Abstract
The effects of chromosomal position and neighboring genomic elements on gene targeting in human cells remain largely unexplored. To study these, we used a shuttle vector system in which murine leukemia virus (MLV)-based proviral targets present at different chromosomal locations and containing mutations in the neomycin phosphotransferase (neo) gene were corrected by adeno-associated virus (AAV)-mediated gene targeting. Sixteen identical target loci present in HT-1080 human sarcoma cells were all successfully corrected by gene targeting. The gene targeting frequencies varied by as much as 10-fold, and there was a clear bias for correction of one of the targets in clones containing two target sites. The targeting frequency at each site was correlated to the proximity and density of various genomic elements, and we found a significant association of higher targeting frequencies at loci near a subset of dinucleotide microsatellite repeats (r = –0.55, P < 0.05), in particular GT repeats (r = –0.87, P < 0.0001). Additionally, there was a correlation between meiotic recombination rates and targeting frequencies at the target loci (r = 0.52, P < 0.05). There was no correlation between surrounding chromosomal transcription units and targeting frequencies. Our results indicate that certain chromosomal positions are preferred sites for gene targeting in human cells.
INTRODUCTION
Homologous recombination is essential for proper chromosome segregation and the preservation of genetic diversity during meiosis, as well as the repair of different types of DNA lesions during mitosis. Gene targeting, in which a transgene recombines with a homologous chromosomal locus, presumably utilizes the same mechanisms as mitotic chromosomal recombination. While studies have reported the effects of target site transcription (1) and genomic methylation levels (2) on gene targeting frequencies, much less is known about the influence of chromosomal position on gene targeting. A better understanding of how specific genomic elements surrounding the target site affect gene targeting will improve our ability to precisely manipulate the human genome and provide insight into chromosomal recombination mechanisms.
Several studies have explored the recombinogenic potential of specific chromosomal regions in eukaryotic cells. In Saccharomyces cerevisiae, sequences in the HOT1 gene present in the ribosomal RNA cluster stimulate both interchromosomal and intrachromosomal mitotic recombination (3) by regulating RNA polymerase I transcription (4). In addition, the presence of a centromeric region from S. cerevisiae chromosome XIV in an autonomously replicating plasmid stimulated homologous genetic exchange between yeast genomic sequences and those present on the plasmid (5). In mammalian cells, the murine immunoglobulin heavy chain (IgH) μ locus is a hotspot for intrachromosomal homologous recombination (6), and deletion of a 7.1-kb segment from the VH-Cμ intron in the same locus decreased recombination 10-fold (7). There are also variable meiotic recombination rates at different human chromosomal loci (8–10). All these studies suggest that chromosomal position effects influence homologous recombination between chromosomes.
Chromosomal position effects on gene targeting frequencies have also been examined. In S. cerevisiae, the same targeting frequency was observed when the target gene was located at different chromosomal positions, arguing against a strong position effect (11,12). In mammalian cells, the data are not as clear. One of the earliest reports of gene targeting in mammalian cells investigated the correction of a defective neomycin phosphotransferase (neo) gene residing in the chromosome of mouse L cells by using DNA microinjection. In one targeted cell line containing four copies of the defective gene integrated at four different unknown sites, three of the four alleles present were targeted, but only four transformants were analyzed (13). Another study investigated the targeting of defective thymidine kinase (tk) genes present in 10 mouse L cell lines, all of which had the transgene based on Southern blot analysis. Interestingly, only one of the cell lines could be corrected by gene targeting. While this may have been due to position effects, neither the number, nor the chromosomal location, of the target tk genes was determined, making it difficult to draw definitive conclusions (14). When multiple targets, both endogenous and ectopic, were present in the same human cell line, there was a bias for preferential targeting at one of the alleles (15), again suggesting that position effects played a role, but this study was limited to a single cell line. Here, we sought to more definitively address the role of position effects on gene targeting in mammalian cells, by comparing the targeting frequencies of multiple, identical target loci present at different, known chromosomal locations.
Although the specific genetic elements responsible for chromosomal position effects are unknown, several lines of evidence suggest that sequence repeats can influence recombination. In mammalian cells, centromeres and telomeres consisting of tandemly repeated DNA sequences, were shown to be highly recombinogenic (16). Short, tandemly repeated sequences present throughout the human genome may also enhance recombination. The repeats d(CG·CG)n and d(GT·AC)n stimulated intramolecular homologous recombination in SV40 viral DNA by 10- to 15-fold and 3- to 5-fold, respectively (17), and d(GA·TC)n enhanced homologous recombination in SV40 minichromosomes by almost two orders of magnitude (18). The repeat d(TG·CA)n stimulated intramolecular homologous recombination between two nonreplicating plasmids introduced into human cells (19). Furthremore, a hypervariable minisatellite DNA sequence stimulated homologous recombination by up to 13.5-fold between two nonreplicating plasmids that reconstituted a wild-type neo gene in mammalian cells (20). Notably, the minisatellite was located 200–1000 bp away from the recombination site (20). Despite numerous studies demonstrating the recombinogenic potential of repeated DNA sequences, evidence for their effects on gene targeting is lacking.
In this study, we have directly addressed whether the chromosomal position of a target site and its surrounding genomic features influence gene targeting by using adeno-associated virus (AAV) vectors to precisely correct mutations in target genes present at different, known chromosomal locations. AAV gene targeting vectors contain single-stranded DNA homologous to the chromosomal target site flanked by viral inverted terminal repeats (21,22). Up to 1% of unselected human cells exposed to AAV vectors undergo gene targeting under optimal conditions (23), which is orders of magnitude higher than the targeting frequencies typically obtained with conventional methods based on transfection or electroporation (24,25). This allowed us to accurately compare targeting frequencies at multiple different target loci. In addition, despite the high frequencies of AAV-mediated gene targeting, it shares features with conventional plasmid-based recombination, including stimulation by double-strand breaks (26) and preferential introduction of insertion mutations (27), making it an appropriate model for recombination in general. Here, we report the frequencies of AAV-mediated gene targeting at distinct chromosomal loci and correlate these frequencies with proximity to and density of various genomic elements surrounding the targets.
MATERIALS AND METHODS
Plasmids
The MLV vector plasmid pLHSN37Δ4O is based on plasmid pLHSNO (28) and contains the following sequences: pLXSHD retroviral vector backbone (29); hygromycin phosphotransferase (hph) gene; neomycin phosphotransferase (neo) gene with a simian virus 40 (SV40) early and bacterial Tn5 promoter (30); and p15A plasmid replication origin (31). A 4-bp deletion was introduced into pLHSNO at bp 37 of the neo gene by standard techniques (32) and confirmed by DNA sequencing. AAV2 vector plasmid pA2HSN5′ contains pLHSNO sequences including 309-bp 5′ to the hph gene, the hph gene, the SV40 early and Tn5 promoters, and the 5′ portion of the neo gene (truncated at the NaeI site at bp 629 of the coding sequence), inserted into a backbone based on pAAV-hrGFP (Avigen, Alameda, CA, USA). Plasmid pCI-VSV-G was obtained from Garry Nolan (Stanford University). Plasmid DNA was purified using a Plasmid Maxi kit (Qiagen, Valencia, CA, USA).
Cells and cell culture
All cells were grown at 37°C in 5% CO2 in Dulbecco’s modified Eagle’s medium containing 4 g glucose/l (Invitrogen, Carlsbad, CA, USA), 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin and 1.25 μg/ml amphotericin. HT-1080 human fibrosarcoma cells (33), Phoenix-GP cells (34) and 293 T (35) cells have been described previously.
To generate cells containing proviral target sites, HT-1080 cells were seeded on Day 1 at 6 × 105 cells/dish in three 6-cm-diameter dishes, infected with MLV vector LHSN37Δ4O at a multiplicity of infection (MOI) of 0.1 transducing units/cell (assuming one population doubling since plating) in the presence of 4 μg/ml of Polybrene (Sigma-Aldrich, St. Louis, MO, USA) on Day 2, and selected with 0.2 mg/ml hygromycin B (Calbiochem, San Diego, CA, USA) beginning on Day 3. Selection was continued until all cells in control dishes had detached (9–12 days), and drug-resistant clones of HT-1080 cells were isolated with cloning rings. LHSN37Δ4O proviruses present in these clones were detected on Southern blots digested with EcoRI and hybridized with an hph-specific probe.
Vector preparations
MLV vector LHSN37Δ4O was made by calcium phosphate transfection of Phoenix-GP cells with pCI-VSV-G and vector plasmid pLHSN37Δ4O (1:1 ratio), and the preparation was then concentrated ∼100-fold by ultracentrifugation, as described previously (36). The titer was determined on HT-1080 cells by seeding 3 × 105 cells/dish in five 6-cm-diameter dishes on Day 1, infecting the cells with various volumes (0, 0.1, 1, 10 or 50 μl) of MLV vector LHSN37Δ4O in the presence of 4 μg/ml of Polybrene on Day 2, splitting the dishes into various dilutions (0.1%, 1% or 10%) on 10-cm-diameter dishes on Day 3, and beginning selection with 0.2 mg/ml hygromycin B on Day 4. Selection was continued until all cells in control dishes had detached (9–12 days), and the titer was calculated as the number of hygromycin-resistant colony forming units per ml of vector.
AAV vector AAV2-HSN5′ (serotype 2) was made by calcium phosphate transfection of 293 T cells with pDG (37) and pA2HSN5′, as described previously (38). The AAV vector titer was based on the amount of full-length single-stranded vector genomes detected on Southern blots quantified by PhosphorImager analysis (Molecular Dynamics, Sunnyvale, CA, USA).
Gene targeting
To measure neo gene correction frequencies, HT-1080-derived clones containing MLV target proviruses were seeded on Day 1 at 5 × 104 cells/well in 24-well dishes and infected with AAV2-HSN5′ on Day 2 at an MOI of 10 000 genomes/cell (assuming one population doubling since plating). On Day 3, the cells in each well were treated with trypsin, counted, and 0.25% and 99.75% dilutions were plated into separate 10-cm-diameter dishes. On Day 4, G418 (0.7 mg active compound/ml) was added to the medium of all 99.75% dishes. Cells were cultured with media changes every 3–4 days until all cells in control dishes had detached (10–12 days), and dishes were then stained with Coomassie brilliant blue G. The total number of colony forming units per original well was determined by colony counts of the unselected 0.25% dishes, and G418-resistant colonies were counted on the 99.75% dishes. Targeting frequencies were expressed as the number of G418-resistant colonies/total number of colonies obtained. Exposure to vector was not cytotoxic, since plating efficiencies (colony counts of the 0.25% dishes/cell number counted on day of plating) were ∼70% with or without AAV vector infection. Experiments to measure neo gene correction frequencies were performed in triplicate. Reversion frequencies for individual clones were measured by plating ∼107 uninfected cells in medium containing G418 (0.7 mg active compound/ml). The neo reversion frequency for all HT-1080-derived clones except clone 49 was <10–7 and the neo reversion frequency for clone 49 was 1 in 107.
RNA studies
HT-1080-derived clones containing LHSN37Δ4O proviruses were preselected with 0.2 mg/ml hygromycin B, and total RNA was isolated using the RNeasy Mini kit (Qiagen, Valencia, CA, USA). Northern blot analysis was performed with neo- and glyceraldehyde 3-phosphate dehydrogenase (GAPDH)-specific probes by using standard techniques (32). Neo transcript levels were quantified by PhosphorImager analysis (Molecular Dynamics, Sunnyvale, CA, USA) and corrected for differences in loading.
Trichostatin A studies and chromatin immunoprecipitation assay
To test the effect of trichostatin A (TSA) on neo gene correction frequencies, HT-1080-derived clones containing MLV target proviruses were treated with or without 125 nM TSA (Sigma–Aldrich, St. Louis, MO, USA) from 4.5 h after infection with AAV2-HSN5′ until the next day, when 0.25% and 99.75% dilutions were plated into separate 10-cm-dishes. The targeting experiments were otherwise performed as described above and done in triplicate. The TSA concentration used was determined in a kill-curve assay of TSA concentrations ranging from 0 to 1000 nM on HT-1080 cells.
The chromatin immunoprecipitation (ChIP) assay on HT-1080-derived clones, treated with or without 125 nM TSA for 24 h, was performed using the Imprint Chromatin Immunoprecipitation kit (Sigma–Aldrich, St. Louis, MO, USA) according to the manufacturer’s protocol. Briefly, histones were cross-linked to DNA for 10 min at 25°C by adding 1% formaldehyde to the culture medium. Nuclei were sonicated on ice to shear chromatin into 200- to 1000-bp fragments using a Virsonic 475 (VirTris, Gardiner, NY, USA) microprobe set at power level 3. A fraction of each DNA sample was saved as input to calculate the total amount before immunoprecipitation. Immunoprecipitation was performed on the remainder of the sample by incubating DNA with rabbit anti-acetyl-histone H3 (Lys9) polyclonal antibody (Millipore, Temecula, CA, USA) for 90 min at 25°C. After reverse cross-linking, DNA isolated from each input and immunoprecipitated sample was used for analysis by quantitative PCR (qPCR). Mock immunoprecipitations were performed with mouse IgG. qPCR was performed using the StepOnePlus Real-Time PCR System (Applied Biosystems, Warrington, UK). Individiual 20-μl reactions were prepared with SYBR Green PCR Master Mix (Applied Biosystems, Warrington, UK), 200 nM final concentration of primers specific for the neo gene (forward: 5′-GCCCGGTTCTTTTTGTCAAG-3′; reverse: 5′-CTGCCTCGTCCTGCAGTTC-3′) and 3 μl of either input or immunoprecipitated sample as DNA template. Six 2-fold dilutions of control genomic DNA were used to generate a standard curve, and three replicates were used for each sample. The amount of DNA in each sample was calculated using the standard curve, and the amount of immunoprecipitated DNA relative to input was calculated as [(Amount of ChIP DNA)/Amount of input DNA)] × 100.
Shuttle vector rescue in bacteria and mapping of integration sites
Genomic DNA was isolated from gene-targeted, G418-resistant, HT-1080-derived clonal cell lines, and the shuttle vector target sites along with flanking chromosomal DNA were rescued as bacterial plasmids as described previously (39), except for the following modifications. Genomic DNA (10 μg) containing integrated, targeted LHSN37Δ4O proviruses was digested with EcoRI and MfeI for 4 h at 37°C, circularized with T4 DNA ligase for 2 h at 25°C, extracted with phenol and chloroform, and precipitated with ethanol. The DNA pellets were resuspended in 10 μl of H2O and Escherichia coli strain DH10B (40) was transformed by electroporation with ∼1 μg (1 μl) of DNA. Transformed bacteria were selected on agar containing 50 μg kanamycin/ml. Plasmid DNA was purified using a Plasmid Mini kit (Qiagen, Valencia, CA, USA) and sequenced using primer 5′-GTTCGCTTCTCGCTTCTGTT-3′ specific for the 3′ long terminal repeat (LTR). Unambiguous sequences were obtained for each rescued target site from bp –150 to at least 300 bp beyond the 3′ LTR chromosomal junction. The resulting junction sequences were aligned to build 36.1 (March 2006 assembly) of the human genome using BLAT (41), and their chromosomal positions were identified.
Calculating the proximity to and density of chromosomal features around integration sites
The locations of RefSeq transcripts, CpG islands, simple repeats, microsatellites (including all dinucleotide repeat subsets), short interspersed nuclear elements (SINEs), long interspersed nuclear elements (LINEs), DNA transposons, LTR retrotransposons and RNA repeats were determined by using tables available from the University of California Santa Cruz (UCSC) database (42). Proximity was measured as the distance from bp 37 of the neo gene in the MLV target, where a 4-bp deletion was present in MLV-LHSN37Δ4O, to the center of the nearest genomic element, with the exception of RefSeq transcripts, where distance was measured to the transcription start site. Density was measured as the number of base pairs comprised by each type of genomic element within a given interval extending in both directions from the chromosomal position where the target integrated (±1, 10 or 100 kb). The analysis was performed with programs available on the Galaxy website (http://galaxy.psu.edu) (43) and processed using Microsoft Excel. Sex-averaged meiotic recombination rates (in centiMorgans/Mb) from the deCODE, Marshfield, and Genethon genetic maps were obtained for each target locus using the UCSC Genome Browser.
Statistical analysis
Correlations were determined by calculating the Pearson correlation coefficient, r, and targeting frequencies were compared to each other using the one-way analysis of variance (ANOVA) test. Two-tailed P-values <0.05 were considered significant.
RESULTS
Targeting system
Provirus target sites were introduced into HT-1080 human sarcoma cells by transduction with MLV vector LHSN37Δ4O, which contains a nonfunctional neomycin phosphotransferase (neo) gene with a 4-bp deletion at bp 37 of the coding sequence (Figure 1A). LHSN37Δ4O expresses neo from both mammalian SV40 early and prokaryotic Tn5 promoters, allowing selection in both mammalian and bacterial cells with G418 and kanamycin, respectively. The p15A plasmid origin was included to allow replication of rescued target sites as bacterial plasmids. LHSN37Δ4O also contains a hygromycin phosphotransferase gene (hph) under the control of the MLV LTR promoter. Cells transduced with the MLV vector can be selected for by growth in hygromycin, and the mutant neo genes in the integrated vector proviruses can be corrected by gene targeting with AAV vectors and scored by selection in G418. The AAV2-HSN5′ vector used for gene correction contains 3233 bp of sequence homology to the target locus and a truncation in neo at bp 629 of the coding sequence that disrupts gene function (Figure 1A). Transduced, hygromycin-resistant HT-1080 clones were screened by Southern blot hybridization to determine the vector provirus copy number in each. Ten clones containing a single copy (Figure 1B) and three containing two copies (Figure 1C) of LHSN37Δ4O were used in targeting experiments.
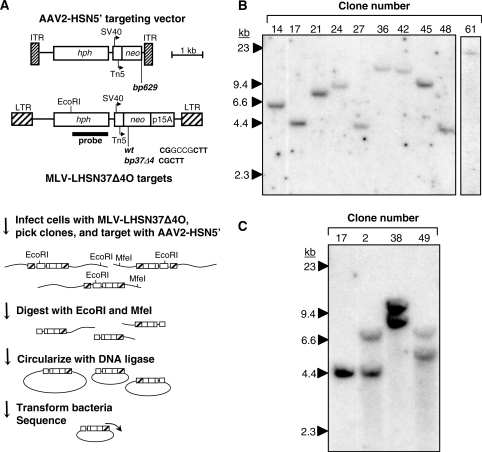
Figure 1.
Shuttle vector rescue of targeted loci. (A) Maps of AAV targeting vector AAV2-HSN5′ containing a neo gene truncated at bp 629 and an MLV vector LHSN37Δ4O provirus containing a 4-bp deletion in neo at bp 37. The locations of the AAV inverted terminal repeats (ITRs), simian virus 40 (SV40) and Tn5 promoters, transcriptional start sites (arrows), hph and neo genes, p15A replication origin, and retrovirus long terminal repeats (LTR) are shown. The fragment used to probe Southern blots is indicated. The strategy used for recovering targeted loci is shown below. (B) Southern blot of genomic DNA from HT-1080-derived clonal cell lines containing a single copy of the LHSN37Δ4O provirus, digested with EcoRI and probed for hph sequences. The positions of size standards are shown on the left. (C) Southern blot of genomic DNA from HT-1080-derived clonal cell lines containing two copies of the LHSN37Δ4O provirus, with clone 17 as a single-copy control, digested and probed as in (B).
The chromosomal positions of targets were determined by recovering integrated vector proviruses along with flanking human DNA as bacterial plasmids (Figure 1A). To facilitate provirus rescue, the neo genes in target loci were first corrected by AAV-mediated gene targeting and isolation of G418-resistant clones (see below). Then, genomic DNA was digested with EcoRI and MfeI, circularized with DNA ligase, and rescued by electroporation of E. coli and kanamycin selection. The 3′ LTR chromosomal junction fragments were sequenced and the target loci mapped to their locations in the human genome (Tables 1 and 2). All the proviruses present in the 13 HT-1080 clones used in our study were recovered and mapped, and their predicted hph-hybridizing restriction fragment sizes (Tables 1 and 2) corresponded to those observed on Southern blots (Figure 1B and C). Over 50% of the target loci are within RefSeq transcription units, reflecting the MLV bias for integration near transcription start sites and within genes (44).
Table 1.
Targeting frequencies and site locations in single-target clones
| Clone | Targeting frequency | Chromosomal location | Nearest RefSeq gene | Location of integration site | Distance to transcription start sitea | Predicted size of hph-hybridizing fragmentb |
|---|---|---|---|---|---|---|
| 14 | 2.19 × 10−4 | Chr 2: 157,621,301 | GALNT5 | Intergenic | −201.3 kb | 5.8 kb |
| 17 | 3.04 × 10−4 | Chr 6: 45,515,506 | RUNX2 | Intron 5 | +17.6 kb | 4.3 kb |
| 21 | 1.55 × 10−3 | Chr 12: 78,950,944 | PPP1R12A | Intergenic | +259.5 kb | 7.2 kb |
| 24 | 2.69 × 10−4 | Chr 7: 129,782,991 | CPA5 | Intron 6 | +11.1 kb | 8.6 kb |
| 27 | 1.81 × 10−4 | Chr 9: 16,326,741 | BNC2 | Intergenic | −72.8 kb | 4.1 kb |
| 36 | 5.77 × 10−4 | Chr 6: 150,220,289 | LRP11 | Intron 6 | −31.0 kb | 13.1 kb |
| 42 | 1.07 × 10−3 | Chr 2: 15,149,860 | NAG | Intergenic | −74.6 kb | 12.6 kb |
| 45 | 1.06 × 10−3 | Chr 2: 207,966,780 | CREB1 | Intergenic | −136.1 kb | 9.6 kb |
| 48 | 2.21 × 10−4 | Chr 14: 37,194,933 | BC038110 | Intron 2 | +66.0 kb | 4.2 kb |
| 61 | 9.44 × 10−4 | Chr 19: 18,560,834 | LOC55049 | Exon 2 | +0.3 kb | 18.7 kb |
aPositive and negative distances indicate that the integration site is downstream or upstream, respectively, relative to the nearest RefSeq transcription start site.
bSee Figure 1B for bands of corresponding sizes on Southern blots.
Table 2.
Targeting frequencies and site locations in double-target clones
| Clone | Overall targeting frequency | Proportion targeted at each sitea | Partial targeting frequency | Chromosomal location | Nearest RefSeq gene | Location of integration site | Distance to transcription start siteb | Predicted size of hph-hybridizing fragmentc |
|---|---|---|---|---|---|---|---|---|
| 2 | 1.15 × 10−3 | A: 13/20 | 7.46 × 10−4 | Chr 16: 19,636,191 | IQCK | Intron 1 | −0.9 kb | 4.4 kb |
| B: 7/20 | 4.01 × 10−4 | Chr 9: 12,804,291 | LOC286343 | Intron 1 | −39.3 kb | 7.1 kb | ||
| 38 | 2.10 × 10−3 | A: 18/20 | 1.89 × 10−3 | Chr 3: 173,242,513 | FNDC3B | Intron 1 | −1.5 kb | 8.4 kb |
| B: 2/20 | 2.10 × 10−4 | Chr 16: 30,576,276 | PRR14 | Intergenic | −6.5 kb | 10.3 kb | ||
| 49 | 1.32 × 10−3 | A: 16/21 | 1.00 × 10−3 | Chr 5: 153,581,833 | GALNT10 | Intron 1 | −31.3 kb | 5.5 kb |
| B: 5/21 | 3.14 × 10−4 | Chr 4: 167,577,088 | SPOCK3 | Intergenic | +314.0 kb | 7.5 kb |
aRepresents the number of rescued target plasmids from site A or B over the total analyzed for each clone.
bPositive and negative distances indicate that the integration site is downstream or upstream, respectively, relative to the nearest RefSeq transcription start site.
cSee Figure 1C for bands of corresponding sizes on Southern blot.
Gene targeting frequencies in cell lines containing target vector proviruses
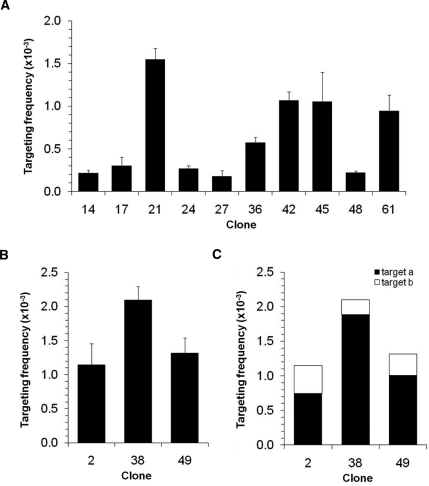
The 10 HT-1080-derived clones containing a single target provirus were infected with the AAV2-HSN5′ targeting vector, plated at different dilutions, then cultured in the presence or absence of G418 to determine the percentage of G418-resistant colony-forming units. All neo target site mutations were correctable (Figure 2A), with a variation of about 10-fold in targeting frequencies among clones, ranging from 1.81 × 10–4 for clone 27 to 1.55 × 10–3 for clone 21 (Table 1). The differences in targeting frequencies measured at the 10 sites were statistically significant (P < 0.0001; ANOVA). In each case, we isolated targeted subclones and confirmed by sequencing that the neo genes had been accurately corrected (data not shown). Although several of the targets were within genes (Table 1), the 3 clones with the highest targeting frequencies were not (clones 21, 42 and 45).
Figure 2.
Gene targeting frequencies of clonal cell lines. (A) Frequencies in clones containing one LHSN37Δ4O target locus infected with AAV2-HSN5′ at an MOI of 10 000 vector particles/cell. (B) Frequencies in clones containing two LHSN37Δ4O target loci infected as in (A). (C) Partial gene targeting frequencies for each target (a or b) present in cell lines containing two targets.
Three HT-1080 clones contained two target vector proviruses, so in order to determine their individual targeting frequencies the proportion of targeted proviruses rescued from each site was multiplied by the overall, clonal targeting frequency (Table 2). As with the cell lines containing a single target provirus, there was up to 10-fold variation in the targeting frequencies at the distinct target loci present in a single cell. Importantly, these findings demonstrate that the targeting frequency variation was not due to unrelated cell-specific effects that might affect results when comparing cell lines with single-target sites. Despite this variation at individual target loci, the overall targeting frequencies of all three clones with two targets were not statistically significantly different (P = 0.07; ANOVA) and were remarkably similar to each other (1.15 × 10–3 to 2.10 × 10–3) and to the single copy clone with the highest targeting frequency (clone 21: 1.55 × 10–3), suggesting that there may be some cellular limits on the maximal targeting frequency that can be achieved.
Target site transcript levels and gene targeting frequencies
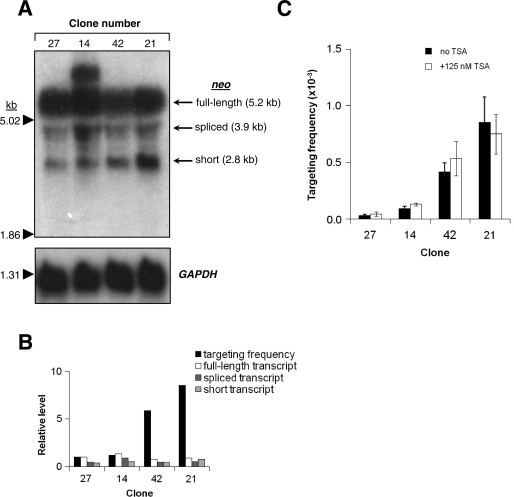
There is evidence that gene targeting might be most efficient at highly expressed chromosomal loci (1,45). To test whether differential transcription of target loci can account for the observed differences in gene targeting frequencies, we performed northern blot analysis to measure neo transcript levels in the single-target clones with the two lowest (clones 27 and 14) and two highest (clones 42 and 21) targeting frequencies (Figure 3A). Three separate neo transcripts exist in each of the four clones: a full-length transcript of 5.2 kb starting at the 5′ LTR promoter, a spliced transcript of 3.9 kb also driven from the LTR promoter and a short transcript of 2.8 kb driven from the SV40 promoter. In all clones studied, the most abundant transcript was the full-length one. Although there was some variation in transcript levels, these did not correlate with differences in targeting frequencies (Figure 3B). Of note, clone 14 has an additional neo-hybridizing transcript of ∼8 kb in length, which is likely due to read-through transcription from a genomic promoter located upstream of the provirus. Three individual expressed sequence tags (ESTs) are located between 5.9 and 1.2 kb upstream of the proviral integration site.
Figure 3.
Northern blot analysis and effect of trichostatin A (TSA) on gene targeting. (A) Northern blot of total RNA extracted from the HT-1080-derived clonal cell lines containing a single copy of the LHSN37Δ4O provirus having the two lowest (clones 27 and 14) and two highest (clones 42 and 21) gene targeting frequencies. The blot was probed for neo transcripts and for GAPDH transcripts to check loading. The positions of size standards are shown on the left, and the three expected neo transcript forms (full-length, spliced or short) and corresponding sizes are indicated on the right. (B) Comparison of targeting frequencies (calculated relative to targeting frequency of clone 27) and neo transcript levels (calculated relative to full-length transcript of clone 27) for the four clones. (C) Gene targeting frequencies in four clones treated with or without 125 nM TSA from 4.5 h after infection with AAV2-HSN5′ at an MOI of 10 000 vector particles/cell until splitting.
Chromatin structure and gene targeting frequencies
To test whether chromatin structure, rather than the sequence context of target sites, can affect gene targeting frequencies, we performed experiments in cells treated with TSA, a histone deacetylase inhibitor. Histone deacetylases play an important role in establishing chromatin structure, since they catalyze the removal of acetyl groups from histone proteins, causing chromatin condensation and transcriptional repression. The treatment of cells with TSA during targeting provided a way to reversibly inhibit histone deacetylaes and transiently alter chromatin structure, allowing us to test the effect. No statistically significant differences (P > 0.05; t-test) in targeting frequencies were observed between cells treated with TSA and untreated cells (Figure 3C). To determine if TSA treatment had the expected effect on histone modification at the target loci, we performed a ChIP analysis to measure the change in acetyl-histone H3 Lys 9 (H3K9ac) levels at neo target sites in the presence of TSA. We chose H3K9ac because it is a transcriptionally permissive histone mark, and treatment with TSA has been shown to induce acetylation at H3K9 (46,47). However, our analysis showed that the H3K9ac levels were not increased at neo loci by the TSA treatment (data not shown).
Effects of surrounding genomic features on targeting frequencies
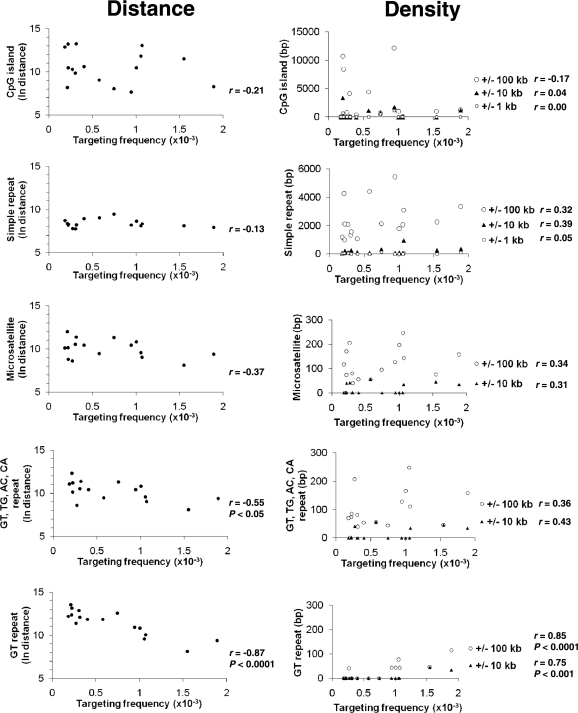
We determined the distances of several types of genomic elements to each target site, including RefSeq transcription start sites, CpG islands, simple repeats, microsatellites, SINEs, LINEs, DNA transposons, LTR retrotransposons and RNA repeats (see ‘Materials and Methods’ section) and calculated correlation coefficients between the targeting frequencies at each of the 16 target loci and their distance from these elements (Table 3). We also analyzed the data in relation to the natural logarithms of the distance to each genomic element, reasoning that the effects of any element on targeting frequency may decrease exponentially with distance (Table 3). The largest correlation coefficients were observed with CpG islands and microsatellite repeats, with gene targeting frequencies inversely correlating with distance from these genetic elements. Similarly, we determined whether the density of surrounding genomic elements influenced targeting by correlating targeting frequencies to the proportion of sequence covered by each type of genomic element in windows extending 1, 10 and 100 kb in either direction at each target site (Table 3). The largest correlations were observed with simple repeats and microsatellites. The data for CpG islands, simple repeats and microsatellites are shown graphically in Figure 4. Although intriguing, none of these correlations were statistically significant (P > 0.1 for all).
Table 3.
Correlation between targeting frequencies and distance to or density of genomic elements
| Genomic element | Distancea |
Densityb |
|||
|---|---|---|---|---|---|
| Interval size |
|||||
| bp |
Natural log bp |
±1 kb |
±10 kb |
±100 kb |
|
| Correlation coefficient, r | Correlation coefficient, r | Correlation coefficient, r | Correlation coefficient, r | Correlation coefficient, r | |
| RefSeq transcripts | 0.01 | −0.15 | 0.15 | −0.03 | 0.02 |
| CpG islandsc | −0.23 | −0.21 | 0.00 | 0.04 | −0.17 |
| Simple repeats | −0.11 | −0.13 | 0.05 | 0.39 | 0.32 |
| Microsatellites (all)c | −0.33 | −0.37 | n/ad | 0.31 | 0.34 |
| SINEs | −0.12 | −0.03 | −0.04 | 0.25 | 0.07 |
| LINEs | −0.07 | −0.08 | 0.17 | −0.01 | −0.05 |
| DNA transposons | 0.03 | 0.05 | −0.17 | 0.10 | 0.17 |
| LTR retrotransposons | −0.23 | −0.18 | −0.18 | 0.29 | 0.26 |
| RNA repeats | 0.21 | 0.19 | n/a | n/a | −0.28 |
aDistance was measured from bp 37 of the neo gene in the MLV target locus to the center of the nearest respective genomic element, with the exception of RefSeq transcripts, where distance was measured to the start site of the nearest RefSeq transcription unit; correlation coefficients were computed using either distance (in bp) or the natural logarithm of distance.
bDensity was measured as the total number of base pairs covered by the corresponding genomic element within a given interval surrounding the target integration site.
cSee Figure 3 for representative scatter plots.
dn/a denotes the absence of specified genomic element within given interval.
Figure 4.
Effects of neighboring genetic elements on gene targeting. In each panel, scatter plots on the left graph the targeting frequency at each target site and the natural logarithm of the distance from bp 37 of the neo gene in each target to the center of the indicated genetic element. Scatter plots on the right graph the density (total number of base pairs covered) of each genomic element within 100 kb (open circle), 10 kb (filled triangle) or 1 kb (open square) on either side of each target locus integration site. The lack of density values within ±1 kb in some panels is due to the absence of the respective genomic element in that interval. Correlation coefficients (r) and P-values (for significant r) are to the right of the scatter plots.
Microsatellites comprise a group of simple sequence repeats that can be broken down into further subsets such as dinucleotide repeats. We categorized dinucleotides into four distinct classes, each comprised of repeats that differed only in starting nucleotide and/or direction: GT, TG, AC and CA; GA, AG, TC and CT; AT and TA; and GC and CG (Table 4). For example, AC is considered equivalent to CA, and both AC and CA are considered equivalent to GT and TG on the complementary strand. The only class of dinucleotide repeats whose proximity was significantly correlated to targeting frequencies was GT, TG, AC and CA (r = –0.55, P < 0.05), meaning that as the distance of a target site from this class of repeats increases, its predicted targeting frequency decreases. In addition, in terms of density, the class of GT, TG, AC and CA dinucleotide repeats had the highest correlation to targeting frequencies within 10 kb (r = 0.43) and 100 kb (r = 0.36), although neither was significant (Figure 4).
Table 4.
Correlation between targeting frequencies and distance to or density of dinucleotide repeats
| Dinucleotide repeata | Distanceb |
Densityb |
|||
|---|---|---|---|---|---|
| Interval size |
|||||
| bp |
Natural log bp |
±1 kb |
±10 kb |
±100 kb |
|
| Correlation coefficient, r | Correlation coefficient, r | Correlation coefficient, r | Correlation coefficient, r | Correlation coefficient, r | |
| GT, TG, AC and CA repeatsc | −0.44 | −0.55d | n/ae | 0.43 | 0.36 |
| GT | −0.62f | −0.87g | n/a | 0.75h | 0.85i |
| TG | 0.26 | 0.26 | n/a | −0.21 | −0.12 |
| AC | −0.19 | −0.15 | n/a | 0.06 | −0.09 |
| CA | −0.15 | −0.16 | n/a | n/a | 0.08 |
| GA, AG, TC and CT repeats | −0.20 | −0.05 | n/a | n/a | −0.02 |
| GA | −0.11 | −0.01 | n/a | n/a | n/a |
| AG | 0.00 | −0.01 | n/a | n/a | 0.16 |
| TC | −0.14 | −0.12 | n/a | n/a | 0.20 |
| CT | −0.17 | 0.00 | n/a | n/a | −0.26 |
| AT and TA repeatsc | 0.49 | 0.40 | n/a | −0.24 | −0.07 |
| AT | 0.39 | 0.30 | n/a | n/a | 0.44 |
| TA | 0.36 | 0.41 | n/a | −0.24 | −0.24 |
aMicrosatellites GT, TG, AG, CA, GT, GA, AG, TC, CT, AT and TA represent sequences of at least 15 perfect dinucleotide repeats.
bDistance and density were computed as in Table 3.
cSee Figure 3 for representative scatter plots.
dRepresents significant correlation (P < 0.05).
en/a denotes the absence of specified genomic element within given interval.
fRepresents significant correlation (P = 0.01).
gRepresents significant correlation (P < 0.0001).
hRepresents significant correlation (P < 0.001).
iRepresents significant correlation (P < 0.0001).
We also considered each type of dinucleotide repeat on its own and investigated whether their distance to or density surrounding target loci correlates to targeting frequencies (Table 4). The dinucleotides used in the analysis (GT, TG, AC, CA, GA, AG, TC, CT, AT and TA) represented sequences of at least 15 perfect repeats. We did not include GC and CG repeats in our analysis because of the absence of sequences of at least 15 perfect dinucleotide repeats within 1 Mb of MLV target vector integration sites (Supplementary Table S1). Among individual dinucleotide repeats, only the proximity and density of GT repeats was found to correlate significantly to targeting frequencies, as seen in Figure 4E. As the distance between a target site and a GT repeat increases, the targeting frequency at that site tends to decrease (r = −0.87, P < 0.0001). Moreover, as the density of GT dinucleotide repeats increases within 10 kb or 100 kb on either side of a target locus, the targeting frequency at that locus tends to increase (r = 0.75, P < 0.001 and r = 0.85, P < 0.0001, respectively). Interestingly, the dinucleotide repeats considered to be equivalent to GT repeats, namely TG, AC and CA repeats, did not have significant correlations to targeting frequencies for either distance or density.
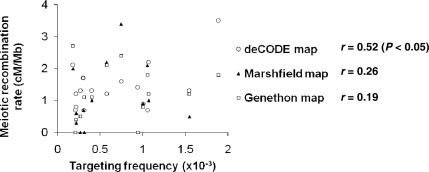
Meiotic recombination rates at target loci
To determine whether any relationship exists between gene targeting and meiotic recombination, we correlated the measured targeting frequencies at the loci we tested with the sex-averaged meiotic recombination rates at the same loci as measured by the deCODE (8), Marshfield (9) and Genethon (10) genetic maps (Figure 5). The meiotic recombination rates from the deCODE map were the only ones that correlated significantly with gene targeting frequencies (r = 0.52, P < 0.05). The most likely explanation for the significant correlation found between targeting frequencies and deCODE map recombination rates is that the deCODE map, representing 1257 total meioses, provides recombination rates at a higher resolution than the Marshfield and Genethon maps, based on XX and YY meioses, respectively.
Figure 5.
Correlation between gene targeting frequencies and the sex-averaged meiotic recombination rates. The scatter plot graphs meiotic recombination rates from the deCODE (open circle), Marshfield (filled triangle) and Genethon (open square) genetic maps at each provirus target site in centiMorgans/Megabase. Correlation coefficients (r) and P-values (for significant r) are to the right of the corresponding legends.
DISCUSSION
Our results clearly establish a chromosomal position effect on gene targeting in human cells. The experiments we performed in clones containing a single mutant neo provirus per cell showed statistically significant differences in frequencies among the targeted loci (P < 0.0001), with as much as 10-fold variability. In clones containing two neo proviruses located at different positions, each provirus had a distinct targeting frequency, eliminating variables such as growth or infectivity rates that could have influenced targeting when comparing different clones. By determining the positions of neighboring genetic elements, we showed that targeting frequencies increase significantly based on proximity to GT dinucleotide repeats.
While several studies involving S. cerevisiae have shown that targeting frequency is not strongly influenced by the surrounding chromosomal context of the sites tested (11,12), this is less clear in mammalian cells. To our knowledge, only three studies prior to our own addressed this issue. In the first of these studies, DNA was added by the calcium phosphate method to mouse L cells to reconstruct a functional thymidine kinase (tk) gene from two defective genes, resulting in one tk+ transformant per 106 cells, but only one of the 10 tk- cell lines was successfully targeted and became tk+ (14). Our results showed successful gene targeting at all 16 loci we tested, which we attribute to the greater sensitivity of our experimental design, with targeting frequencies over 10–4 for all loci. This allowed us to detect targeting at loci that are not preferred sites for homologous recombination. The second study involved the correction of an expressed, defective neo gene residing in the chromosome of mouse L cells by using DNA microinjection (13). However, the targeting frequencies in the cell lines tested were not quantitatively reported, and in the cell line that contained multiple target genes, the relative targeting frequencies at different targets were not determined, making it difficult to draw any conclusions about the existence of a position effect on gene targeting. In the third study, which involved a human cell line containing four endogenous and two ectopic alleles, a 34-fold preference for the targeted correction of one of the ectopic alleles was observed, consistent with a position effect (15). Importantly, none of the prior studies reported the genomic positions of the target loci, so it was not possible to study the effects of neighboring genetic elements on targeting. Our results expand on these studies by demonstrating a chromosomal position effect on targeting using multiple identical target alleles with known structure and chromosomal location, in both single-target and double-target cell lines.
Although there is evidence that transcription enhances extrachromosomal (48) and intrachromosomal (49) homologous recombination in mammalian cells, and that gene targeting might be most efficient at highly expressed chromosomal loci (1,45), target site transcription levels did not account for the differences in targeting frequencies we observed. The neo targets we introduced were all transcribed from the same promoters, regardless of their chromosomal position. Northern blot analysis of neo transcript levels showed that higher targeting frequencies were not associated with higher transcript levels. Moreover, we found that there was no relationship between the presence of a target locus within a gene and its targeting frequency. In addition, neither the proximity to the nearest RefSeq transcription start site, nor the density of RefSeq transcripts within 1, 10 or 100 kb of the targets, correlated significantly to the measured gene targeting frequencies.
We were unable to definitively address whether changes in chromatin structure affected gene targeting frequencies. No significant differences in targeting frequencies occurred after treatment with TSA, a histone deacetylase inhibitor. However, ChIP analysis at neo target loci did not reveal the expected increase in H3K9ac that has been reported with TSA treatment (46,47,50). Treatment with a higher dose of TSA was not possible because we were already using the maximum dose that did not produce significant cell death. It is possible that TSA does not affect acetylation at the four target loci studied. Previous studies have shown that TSA exhibits gene specificity and site selectivity, such that the state of acetylation of certain genes is not altered (50), and only a fraction of the transcriptome is influenced by exposure to the drug (51). Further experiments investigating the effects of other chromatin-modifying agents on gene targeting frequencies might distinguish between these possibilities.
Homologous recombination between chromosomes occurs in both mitosis and meiosis. However, our understanding of spontaneous mitotic recombination is still limited, in large part because mitotic recombination events are infrequent compared to meiotic exchanges. In yeast, for example, mitotic events are about 104 to 105 less frequent than meiotic events (52). Gene targeting is another process that occurs through homologous recombination, with crossovers between an introduced piece of DNA and a chromosomal sequence. We found a significant correlation between gene targeting frequencies and meiotic recombination rates, suggesting that common mechanisms are involved in both processes.
In our experiments, the only genomic element whose proximity to or density surrounding the target site had a significant correlation to gene targeting frequencies was microsatellite repeats, which could be attributed to the specific effects of GT dinucleotide repeats. Our results are in agreement with several lines of experimental evidence implicating GT repeats in recombination, including a prior study showing that GT repeats stimulate homologous recombination in SV40 viral DNA (18), and studies showing that GT repeats introduced at certain yeast loci increase meiotic crossovers (53,54). Moreover, it was shown that the distribution of GT repeats, in contrast to GC, AT and GA repeats, on human chromosome 22 correlated with meiotic recombination frequencies (55). Taken together with our data, this further supports the hypothesis that common mechanisms are involved in meiosis and mitotic gene targeting. Our analyses did not reveal a significant correlation between GC or CG dinucleotide repeats and gene targeting frequencies, despite previous work showing they stimulate homologous recombination between two plasmids (18). In addition, neither the proximity to, nor density of, CpG islands surrounding target sites was significantly correlated to targeting frequency, despite the fact that CpG islands are rich in CG dinucleotides (56,57).
At present, we cannot explain why the proximity and density of GT repeats influence gene targeting while TG, AC and CA repeats do not, despite the fact that they can be considered functionally equivalent, varying only by starting nucleotide and strand orientation. It is possible that more data would reveal significant relationships between targeting frequencies and other repeats, but there could also be a specific effect of GT repeats that we do not understand. This is supported by the similar effect of GT repeats on meiotic recombination (55).
Dinucleotide repeats could influence gene targeting in several ways. The E. coli RecA protein as well as its yeast and human homologs bind dinucleotide repeats (58,59), albeit without specificity for GT dinucleotides. This could promote homologous pairing of targeting constructs and target loci. Preferential unwinding or misaligned pairing at dinucleotide repeats could expose single-stranded regions near chromosomal targets and recruit proteins involved in recombination or repair that also play a role in targeting. Since AAV-mediated gene targeting occurs preferentially in S-phase (60), it is also possible that hairpin formation at dinucleotide repeats in single-stranded regions present at replication forks recruits proteins involved in targeting. These possibilities, as well as the unexplained specificity of GT dinucleotide effects, will require further experiments and a larger data set to resolve.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The U.S. National Institutes of Health (DK55759, AR48328 to D.W.R.) and the University of Washington Medical Scientist Training Program (Poncin Foundation Scholarship to A.M.C.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Roli K. Hirata, Yi Li, Erik Olson, and Raisa Stolitenko for valuable technical advice and assistance.
REFERENCES
- 1.Thyagarajan B, Johnson B, Campbell C. The effect of target site transcription on gene targeting in human cells in vitro. Nucleic Acids Res. 1995;23:2784–2790. doi: 10.1093/nar/23.14.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Domínguez-Bendala J, McWhir J. Enhanced gene targeting frequency in ES cells with low genomic methylation levels. Transgenic Res. 2004;13:69–74. doi: 10.1023/b:trag.0000017176.77847.80. [DOI] [PubMed] [Google Scholar]
- 3.Keil R, Roeder G. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984;39:377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- 4.Voelkel-Meiman K, Keil R, Roeder G. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987;48:1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- 5.Neitz M, Carbon J. Characterization of a centromere-linked recombination hot spot in Saccharomyces cerevisiae. Mol. Cell. Biol. 1987;7:3871–3879. doi: 10.1128/mcb.7.11.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raynard S, Read L, Baker M. Evidence for the murine IgH mu locus acting as a hot spot for intrachromosomal homologous recombination. J. Immunol. 2002;168:2332–2339. doi: 10.4049/jimmunol.168.5.2332. [DOI] [PubMed] [Google Scholar]
- 7.Buzina A, Shulman M. An element in the endogenous IgH locus stimulates gene targeting in hybridoma cells. Nucleic Acids Res. 1996;24:1525–1530. doi: 10.1093/nar/24.8.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong A, Gudbjartsson D, Sainz J, Jonsdottir G, Gudjonsson S, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. A high-resolution recombination map of the human genome. Nat. Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 9.Broman K, Murray J, Sheffield V, White R, Weber J. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am. J. Hum. Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dib C, Fauré S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, et al. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 11.Wilson J, Leung W, Bosco G, Dieu D, Haber J. The frequency of gene targeting in yeast depends on the number of target copies. Proc. Natl Acad. Sci. USA. 1994;91:177–181. doi: 10.1073/pnas.91.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray M, Honigberg S. Effect of chromosomal locus, GC content and length of homology on PCR-mediated targeted gene replacement in Saccharomyces. Nucleic Acids Res. 2001;29:5156–5162. doi: 10.1093/nar/29.24.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas K, Folger K, Capecchi M. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986;44:419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- 14.Lin F, Sperle K, Sternberg N. Recombination in mouse L cells between DNA introduced into cells and homologous chromosomal sequences. Proc. Natl Acad. Sci. USA. 1985;82:1391–1395. doi: 10.1073/pnas.82.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yáñez R, Porter A. A chromosomal position effect on gene targeting in human cells. Nucleic Acids Res. 2002;30:4892–4901. doi: 10.1093/nar/gkf614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaco I, Canela A, Vera E, Blasco M. Centromere mitotic recombination in mammalian cells. J. Cell. Biol. 2008;181:885–892. doi: 10.1083/jcb.200803042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bullock P, Miller J, Botchan M. Effects of poly[d(pGpT).d(pApC)] and poly[d(pCpG).d(pCpG)] repeats on homologous recombination in somatic cells. Mol. Cell. Biol. 1986;6:3948–3953. doi: 10.1128/mcb.6.11.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benet A, Mollà G, Azorín F. d(GA x TC)(n) microsatellite DNA sequences enhance homologous DNA recombination in SV40 minichromosomes. Nucleic Acids Res. 2000;28:4617–4622. doi: 10.1093/nar/28.23.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wahls W, Wallace L, Moore P. The Z-DNA motif d(TG)30 promotes reception of information during gene conversion events while stimulating homologous recombination in human cells in culture. Mol. Cell. Biol. 1990;10:785–793. doi: 10.1128/mcb.10.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wahls W, Wallace L, Moore P. Hypervariable minisatellite DNA is a hotspot for homologous recombination in human cells. Cell. 1990;60:95–103. doi: 10.1016/0092-8674(90)90719-u. [DOI] [PubMed] [Google Scholar]
- 21.Russell D, Hirata R. Human gene targeting by viral vectors. Nat. Genet. 1998;18:325–330. doi: 10.1038/ng0498-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirata R, Russell D. Design and packaging of adeno-associated virus gene targeting vectors. J. Virol. 2000;74:4612–4620. doi: 10.1128/jvi.74.10.4612-4620.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirata R, Chamberlain J, Dong R, Russell D. Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat. Biotechnol. 2002;20:735–738. doi: 10.1038/nbt0702-735. [DOI] [PubMed] [Google Scholar]
- 24.Zwaka T, Thomson J. Homologous recombination in human embryonic stem cells. Nat. Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- 25.Brown J, Wei W, Sedivy J. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 26.Miller D, Petek L, Russell D. Human gene targeting by adeno-associated virus vectors is enhanced by DNA double-strand breaks. Mol. Cell. Biol. 2003;23:3550–3557. doi: 10.1128/MCB.23.10.3550-3557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell D, Hirata R. Human gene targeting favors insertions over deletions. Hum. Gene. Ther. 2008;19:907–914. doi: 10.1089/hum.2008.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue N, Hirata R, Russell D. High-fidelity correction of mutations at multiple chromosomal positions by adeno-associated virus vectors. J. Virol. 1999;73:7376–7380. doi: 10.1128/jvi.73.9.7376-7380.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller A, Miller D, Garcia J, Lynch C. Use of retroviral vectors for gene transfer and expression. Methods Enzymol. 1993;217:581–599. doi: 10.1016/0076-6879(93)17090-r. [DOI] [PubMed] [Google Scholar]
- 30.Southern P, Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J. Mol. Appl. Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- 31.Chang A, Cohen S. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 33.Rasheed S, Nelson-Rees W, Toth E, Arnstein P, Gardner M. Characterization of a newly derived human sarcoma cell line (HT-1080) Cancer. 1974;33:1027–1033. doi: 10.1002/1097-0142(197404)33:4<1027::aid-cncr2820330419>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 34.Kinsella T, Nolan G. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 35.DuBridge R, Tang P, Hsia H, Leong P, Miller J, Calos M. Analysis of mutation in human cells by using an Epstein–Barr virus shuttle system. Mol. Cell. Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burns J, Friedmann T, Driever W, Burrascano M, Yee J. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl Acad. Sci. USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimm D, Kern A, Rittner K, Kleinschmidt J. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum. Gene Ther. 1998;9:2745–2760. doi: 10.1089/hum.1998.9.18-2745. [DOI] [PubMed] [Google Scholar]
- 38.Zolotukhin S, Byrne B, Mason E, Zolotukhin I, Potter M, Chesnut K, Summerford C, Samulski R, Muzyczka N. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
- 39.Rutledge E, Russell D. Adeno-associated virus vector integration junctions. J. Virol. 1997;71:8429–8436. doi: 10.1128/jvi.71.11.8429-8436.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanahan D, Jessee J, Bloom F. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 1991;204:63–113. doi: 10.1016/0076-6879(91)04006-a. [DOI] [PubMed] [Google Scholar]
- 41.Kent W. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karolchik D, Hinrichs A, Furey T, Roskin K, Sugnet C, Haussler D, Kent W. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor J, Schenck I, Blankenberg D, Nekrutenko A. Curr. Protoc. Bioinformatics. 2007. Using galaxy to perform large-scale interactive data analyses. Chapter 10, Unit 10.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu X, Li Y, Crise B, Burgess S. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 45.Macris M, Glazer P. Transcription dependence of chromosomal gene targeting by triplex-forming oligonucleotides. J. Biol. Chem. 2003;278:3357–3362. doi: 10.1074/jbc.M206542200. [DOI] [PubMed] [Google Scholar]
- 46.Meng C, Zhu X, Peng G, Dai D. Promoter histone H3 lysine 9 di-methylation is associated with DNA methylation and aberrant expression of p16 in gastric cancer cells. Oncol. Rep. 2009;22:1221–1227. doi: 10.3892/or_00000558. [DOI] [PubMed] [Google Scholar]
- 47.Zhong S, Goto H, Inagaki M, Dong Z. Phosphorylation at serine 28 and acetylation at lysine 9 of histone H3 induced by trichostatin A. Oncogene. 2003;22:5291–5297. doi: 10.1038/sj.onc.1206507. [DOI] [PubMed] [Google Scholar]
- 48.Nickoloff J, Reynolds R. Transcription stimulates homologous recombination in mammalian cells. Mol. Cell. Biol. 1990;10:4837–4845. doi: 10.1128/mcb.10.9.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nickoloff J. Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol. Cell. Biol. 1992;12:5311–5318. doi: 10.1128/mcb.12.12.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ou J, Torrisani J, Unterberger A, Provençal N, Shikimi K, Karimi M, Ekström T, Szyf M. Histone deacetylase inhibitor Trichostatin A induces global and gene-specific DNA demethylation in human cancer cell lines. Biochem. Pharmacol. 2007;73:1297–1307. doi: 10.1016/j.bcp.2006.12.032. [DOI] [PubMed] [Google Scholar]
- 51.Gius D, Cui H, Bradbury C, Cook J, Smart D, Zhao S, Young L, Brandenburg S, Hu Y, Bisht K, et al. Distinct effects on gene expression of chemical and genetic manipulation of the cancer epigenome revealed by a multimodality approach. Cancer Cell. 2004;6:361–371. doi: 10.1016/j.ccr.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 52.Pâques F, Haber J. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Treco D, Arnheim N. The evolutionarily conserved repetitive sequence d(TG.AC)n promotes reciprocal exchange and generates unusual recombinant tetrads during yeast meiosis. Mol. Cell. Biol. 1986;6:3934–3947. doi: 10.1128/mcb.6.11.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gendrel C, Boulet A, Dutreix M. (CA/GT)(n) microsatellites affect homologous recombination during yeast meiosis. Genes Dev. 2000;14:1261–1268. [PMC free article] [PubMed] [Google Scholar]
- 55.Majewski J, Ott J. GT repeats are associated with recombination on human chromosome 22. Genome Res. 2000;10:1108–1114. doi: 10.1101/gr.10.8.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bird A. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 57.Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- 58.Dutreix M. (GT)n repetitive tracts affect several stages of RecA-promoted recombination. J. Mol. Biol. 1997;273:105–113. doi: 10.1006/jmbi.1997.1293. [DOI] [PubMed] [Google Scholar]
- 59.Biet E, Sun J, Dutreix M. Conserved sequence preference in DNA binding among recombination proteins: an effect of ssDNA secondary structure. Nucleic Acids Res. 1999;27:596–600. doi: 10.1093/nar/27.2.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trobridge G, Hirata R, Russell D. Gene targeting by adeno-associated virus vectors is cell-cycle dependent. Hum. Gene Ther. 2005;16:522–526. doi: 10.1089/hum.2005.16.522. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.