–1) and could not be determined from the experimental data, indicating that peptide E is unable to dissociate the ED, and that the 0.045 s–1 and 0.016 s–1 values mainly correspond to kf1 and kf2, respectively.
Noticeably, substitution of the Rh6G and Dabcyl dyes at the 5′- and 3′-ends of cTAR by TMR and Fl, respectively, had a marginal influence on the kinetic parameters (Table 1), indicating that the peptide E-promoted cTAR/dTAR annealing kinetics does not depend on the nature of the dyes.
Table 1.
Kinetic parameters of cTAR/dTAR and cTAR/TAR annealing in the presence of core peptidesa
| Oligo- nucleotide | Fluorophores | Complementary sequence | Peptide | Ratio of peptide/ oligo | a | KM1 | KM2 | kf1 | kf2 | kass1 | kass2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| (M–1) | (M–1) | (s–1) | (s–1) | (M–1 s–1) | (M–1 s–1) | ||||||
| × 10–6 | ×10–6 | ×102 | × 102 | × 10–4 | × 10–4 | ||||||
| cTAR | 3′Rh6G – 5′Dabcyl | dTAR | E | 1.4 | 0.70 (±0.01) | 4 (±2) | 1.4 (±0.7) | 4.5 (±0.9) | 1.6 (±0.2) | 18 (±2) | 2.2 (±0.1) |
| cTAR | 3′Fl – 5′TMR | dTAR | E | 1.4 | 0.79 (±0.02) | 1.5 (±0.9) | 1.1 (±0.7) | 6.4 (±2) | 1.1 (±0.2) | 10 (±2) | 1.2 (±0.1) |
| cTAR | 3′Rh6G – 5′Dabcyl | dTAR | E | 1 | 0.60 (±0.01) | 1.1 (±0.4) | 0.55 (±0.1) | 5.3 (±0.6) | 1.7 (±0.2) | 6 (±2) | 1 (±0.02) |
| cTAR | 3′Rh6G – 5′Dabcyl | dTAR-TL | E | 1.4 | 0.60 (±0.01) | 3.1 (±0.8) | 1.3 (±0.6) | 5 (±1) | 1 (±0.1) | 16 (±1) | 1.3 (±0.06) |
| cTAR | 3′Rh6G – 5′Dabcyl | TAR | E | 1.4 | 0.35 (±0.02) | 1.6 (±0.5) | 1.2 (±0.8) | 2 (±0.1) | 0.13 (±0.01) | 3 (±0.05) | 0.16 (±0.008) |
| (14-39) cTAR | 3′Rh6G – 5′Dabcyl | dTAR | E | 1.4 | – | 0.33 (±0.02) | 0.07 (±0.03) | 0.030 (±0.003) | |||
| cTAR 1,2 | 3′Rh6G – 5′Dabcyl | dTAR | E | 1.4 | -b | -b | 0.4 (±0.2) | -b | 0.02 (±0.01) | b | 0.008 (±0.002) |
| cTAR | 3′Rh6G – 5’Dabcyl | dTAR | F | 1.4 | 0.85 (±0.02) | 3 (±1) | 2.7 (±0.7) | 17 (±3) | 4.2 (±0.6) | 51 (±3) | 11.3 (±0.4) |
| cTAR | 3′Rh6G – 5′Dabcyl | dTAR | D1 | 1.4 | 0.80 (±0.02) | 2 (±1) | 1.5 (±0.6) | 4.1 (±0.4) | 0.6 (±0.05) | 8 (±0.8) | 1 (±0.03) |
aThe association equilibrium constants (KM1 for fast and KM2 for slow components) and interconversion rate constants (kf1 for fast and kf2 for slow component) are calculated from Equation (4), as described in Figure 4; kass1 and kass2 are given by: kassi = KMi × kfi, while a corresponds to the amplitude of the fast component.
b A faster kinetic pathway with an amplitude of <5% is observed, but not reported.
In contrast to the kobs values, the amplitudes of the two components (Table 1) were independent of dTAR concentration (data not shown), suggesting that peptide E-promoted annealing reaction involves two parallel pathways. To confirm this, we repeated the annealing reaction at a peptide/ODN molar ratio of 1:1. At this lower ratio, the amplitudes of the two components were also independent of dTAR concentration, but with a decreased value for the fast component as compared to the 1.4:1 ratio, in line with the hypothesis of two parallel pathways. Moreover, while the KM1 and KM2 values were three to four times lower than at the higher ratio, the interconversion rate constants were similar (Table 1), suggesting that peptide E mainly affects the pre-equilibrium step.
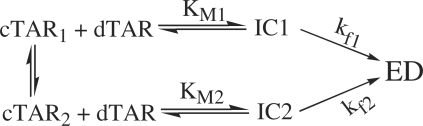
On the basis of our kinetic data, a reaction mechanism with two parallel kinetic pathways involving two different cTAR/peptide E complexes (cTAR1 and cTAR2) can be proposed:
Scheme 1.
where IC1 and IC2 are the intermediate complexes of the fast and slow pathways, respectively, and kf1 and kf2 are the corresponding interconversion rate constants. Further insights into nature of the two pathways were obtained from the temperature dependence of the kobs values, using the Arrhenius equation:
| (5) |
where the rate constant ki is given by kobsi/[dTAR], Ai is the pre-exponential Arrhenius factor, Ea,i is the activation energy, R is the universal gas constant and T is the temperature (in Kelvin).
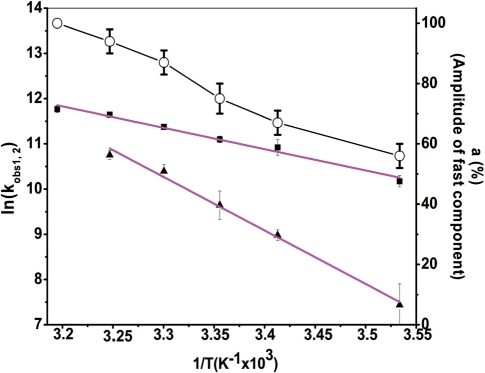
Both reaction rates as well as the amplitude of the fast pathway increased with temperature (Figure 5). Positive enthalpy values for the transition state of 9.5(±0.6) kcal/mol and 24(±1) kcal/mol for the fast and slow pathways, respectively, were obtained. These values indicate that cTAR/dTAR annealing promoted by the core peptide involves pre-melting of ∼2 and 5 bp, for the fast and slow pathways, respectively (46,47). Moreover, since the fast pathway represents 100% at 40°C (Figure 5), it is necessarily associated with ED formation and thus differs from the unproductive fast pathway observed for the cTAR/dTAR annealing reaction promoted by the HIV-1 NC (11-55) mutant (30).
Figure 5.
Temperature dependence of cTAR/dTAR annealing kinetics in the presence of peptide E at ratio 1.4:1. The reaction was performed with 10 nM doubly labeled cTAR and 300-nM non-labelled dTAR. The natural logarithm of the rate constant values for the fast (black squares) and slow (black triangles) components as well as the amplitude of the fast component (open circles) are indicated at six different temperatures. The solid magenta lines are the best fit to Equation (5) with Ea1 = 9.5(±0.6) kcal/mol and Ea2 = 24(±1) kcal/mol, for the fast and slow components, respectively. It was checked that no aggregation was associated with the changes in temperature.
Effects of oligonucleotide sequence and stability on the annealing kinetics promoted by the core peptide E
To further characterize how the core peptide influenced the annealing kinetics, we investigated the impact of the oligonucleotide sequence and stability on the reaction.
First, dTAR was substituted by the dTAR T-L mutant where the 6 nt of the loop were changed to T residues (Figure 6), thus preventing base-pairing with the cTAR loop. These nucleotide substitutions did not significantly change the kinetic parameters of the two pathways (Table 1) and ED formation (Figure 7), indicating that loop–loop interactions do not play a significant role in peptide E-promoted cTAR/dTAR annealing reaction.
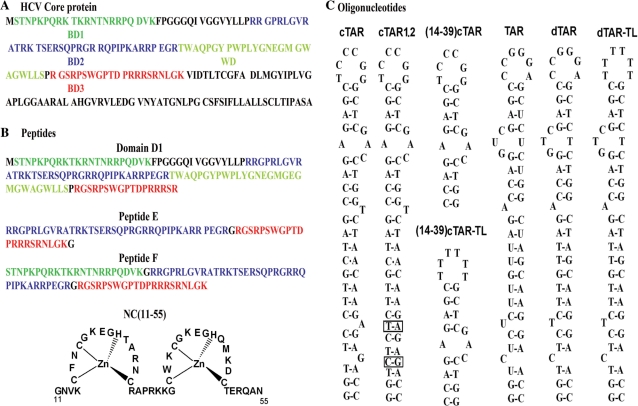
Figure 6.
Sequences of the HCV core protein (A) and the peptides (B) and oligonucleotides (C) used in this study. The N-terminal D1 domain of the core protein (B) comprises three regions rich in basic residues (BD1 to BD3) and a region rich in tryptophan residues (WD). Peptides E and F correspond to the association of two (BD2 and BD3) and all three (BD1, BD2 and BD3) basic clusters, respectively. The NC(11–55) peptide corresponds to the zinc finger domain of the HIV-1 nucleocapsid protein. The cTAR and TAR RNA sequences (c) are from the HIV-1 MAL strain. The secondary structures of the oligonucleotides were predicted from the structure of TAR (36) and the mfold program (http//www.bioinfo.rpi.edu/applications/mfold/old/dna/form1.cgi).
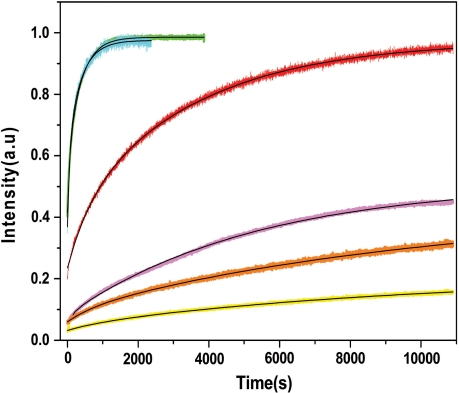
Figure 7.
Kinetics of peptide E-promoted annealing of cTAR derivatives with TAR or dTAR derivatives. Kinetic traces of 10 nM doubly labeled cTAR derivatives with 300 nM nonlabeled TAR or dTAR derivatives [cTAR/dTAR (cyan), cTAR/dTAR-TL (green), cTAR/TAR RNA (red), (14-39)cTAR/dTAR (magenta), (14-39)cTAR-TL/dTAR (orange) and cTAR1,2/dTAR (yellow)]. Black lines correspond to fits of the kinetic curves with Equation (2) and the parameters in Table 1. Peptide E was added at a peptide/oligonucleotide ratio of 1.4:1 in all the cases and experimental conditions were as in Figure 3.
To investigate the role of the cTAR stem in the annealing reaction, we used the cTAR1,2 derivative where bases complementary to the bulged bases at positions 49 and 52 have been introduced in order to stabilize the lower half of the stem (Figure 6). This mutant was previously shown to be about four times more stable than cTAR (44). Peptide E did not cause a fluorescence change of the doubly labeled cTAR1,2 (data not shown), indicating that peptide E was unable to destabilize the mutated stem. Moreover, the annealing of this mutant to dTAR in the presence of peptide E was extremely slow (Figure 7) with one kinetic pathway and a 10-fold decrease in the values of the pre-equilibrium constant, KM, and the interconversion rate constant, kf (Table 1). Thus, in a manner similar to NCp7 (30,35,48), peptide E nucleates cTAR/dTAR annealing probably through the cTAR and dTAR ends. This is confirmed by the increased value of the transition state enthalpy (30(±2) kcal/mol), indicating that the rate-limiting step of the peptide-promoted cTAR1,2/dTAR annealing involves melting of the stabilized cTAR1,2 stem (46). Moreover, the decreased stability of the cTAR1,2/dTAR intermediate as compared with the cTAR/dTAR one, most probably results from the base pair mismatches in the intermediate due to the additional bases in cTAR1,2 stem, further supporting the notion that the annealing reaction is nucleated through the stem ends.
Substitution of dTAR by TAR RNA also caused a significant decrease of the annealing reaction promoted by peptide E (Figure 7), due to a decrease in the amplitude of the fast component together with a 10-fold decrease of the interconversion rate, kf2 of the slow pathway (Table 1). This reduced annealing rate is likely caused by the higher stability of the TAR stem (29), suggesting that as with HIV-1 NCp7 (30,33,35), the cTAR/TAR annealing reaction promoted by peptide E is probably starting from the stem. This conclusion is further supported by the similar values of the transition state enthalpies (data not shown) and the equilibrium binding constants of the reaction intermediates (Table 1) in the cTAR/dTAR, cTAR/dTAR T-L and cTAR/TAR annealing reactions, indicating that the ICs in all these systems are stabilized by a similar number of base pairs.
To further confirm the critical role of the cTAR stem end, we investigated the annealing kinetics with the (14-39)cTAR derivative, also called mini-cTAR (49), in which the lower half of cTAR stem has been deleted (Figure 6). This cTAR mutant was previously shown to be about twice more stable than the entire cTAR (50). The annealing of (14–39)cTAR/dTAR in the presence of core peptide E was much slower than that of cTAR/dTAR (Figure 7), and exhibited only one kinetic pathway, confirming that the peptide E-promoted cTAR/dTAR annealing is nucleated through the cTAR ends. Substitution of (14-39)cTAR by (14-39)cTAR-TL, in which the 6 nt of the (14-39)cTAR loop were substituted with T residues (Figure 6), further decreased the annealing reaction rate (Figure 7). This suggests that as for NCp7 (51), the (14-39)cTAR/dTAR annealing reaction promoted by peptide E may partially be nucleated through kissing loop intermediates. This conclusion was further supported by the value of the transition state enthalpy 15(±5) kcal/mol, that perfectly matches the value found with the NCp7-promoted annealing of mini-cTAR (51).
Taken together, our data indicate that both kinetic pathways of the peptide E-promoted cTAR/dTAR annealing reaction are nucleated through the stems.
Promotion of cTAR/dTAR annealing by peptide F and D1 domain
Next, we investigated cTAR/dTAR annealing in the presence of peptide F and the D1 domain to determine whether they promote the annealing reaction through the same mechanism as peptide E (Figure 6B). The annealing kinetics in their presence was also biphasic with a hyperbolic dependence of kobs1,2 values and a constant amplitude value of about 0.8, strongly suggesting that the three core peptides promote cTAR/dTAR annealing by the same mechanism. While the three peptides provide similar values for the pre-equilibrium binding constants, they differ in the interconversion rate constants. Indeed, the interconversion rates of the intermediates to ED were about 3- to 7-fold faster with peptide F than that with peptide E and the D1 domain (Table 1).
Interestingly, the temperature dependence of the kobs values with peptide F and domain D1 yielded enthalpy values that were similar to those with peptide E (data not shown), confirming that the three peptides follow the same mechanism by nucleating ICs with a similar number of base pairs.
Comparison of the three peptides suggests that the third basic domain (BD3) favors the interconversion step while the additional sequences in D1, notably the tryptophan-rich domain, WD, have a counter effect.
Comparison of HCV core peptide E and HIV-1 NCp7 in promoting cTAR/dTAR annealing
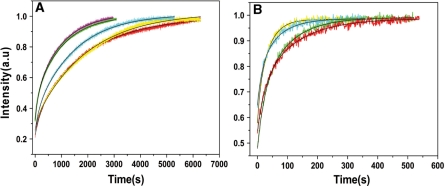
Since both peptide E and NCp7 promote cTAR/dTAR annealing, this prompted us to compare their efficiencies in this activity. Because wild-type NCp7 strongly aggregates oligonucleotides (30,42,52,53), we used the truncated NC (11-55) peptide (Figure 6B), which mimics the activities of NCp7 without aggregation (30,38). Comparison of peptide E with NC(11-55) at several molar ratios revealed that 1 and 1.4 molecules of peptide E provided the same annealing activity as that of 4 and 6 NC (11-55) molecules (Figure 8A), respectively. This suggests that one peptide E molecule exhibits nearly the same chaperone activity as 4 molecules of NC(11-55). Furthermore, the kinetic traces in the presence of both NC(11-55) added at a ratio of 4 peptides per oligonucleotide and peptide E added at ratios of 1 and 1.4 peptides per oligonucleotide were similar to the kinetic traces obtained with ratios of 8 and 10 NC(11-55) per oligonucleotide, respectively (Figure 8B). This confirmed that one peptide E exhibits the same chaperone activity as four NC(11-55) and favored the notion that the two peptides can chaperone in concert the annealing of the two complementary ODNs.
Figure 8.
Comparison of the chaperoning properties of peptide E and HIV-1 NC(11-55). Kinetic traces were recorded with 10 nM doubly labeled cTAR and 100 nM nonlabeled dTAR. (A) Kinetic traces in the presence of NC (11-55) added at a peptide/oligonucleotide molar ratio of 4 (red), 5 (cyan) and 6 (magenta) or in the presence of peptide E added at a molar ratio of 1:1 (yellow) and 1.4:1 (green). (B) Kinetic traces with NC (11-55) added at a peptide/oligonucleotide molar ratio of 4:1 in the presence of peptide E, added at a peptide/oligonucleotide molar ratio of 1:1 (red) and 1.4:1 (cyan) compared with only NC (11–55) added at a molar ratio of 8:1 (green) and 10:1 (yellow) respectively. Black lines correspond to fits of the kinetic curves with Equation (2).
DISCUSSION
The aim of the present study was to characterize the mechanism by which the basic domain of the HCV core chaperones the annealing of two canonical complementary stem–loop sequences, namely the HIV-1 cTAR and dTAR DNA, which have been extensively used to study the chaperone properties of retroviral NC proteins (30,32–35,54–56). This study was performed with the D1 core domain and two peptides that encompass the basic clusters critical for the chaperone properties of the core (20,28). To maintain the nonaggregating conditions required for the present fluorescent experiments, we used a limited peptide to oligonucleotide molar ratio of 1.4:1.
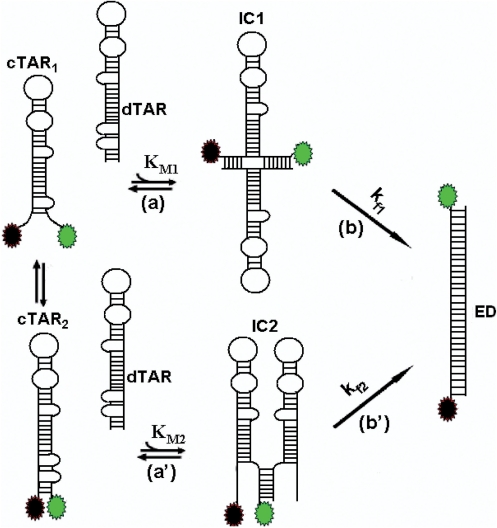
In contrast to retroviral NCs, such as HIV-1 NCp7 and MuLV NCp10 (29,32), the core peptides exhibit only a slight destabilizing activity, suggesting that this component plays a limited role in the core-promoted annealing reaction. Nevertheless, these peptides efficiently activate the cTAR/dTAR annealing reaction as shown by the up to three orders of magnitude difference in the values of the overall kinetic constants (kass = KM × kf) in the presence of the core peptides (Table 1) as compared to the bimolecular rate constant (85 M–1 s–1) in their absence (30). The promotion of cTAR/dTAR annealing by the core peptides was found to start from the cTAR end and to proceed through two parallel kinetic pathways that include a fast pre-equilibrium intermediate and a rate-limiting conversion into the final ED. The fast and slow pathways likely differ by the number of base pairs (≈2 and 5, respectively) that should be melted in the original cTAR secondary structure to nucleate the ICs. Since thermally driven fraying of cTAR termini occurs spontaneously at room temperature (29,37,44), the fast pathway is probably associated with a cTAR species where the 3 bp of the terminal stem are melted, while the slow pathway may be associated with the fully structured oligonucleotide (Figure 9). Therefore, formation of IC1 in the fast pathway is thought to involve the melting of the 2 bp from the penultimate double-stranded segment while the 5 bp of both terminal and penultimate double stranded DNA segments should melt to form IC2 in the slow pathway. As previously shown with HIV-1 NCp7 (35) and Tat (submitted), IC1 may result from the annealing of both 3′ and 5′ terminal strands of cTAR with the complementary terminal strands of dTAR and be stabilized by 12 intermolecular base pairs. Since the IC2 complex in the slow pathway is less stable than IC1, it likely involves a smaller number of intermolecular base pairing. A possible explanation is that IC2 results from an invasion mechanism, in which only one cTAR strand anneals with the complementary dTAR strand (54,56), allowing the formation of 5–7 intermolecular base pairs. Both IC1 and IC2 are further converted into the ED, which most probably relies on the conformational rearrangement and melting of the stable upper part of both TAR species. In this respect, the 3- to 4-fold faster interconversion rate of IC1 as compared to IC2 probably results from its larger number of intermolecular base pairs and/or more favorable conformation for the subsequent conversion into the ED.
Figure 9.
Proposed mechanism for core-promoted cTAR/dTAR annealing. The annealing reaction at sub-saturating protein concentrations is thought to involve two pathways that rely on thermal fraying of cTAR. This fraying leads to a fast equilibrium (microsecond range) between the fully closed cTAR species (cTAR2) and a partially melted cTAR species (cTAR1) (37). The upper pathway (steps a and b) and the lower pathway (steps a′ and b′) are associated with the fast and the slow kinetic components, respectively. In both pathways, the cTAR species nucleate an intermediate complex (IC) that subsequently, converts in a rate-limiting step to the final extended duplex (ED). In both pathways, the ICs are nucleated through the stem termini.
Interestingly, the degree of coating of cTAR and dTAR by the peptides was found to strongly affect the equilibrium binding constant of the ICs but not their interconversion rate. Moreover, the amplitude of the fast component increases with the coating degree, suggesting that a single pathway can probably take place at a saturating concentration of core peptide, as in the case of retroviral proteins (35,49,51). In the proposed model reaction (Figure 9), the amplitude of the fast component is a complex function of the fast equilibrium between cTAR1 and cTAR2 species, their stability, their initial melting by the peptides and the fluorescence intensity of the intermediates. Accordingly, the amplitude of the fast component decreased, when either of the two partners, cTAR or dTAR, was substituted by a more stable one (∼35% for cTAR/TAR and <5% cTAR1,2/dTAR) as compared to 70% for cTAR/dTAR. On the other hand, a decrease in stability of partners results in an increase of amplitude (100% at 40°C for cTAR/dTAR). Finally, the core chaperone mechanism was found to be similar for peptide E, peptide F and D1 domain, confirming that the basic clusters are the main determinants in this mechanism (20,28).
Comparison with the truncated HIV-1 NC(11-55) peptide revealed that one core peptide molecule exhibits the same chaperone activity as four NC(11-55) molecules in cTAR/dTAR annealing (Figure 8). A similar conclusion applies for cTAR/TAR RNA annealing with the full-length NCp7, since the KM values of the ICs with peptide E (about 106 M–1) are similar to the corresponding KM value obtained with NCp7 added at a ratio of six proteins per oligonucleotide (35). This suggests that a 1.4 equivalent of peptide E could be as active as 6 NCp7 equivalents in the present experimental conditions. This is likely a consequence of the stronger ‘nucleic acid aggregating’ properties of the core peptides, in line with the efficient oligonucleotide aggregation observed by FCS (Figure 1). This propensity of the core peptides to neutralize the negatively charged nucleic acids and to promote their aggregation is probably related to the highly flexible nature of the core peptides (19,57) as compared to the folded NCp7 structure with two zinc fingers (58,59). In addition, NCp7 and the core peptides likely differ on how they promote the interconversion reaction. For cTAR/TAR, the interconversion rate constant, in the absence of any peptide is about 1.5 × 10–4 s–1, while it is about 3-fold higher in the presence of NCp7, at any protein/oligonucleotide ratio (35). For the core peptides, the corresponding values for the fast and slow pathway are respectively, two and one order of magnitude higher than in the absence of protein. Thus, in contrast to NCp7, the core peptides strongly favor the interconversion reaction, which is thought to be rate-limited by a conformational change involving cooperative melting of a large fragment of one or both hairpin stems. Moreover, while NCp7 also promotes cTAR/TAR annealing through kissing loop intermediates when added at sub-saturating concentrations to ODNs (35), the core peptides mainly chaperone the annealing through the stems.
Taken together, our data suggest that though at sub-saturating conditions, the core peptides only marginally destabilize the secondary structure of oligonucleotides, they are more efficient than the truncated HIV-1 NC(11-55) peptide in nucleating IC formation through their nucleic acid aggregation properties and promoting the subsequent IC conversion into ED. Moreover, in contrast to NC(11-55) and native NCp7 (35) which requires high peptide/ODN ratios for optimal activity, the core peptides already show strong nucleic acid annealing activity at low peptide/ODN ratios, suggesting that the core protein can exert its strong chaperone activity at low fractional saturation of nucleic acids. Importantly, since the core protein and NCp7 are structurally different proteins from two different virus families, the conserved nucleic acid chaperone properties suggest that they are required in a large panel of viruses, in line with the recently demonstrated conservation of RNA chaperoning in Flaviviridae core proteins (60). The potent chaperone properties of the core peptides in the TAR oligonucleotides model used in this study further suggest that the core protein is a nucleic chaperone with broad sequence specificity. These properties may be critical for genomic RNA dimerization in HCV replication and RNA packaging, as well as for facilitating recombination between various HCV genotypes and subtypes to increase viral variability (61–63).
FUNDING
The French National Agency for Research on AIDS and Viral Hepatitis (CSS n°4 to K.K.S.). Funding for open access charge: The funds from the French Agency against AIDS could partly support the publication charges.
Conflict of interest statement. None declared.
REFERENCES
- 1.Choo Q.L., Kuo G., Weiner A.J., Overby L.R., Bradley D.W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Recommendations from the National Institutes of Health consensus development conference statement: management of hepatitis C: 2002. Hepatology. 36:1039. doi: 10.1002/hep.510360502. [DOI] [PubMed] [Google Scholar]
- 3.Lauer G.M., Walker B.D. Hepatitis C virus infection. N. Eng. J. Med. 2001;345:41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 4.Moradpour D., Penin F., Rice C.M. Replication of hepatitis C virus. Nat. Rev. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 5.Honda M., Beard M.R., Ping L.H., Lemon S.M. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blight K.J., Rice C.M. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. Journal of virology. 1997;71:7345–7352. doi: 10.1128/jvi.71.10.7345-7352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolykhalov A.A., Feinstone S.M., Rice C.M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. Journal of virology. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friebe P., Bartenschlager R. Genetic analysis of sequences in the 3' nontranslated region of hepatitis C virus that are important for RNA replication. Journal of virology. 2002;76:5326–5338. doi: 10.1128/JVI.76.11.5326-5338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Penin F., Dubuisson J., Rey F.A., Moradpour D., Pawlotsky J.M. Structural biology of hepatitis C virus. Hepatology. 2004;39:5–19. doi: 10.1002/hep.20032. [DOI] [PubMed] [Google Scholar]
- 10.Ray R.B., Ray R. Hepatitis C virus core protein: intriguing properties and functional relevance. FEMS Microbiol. Lett. 2001;202:149–156. doi: 10.1111/j.1574-6968.2001.tb10796.x. [DOI] [PubMed] [Google Scholar]
- 11.McLauchlan J. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J. Viral Hepat. 2000;7:2–14. doi: 10.1046/j.1365-2893.2000.00201.x. [DOI] [PubMed] [Google Scholar]
- 12.Hijikata M., Kato N., Mori S., Ootsuyama Y., Nakagawa M., Sugimura T., Ohkoshi S., Kojima H., Meguro T., Taki M., et al. Frequent detection of hepatitis C virus US strain in Japanese hemophiliacs. Jpn J Cancer Res. 1990;81:1195–1197. doi: 10.1111/j.1349-7006.1990.tb02676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemberg M.K., Martoglio B. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol. Cell. 2002;10:735–744. doi: 10.1016/s1097-2765(02)00655-x. [DOI] [PubMed] [Google Scholar]
- 14.McLauchlan J., Lemberg M.K., Hope G., Martoglio B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 2002;21:3980–3988. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moradpour D., Englert C., Wakita T., Wands J.R. Characterization of cell lines allowing tightly regulated expression of hepatitis C virus core protein. Virology. 1996;222:51–63. doi: 10.1006/viro.1996.0397. [DOI] [PubMed] [Google Scholar]
- 16.Barba G., Harper F., Harada T., Kohara M., Goulinet S., Matsuura Y., Eder G., Schaff Z., Chapman M.J., Miyamura T., et al. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc. Natl Acad. Sci. USA. 1997;94:1200–1205. doi: 10.1073/pnas.94.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hope R.G., McLauchlan J. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol. 2000;81:1913–1925. doi: 10.1099/0022-1317-81-8-1913. [DOI] [PubMed] [Google Scholar]
- 18.Schwer B., Ren S., Pietschmann T., Kartenbeck J., Kaehlcke K., Bartenschlager R., Yen T.S., Ott M. Targeting of hepatitis C virus core protein to mitochondria through a novel C-terminal localization motif. J. Virol. 2004;78:7958–7968. doi: 10.1128/JVI.78.15.7958-7968.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boulant S., Vanbelle C., Ebel C., Penin F., Lavergne J.P. Hepatitis C virus core protein is a dimeric alpha-helical protein exhibiting membrane protein features. J. Virol. 2005;79:11353–11365. doi: 10.1128/JVI.79.17.11353-11365.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivanyi-Nagy R., Kanevsky I., Gabus C., Lavergne J.P., Ficheux D., Penin F., Fosse P., Darlix J.L. Analysis of hepatitis C virus RNA dimerization and core-RNA interactions. Nucleic Acids Res. 2006;34:2618–2633. doi: 10.1093/nar/gkl240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boulant S., Montserret R., Hope R.G., Ratinier M., Targett-Adams P., Lavergne J.P., Penin F., McLauchlan J. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J. Biol. Chem. 2006;281:22236–22247. doi: 10.1074/jbc.M601031200. [DOI] [PubMed] [Google Scholar]
- 22.Shavinskaya A., Boulant S., Penin F., McLauchlan J., Bartenschlager R. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 2007;282:37158–37169. doi: 10.1074/jbc.M707329200. [DOI] [PubMed] [Google Scholar]
- 23.Giannini C., Brechot C. Hepatitis C virus biology. Cell Death Differ. 2003;10 (Suppl. 1):S27–S38. doi: 10.1038/sj.cdd.4401121. [DOI] [PubMed] [Google Scholar]
- 24.Lerat H., Honda M., Beard M.R., Loesch K., Sun J., Yang Y., Okuda M., Gosert R., Xiao S.Y., Weinman S.A., et al. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology. 2002;122:352–365. doi: 10.1053/gast.2002.31001. [DOI] [PubMed] [Google Scholar]
- 25.Kunkel M., Lorinczi M., Rijnbrand R., Lemon S.M., Watowich S.J. Self-assembly of nucleocapsid-like particles from recombinant hepatitis C virus core protein. J. Virol. 2001;75:2119–2129. doi: 10.1128/JVI.75.5.2119-2129.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baumert T.F., Ito S., Wong D.T., Liang T.J. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J. Virol. 1998;72:3827–3836. doi: 10.1128/jvi.72.5.3827-3836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimoike T., Mimori S., Tani H., Matsuura Y., Miyamura T. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J. Virol. 1999;73:9718–9725. doi: 10.1128/jvi.73.12.9718-9725.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cristofari G., Ivanyi-Nagy R., Gabus C., Boulant S., Lavergne J.P., Penin F., Darlix J.L. The hepatitis C virus Core protein is a potent nucleic acid chaperone that directs dimerization of the viral (+) strand RNA in vitro. Nucleic Acids Res. 2004;32:2623–2631. doi: 10.1093/nar/gkh579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernacchi S., Stoylov S., Piemont E., Ficheux D., Roques B.P., Darlix J.L., Mely Y. HIV-1 nucleocapsid protein activates transient melting of least stable parts of the secondary structure of TAR and its complementary sequence. J. Mol. Biol. 2002;317:385–399. doi: 10.1006/jmbi.2002.5429. [DOI] [PubMed] [Google Scholar]
- 30.Godet J., de Rocquigny H., Raja C., Glasser N., Ficheux D., Darlix J.L., Mely Y. During the early phase of HIV-1 DNA synthesis, nucleocapsid protein directs hybridization of the TAR complementary sequences via the ends of their double-stranded stem. J. Mol. Biol. 2006;356:1180–1192. doi: 10.1016/j.jmb.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 31.Ramalanjaona N., de Rocquigny H., Millet A., Ficheux D., Darlix J.L., Mely Y. Investigating the mechanism of the nucleocapsid protein chaperoning of the second strand transfer during HIV-1 DNA synthesis. J. Mol. Biol. 2007;374:1041–1053. doi: 10.1016/j.jmb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Egele C., Piemont E., Didier P., Ficheux D., Roques B., Darlix J.L., de Rocquigny H., Mely Y. The single-finger nucleocapsid protein of moloney murine leukemia virus binds and destabilizes the TAR sequences of HIV-1 but does not promote efficiently their annealing. Biochemistry. 2007;46:14650–14662. doi: 10.1021/bi7012239. [DOI] [PubMed] [Google Scholar]
- 33.Liu H.W., Cosa G., Landes C.F., Zeng Y., Kovaleski B.J., Mullen D.G., Barany G., Musier-Forsyth K., Barbara P.F. Single-molecule FRET studies of important intermediates in the nucleocapsid-protein-chaperoned minus-strand transfer step in HIV-1 reverse transcription. Biophys. J. 2005;89:3470–3479. doi: 10.1529/biophysj.105.065326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart-Maynard K.M., Cruceanu M., Wang F., Vo M.N., Gorelick R.J., Williams M.C., Rouzina I., Musier-Forsyth K. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J. Virol. 2008;82:10129–10142. doi: 10.1128/JVI.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vo M.N., Barany G., Rouzina I., Musier-Forsyth K. HIV-1 nucleocapsid protein switches the pathway of transactivation response element RNA/DNA annealing from loop-loop “kissing” to “zipper”. J. Mol. Biol. 2009;386:789–801. doi: 10.1016/j.jmb.2008.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baudin F., Marquet R., Isel C., Darlix J.L., Ehresmann B., Ehresmann C. Functional sites in the 5' region of human immunodeficiency virus type 1 RNA form defined structural domains. J. Mol. Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 37.Azoulay J., Clamme J.P., Darlix J.L., Roques B.P., Mely Y. Destabilization of the HIV-1 complementary sequence of TAR by the nucleocapsid protein through activation of conformational fluctuations. J. Mol. Biol. 2003;326:691–700. doi: 10.1016/s0022-2836(02)01430-4. [DOI] [PubMed] [Google Scholar]
- 38.Beltz H., Clauss C., Piemont E., Ficheux D., Gorelick R.J., Roques B., Gabus C., Darlix J.L., de Rocquigny H., Mely Y. Structural determinants of HIV-1 nucleocapsid protein for cTAR DNA binding and destabilization, and correlation with inhibition of self-primed DNA synthesis. J. Mol. Biol. 2005;348:1113–1126. doi: 10.1016/j.jmb.2005.02.042. [DOI] [PubMed] [Google Scholar]
- 39.Clamme J.P., Azoulay J., Mely Y. Monitoring of the formation and dissociation of polyethylenimine/DNA complexes by two photon fluorescence correlation spectroscopy. Biophys. J. 2003;84:1960–1968. doi: 10.1016/S0006-3495(03)75004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson N.L. Fluorescence Correlation Spectroscopy, in Topics in Fluorescence Spectroscopy. NY: Plenum Publishers; 1991. [Google Scholar]
- 41.Cristofari G., Darlix J.L. The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:223–268. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- 42.Stoylov S.P., Vuilleumier C., Stoylova E., De Rocquigny H., Roques B.P., Gerard D., Mely Y. Ordered aggregation of ribonucleic acids by the human immunodeficiency virus type 1 nucleocapsid protein. Biopolymers. 1997;41:301–312. doi: 10.1002/(SICI)1097-0282(199703)41:3<301::AID-BIP5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 43.Bloomfield V.A., He S., Li A.Z., Arscott P.B. Light scattering studies on DNA condensation. Biochem. Soc. Transact. 1991;19:496. doi: 10.1042/bst0190496. [DOI] [PubMed] [Google Scholar]
- 44.Beltz H., Azoulay J., Bernacchi S., Clamme J.P., Ficheux D., Roques B., Darlix J.L., Mely Y. Impact of the terminal bulges of HIV-1 cTAR DNA on its stability and the destabilizing activity of the nucleocapsid protein NCp7. J. Mol. Biol. 2003;328:95–108. doi: 10.1016/s0022-2836(03)00244-4. [DOI] [PubMed] [Google Scholar]
- 45.Bombarda E., Grell E., Roques B.P., Mely Y. Molecular mechanism of the Zn2+-induced folding of the distal CCHC finger motif of the HIV-1 nucleocapsid protein. Biophys. J. 2007;93:208–217. doi: 10.1529/biophysj.106.101378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rouzina I., Bloomfield V.A. Heat capacity effects on the melting of DNA. 1. General aspects. Biophys. J. 1999;77:3242–3251. doi: 10.1016/S0006-3495(99)77155-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cantor C., Schimmel P. Biophysical Chemistry Part 2: Techniques for the Study of Biological Structure and Function. NY: Academic Press; 1980. [Google Scholar]
- 48.Cosa G., Zeng Y., Liu H.W., Landes C.F., Makarov D.E., Musier-Forsyth K., Barbara P.F. Evidence for non-two-state kinetics in the nucleocapsid protein chaperoned opening of DNA hairpins. J. Phys. Chem. 2006;110:2419–2426. doi: 10.1021/jp054189i. [DOI] [PubMed] [Google Scholar]
- 49.Vo M.N., Barany G., Rouzina I., Musier-Forsyth K. Effect of Mg(2+) and Na(+) on the nucleic acid chaperone activity of HIV-1 nucleocapsid protein: implications for reverse transcription. J. Mol. Biol. 2009;386:773–788. doi: 10.1016/j.jmb.2008.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beltz H., Piemont E., Schaub E., Ficheux D., Roques B., Darlix J.L., Mely Y. Role of the structure of the top half of HIV-1 cTAR DNA on the nucleic acid destabilizing activity of the nucleocapsid protein NCp7. J. Mol. Biol. 2004;338:711–723. doi: 10.1016/j.jmb.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 51.Vo M.N., Barany G., Rouzina I., Musier-Forsyth K. Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. J. Mol. Biol. 2006;363:244–261. doi: 10.1016/j.jmb.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 52.Le Cam E., Coulaud D., Delain E., Petitjean P., Roques B.P., Gerard D., Stoylova E., Vuilleumier C., Stoylov S.P., Mely Y. Properties and growth mechanism of the ordered aggregation of a model RNA by the HIV-1 nucleocapsid protein: an electron microscopy investigation. Biopolymers. 1998;45:217–229. doi: 10.1002/(SICI)1097-0282(199803)45:3<217::AID-BIP4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 53.Grohmann D., Godet J., Mely Y., Darlix J.L., Restle T. HIV-1 nucleocapsid traps reverse transcriptase on nucleic acid substrates. Biochemistry. 2008;47:12230–12240. doi: 10.1021/bi801386r. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y., Balakrishnan M., Roques B.P., Bambara R.A. Steps of the acceptor invasion mechanism for HIV-1 minus strand strong stop transfer. J.Biol. Chem. 2003;278:38368–38375. doi: 10.1074/jbc.M305700200. [DOI] [PubMed] [Google Scholar]
- 55.Kim J.K., Palaniappan C., Wu W., Fay P.J., Bambara R.A. Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J. Biol. Chem. 1997;272:16769–16777. doi: 10.1074/jbc.272.27.16769. [DOI] [PubMed] [Google Scholar]
- 56.Roda R.H., Balakrishnan M., Kim J.K., Roques B.P., Fay P.J., Bambara R.A. Strand transfer occurs in retroviruses by a pause-initiated two-step mechanism. J. Biol. Chem. 2002;277:46900–46911. doi: 10.1074/jbc.M208638200. [DOI] [PubMed] [Google Scholar]
- 57.Ivanyi-Nagy R., Davidovic L., Khandjian E.W., Darlix J.L. Disordered RNA chaperone proteins: from functions to disease. Cell. Mol. Life Sci. 2005;62:1409–1417. doi: 10.1007/s00018-005-5100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morellet N., de Rocquigny H., Mely Y., Jullian N., Demene H., Ottmann M., Gerard D., Darlix J.L., Fournie-Zaluski M.C., Roques B.P. Conformational behaviour of the active and inactive forms of the nucleocapsid NCp7 of HIV-1 studied by 1H NMR. J. Mol. Biol. 1994;235:287–301. doi: 10.1016/s0022-2836(05)80033-6. [DOI] [PubMed] [Google Scholar]
- 59.Summers M.F., Henderson L.E., Chance M.R., Bess J.W., Jr, South T.L., Blake P.R., Sagi I., Perez-Alvarado G., Sowder R.C., III, Hare D.R., et al. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ivanyi-Nagy R., Lavergne J.P., Gabus C., Ficheux D., Darlix J.L. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res. 2008;36:712–725. doi: 10.1093/nar/gkm1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Colina R., Casane D., Vasquez S., Garcia-Aguirre L., Chunga A., Romero H., Khan B., Cristina J. Evidence of intratypic recombination in natural populations of hepatitis C virus. J. Gen. Virol. 2004;85:31–37. doi: 10.1099/vir.0.19472-0. [DOI] [PubMed] [Google Scholar]
- 62.Kalinina O., Norder H., Magnius L.O. Full-length open reading frame of a recombinant hepatitis C virus strain from St Petersburg: proposed mechanism for its formation. J. Gen. Virol. 2004;85:1853–1857. doi: 10.1099/vir.0.79984-0. [DOI] [PubMed] [Google Scholar]
- 63.Kalinina O., Norder H., Mukomolov S., Magnius L.O. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 2002;76:4034–4043. doi: 10.1128/JVI.76.8.4034-4043.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]