Abstract
The AddAB helicase and nuclease complex is used for repairing double-strand DNA breaks in the many bacteria that do not possess RecBCD. Here, we show that AddAB, from the Gram-negative opportunistic pathogen Bacteroides fragilis, can rescue the ultraviolet sensitivity of an Escherichia coli recBCD mutant and that addAB is required for survival of B. fragilis following DNA damage. Using single-molecule observations we demonstrate that AddAB can translocate along DNA at up to 250 bp per second and can unwind an average of 14 000 bp, with some complexes capable of unwinding 40 000 bp. These results demonstrate the importance of processivity for facilitating encounters with recognition sequences that modify enzyme function during homologous recombination.
INTRODUCTION
Homologous recombination (HR) is required for repair of DNA damage and restarting stalled or regressed replication forks in both prokaryotes and eukaryotes. Escherichia coli has provided the paradigm for understanding the molecular processes involved in HR. One estimate suggests that spontaneous double-strand breaks (DSBs) occur once per chromosome replication cycle in E. coli, although it has been argued that the rate may be 100-fold lower (1,2). The first step in processing a DSB in E. coli is resection of the DNA by the trimeric RecBCD complex, using a nuclease domain within RecB coupled to two helicase motors with opposite strand polarities (3,4). When RecBCD loads onto a DSB, the DNA is unwound by the combined ATP-dependent action of the two motor proteins pulling the DNA through the complex, with RecC helping to split the duplex using a ‘pin-like’ structure wedged between the strands. The 3′–5′ strand is then preferentially degraded as it passes into the nuclease domain of RecB, while the 5′–3′strand is occasionally cut if it occupies the nuclease site (5,6). Single-molecule experiments upon RecBCD show that the enzyme translocates along DNA at ∼500–1500 bp s−1 unwinding and cleaving it as it goes, until it encounters an 8-bp chi site, after which translocation decreases to ∼150 bp s−1 (7,8). It is postulated that RecD is the lead and faster helicase prior to an encounter with chi, with this motor protein being inactivated following the encounter and RecB then predominating as the primary helicase with a slower onward translocation speed (9). The alteration in translocation is accompanied by a change in nuclease preference to degradation of the 5′–3′strand, which ultimately generates a 3′–5′single-stranded tail that is the substrate for RecB-mediated loading of RecA, followed by strand invasion of the homologous duplex (10).
RecBCD is present in a fraction of the eubacterial superkingdom and is usually replaced by the AddAB/RexAB orthologues (11–15), although Sinha et al. (16) have found a further variation with the discovery of the AdnAB enzyme in the mycobacteria. Biochemical analysis of AddAB from the Gram-positive bacterium Bacillus subtilis (BsAddAB) has shown it to have similar properties to RecBCD (5,6,17–19). In contrast to RecBCD, AddAB is a heterodimer that contains one helicase domain in AddA and two nuclease domains, one in each subunit of the complex. Many AddB also contain an Fe-S cluster (15) essential for the stability of the subunit and binding to double-stranded DNA ends (19). The protein–DNA complex unwinds double-stranded DNA and degrades both strands until it encounters a 5-bp chi site, at which point it stops degrading the 3′–5′ strand and so produces a substrate appropriate for loading of RecA.
Bacteroides fragilis is a member of the normal human gastro-intestinal tract microbiota and the Gram-negative obligate anaerobe most frequently isolated from opportunistic infections, such as peritonitis (20). During the genome sequencing of B. fragilis NCTC9343, we identified a gene (BF2192) encoding a protein with 23% identity to AddA from the Gram-positive bacterium Lactococcus lactis (21). The predicted polypeptide in B. fragilis contains a RecB-like nuclease motif (ValAspTyrLys) at the C-terminus and also contains conserved amino acids (aa) within the seven motor motifs that characterize the SF1 helicases (22,23). Adjacent to BF2192 is a gene, BF2191, predicted to encode a protein with 16% identity to B. subtilis AddB. The B. fragilis AddB is 209 aa shorter than the B. subtilis homologue, but contains three C-terminal cysteine residues (C946, C949 and C955) and a fourth cysteine (C674), flanking a nuclease motif (ValAspTyrLys), which is consistent with the presence of an Fe-S cluster as found in the B. subtilis AddB (19).
The presence of similar functions in bacteria that utilize different protein components raises a fundamental question about the roles of two versus one helicase motor in DSB repair. Do RecBCD and AddAB complexes have similar or dissimilar processivities and rates of translocation? To address this question, we have performed single-molecule observations on the AddAB homologues we identified in B. fragilis (BfAddAB) using high-throughput total internal reflection fluorescence microscopy (24) to simultaneously visualize numerous surface-tethered, hydrodynamically-stretched lambda DNA and the action of BfAddAB as a function of [ATP], shear rate and degree of staining with YOYO-1 fluorophore.
MATERIALS AND METHODS
Reagents
All chemicals were purchased from Sigma–Aldrich Company Ltd. (Dorset, UK), unless otherwise stated.
Bacterial strains, cultivation and plasmid construction
Escherichia coli strains for UV sensitivity tests were derivatives of MG1655. The ΔrecBCD allele was introduced by P1 transduction from JJC1056 (kindly gifted by David Leach, Edinburgh University). Serial dilutions of stationary phase cultures were spotted (10 µl) onto plates and irradiated with UV light at 80 J/m2 followed by incubation in the dark. Escherichia coli strains were cultured on LB at 37°C. Bacteroides fragilis NCTC9343 was cultivated on supplemented brain heart infusion (BHI-S), in a Mini-MACS anaerobic workstation (Don Whitley Scientific Ltd, UK), at 37°C in an atmosphere of 80% nitrogen, 10% carbon dioxide and 10% hydrogen. Plasmids expressing addAB, addA and addB were constructed by PCR amplification of the genes followed by ligation into the SacI and XbaI sites of pTRC99a (GE Healthcare, UK), allowing IPTG control of expression by the hybrid tac promoter. Deletions of addAB in B. fragilis were constructed by amplification of 500 bp flanking each side of the genes, followed by fusion with an ermF cassette, ligation into pGB909 and conjugation from E. coli S17 λpir as described by Patrick et al. (25).
Protein preparation and in vitro unwinding assay
Proteins were purified from the MG1655 ΔrecBCD strain containing the plasmid expressing addAB and a second plasmid encoding rare E. coli tRNA codons. This strain was grown in 2 l LB to an optical density of 0.5, followed by overnight induction at 18°C with 1 mM IPTG. Cell lysis and protein purification was done according to a method described in Chedin et al. (17). Purification used DEAE ion exchange, hydroxyapatite and heparin affinity chromatography. BfAddAB protein was concentrated using 30,000 MWCO ultrafiltration columns (Vivascience, UK) to yield a 17 µM AddAB stock solution to which glycerol was added to 50% v/v and stored at −20°C. Based on the content of aromatic amino acids and cysteine residues in the BfAddAB protein an absorption coefficient ε280 nm = 240 380 M−1 cm−1 was calculated for the BfAddAB dimer (26).
MALDI-TOF mass spectrometry of peptides derived from BfAddAB via in-gel trypsin proteolysis was used to confirm that the isolated protein complex comprised AddA and AddB proteins by comparison with entries in the NCBInr data base using MS-Fit search engine. The mass spectra were acquired on a Voyager DE-STR MALDI-TOF Mass Spectrometer (Applied Biosystems Inc, USA).
DNA agarose gel electrophoresis was used to assay Mg2+-ATP-dependent digestion of lambda DNA by BfAddAB. Fifty micrograms per mililitres of biotinylated lambda DNA at 5 nM concentration was reacted with 170 nM AddAB in 40 mM NaHCO3 (pH 8.0) buffer containing 35 mM DTT, 13% w/v sucrose, 1 mM ATP and 2 mM magnesium acetate. The DNA was stained with ethidium bromide and visualized using a UV transilluminator, with images acquired using a CCD camera and processed with IrfanView.
TIRF microscopy
The objective-TIRF setup was built around a Nikon Eclipse TE2000-U microscope in conjunction with a 60 × (NA 1.49) Apo-TIRF oil objective (Nikon, Japan). Zeiss oil (Germany) for fluorescence microscopy was used as immersion liquid. A collimated Ar ion laser beam (Spectra Physics, USA) was sent through a 488-nm line filter (Chroma Technology Corp, USA) and varying combinations of neutral density (ND) filters to spectrally clean and attenuate the laser light. A 10 × beam expander (Linos Photonics Ltd, UK) was used to spread the laser beam which was then focussed by a f = 400-mm plano-convex lens onto the objective back-focal plane. The focusing lens and the upper periscope mirror were mounted onto a vertically actuatable one-dimensional stage. This allowed the systematic displacement of the laser beam parallel to the optical axis of the objective. In this manner, one could establish epi-fluorescence, oblique illumination or evanescent-field excitation. The excitation beam was reflected into the objective by a 495-nm dichroic mirror. A 50-nm bandwidth 530-nm bandpass filter, which removes the Raman band of water, and a 500-nm longpass filter were used to collect the emitted fluorescence light. Digital images were acquired with a Luca EM-CCD camera (ANDOR, UK). Cooling maintained a stable temperature of –20°C at the detector. Exposure time was 0.2 s. A USB-6218 shutter (National Instruments, USA) was run at a frequency of 0.5 Hz and a 10% duty cycle, thereby producing 0.2-s pulse trains matching the camera exposure time. Laser powers were measured with a Fieldmaster (Coherent, USA). Using ND filters of 1.6 and 2.0, laser powers of 28.7 µW and 9.15 µW were obtained.
Preparation of microfluidic flowcells
Microfluidic cells were fabricated from Perspex (block 70 × 40 × 6 mm) with a one-mm channel milled out to a depth of 0.07 ± 0.01 mm. The inlet and outlet holes were drilled 1 mm in diameter and attached via dispenser tips to polypropylene tubing. The flowcell was sealed by gluing a coverglass over the channels. A KDS200 syringe pump (KD Scientific Inc. USA) was then attached to the flowcell.
Buffer A contained 100 mM NaCl, 10 mM Tris–HCl (pH 8.0) and 1 mM EDTA. Buffer B1 was 10 mM Tris–HCl (pH 8.0), 1 mM EDTA. Buffer B2 was 40 mM Tris–HCl (pH 7.8), 1 mM MgCl2 1 mM DTT and 0.2 mg/ml bovine serum albumin (BSA). Buffer C1 contained 40 mM NaHCO3 (pH 8.0), 2 mM magnesium acetate, 0.04 mg/ml catalase, 0.5 mg/ml glucose oxidase, 4 mg/ml β-mercaptoethanol, 12.5% w/v glucose and 17 nM BfAddAB. Buffer C2 used to trigger BfAddAB enzyme activity was C1 supplemented with ATP in different concentrations ranging from 0.01 to 5 mM. For 2 and 5 mM ATP conditions, an excess of 1-mM magnesium acetate over ATP was used. All buffers and solutions were sterilized by passing them through 0.2-µm filters. Buffers and ultrapure water were autoclaved before adding temperature-sensitive supplements such as BSA or NeutrAvidin (Pierce Biotechnologies, USA).
Prior to experiments, flowcells with one inlet and one outlet were rinsed with 9-ml ultrapure water followed by 2-ml 2% v/v Hellmanex detergent (Hellma, Germany). Thereafter, the flowcells were rinsed a second time with 9-ml water, then flushed with 2-ml buffer A and subsequently incubated for 30 min with 0.5 ml buffer A containing NeutrAvidin (20 µg/ml). Finally, the flowcells were rinsed again with 9 ml buffer A and 2 ml buffer B2 to remove unbound NeutrAvidin, and 0.2 mg/ml BSA in buffer B2 was introduced and incubated for 30 min producing a flowcell whose inner surface is coated with individual NeutrAvidin molecules and larger BSA patches for surface passivation. Prior to loading the biotinylated DNA samples, the flowcells were purged with 2 ml buffer B2 followed by 0.5 ml buffer C1. The biotinylated lambda–DNA preparations were applied and incubated for at least 30 min before purging the flowcell with 1 ml buffer C1 which removed DNA that was not bound to the surface. Buffer C2 was used to trigger the enzymatic activity of BfAddAB. All single-molecule experiments were performed at 22°C. All rinsing and incubation steps prior to the application of the DNA samples were carried out at flow rates of 1 ml/min. DNA sample loading was at a flow rate of 0.1 ml/min.
Instead of referring to flow rates we introduce the shear rate, which is a flowcell geometry-independent measure defined as
where Q is the volumetric flow rate, h0 and ω0 are the height and width of the rectangular flowcell, respectively (27). Thus, values of 306, 510 and 714 s−1 apply to 0.03, 0.05 and 0.07 ml min−1 flow rates in our flowcell.
Sample preparation for single-molecule TIRF experiments
Fifty microlitres of ultra-pure water was mixed with 20 µl lambda-DNA, 8 µl (10×) T4 DNA ligase buffer (New England Biolabs, USA) and 2 µl (200 µM) of an oligonucleotide with sequence 5′-GGGCGGCGACCT-3′, tagged with biotin-TEG 568 at its 3′-end (Eurogentec, Belgium) which is complementary to the 5′-AGGTCGCCGCCC-3′ cohesive site of lambda. This mixture was heated at 75°C for 30 min in a water bath and allowed to cool overnight. Two microlitres of T4 DNA ligase was added and the solution was incubated for 2 h at 16–20°C after which the enzyme was heat inactivated for 10 min at 65°C. This preparation was purified twice on MicroSpin S-400 HR columns (Amersham Biosciences, USA) to remove ATP and excess oligonucleotide. Buffer B1 was added to obtain five 50-µl biotinylated lambda–DNA stocks kept frozen at –20°C. For final sample preparation, 50 µl of the DNA stock was dissolved in 2 ml buffer C1 and stained with 10 µl or 25 µl 10-µM YOYO-1 iodide (Invitrogen, UK) and incubated at 0°C in the dark for 5 min. The estimated staining ratio was roughly 1/20 or 1/50 dye molecules per DNA base pair. For BfAddAB kinetic experiments, aliquots of the enzyme stock solutions were added to the YOYO-1 stained DNA preparations to produce final enzyme concentrations of 17 nM and left for at least 30 min further incubation at 0°C in the dark.
Imaging data analysis
Raw data were exported as .tif (video stream) files from the ANDOR software. Kymographs were produced in Image J by line selection of individual stretched lambda–DNA molecules and excision from each video frame and recombination into one image. From all analysed kymographs, the apparent start length of the DNA molecule was determined (still in pxl units) and plotted as a histogram for one condition. As this value varied with the amount of intercalation dye and shear rate, a Gaussian fit determined the optimum DNA molecule length which is known and set to be 48 502 bp. Based on this normalization, the enzyme kinetic parameters were calculated from these kymograph images by analysing the extent to which the DNA molecules gradually shortened as a function of time. Both these parameters were first determined in pixel values from the two different image dimensions, but then converted back into a time difference (seconds) and a length (micrometres) which was further converted into a number of DNA basepairs. Two MatLab (MathWorks) routines were written to present kymographs with a length and time scale and to extract DNA lengths versus time. The latter routine calculated the mean pixel value in each kymograph image and set all initial pixel values smaller than this mean value to zero. The first and last two subsequent non-zero values in one time line were taken as molecule start and end, respectively. Adjacent averaging of five DNA molecule lengths smoothed the data. Origin (Microcal) and Grafit (Erithacus Software Ltd) were used for fitting and presenting the kinetic data.
Bulk assays of BfAddAB activity
To monitor the DNA helicase and ATPase activity of the BfAddAB enzyme in bulk, two different photometric assays were carried out using a similar buffer to that used for the single-molecule experiments, i.e. 40 mM NaHCO3 (pH 8.0), at least 2 mM magnesium acetate (for ATP concentrations ≤1 mM; in case of 2 and 5 mM ATP an excess of 1 mM magnesium acetate over ATP was used), 0.04 mg/ml catalase, 0.5 mg/ml glucose oxidase and 4 mg/ml 2-mercaptoethanol.
The pUC18 plasmid (NEB) was digested for 1 h at 37°C with EcoRI endonuclease (NEB) to produce linearized DNA. An aliquot of this stock was diluted in buffer to a final concentration of 1.2 or 2.5 nM DNA ends. This solution was chilled to 0°C for 5 min and then BfAddAB was added to a final concentration of 40 nM.
The helicase activity of the BfAddAB enzyme was monitored through the displacement of the DNA intercalating dye YOYO-1 iodide from the unwound dsDNA. In this assay, 33 nM YOYO-1 iodide was added to the pUC18 DNA 5 min before the enzyme. This gave an approximate staining ratio of 1 dye molecule per 20 DNA base pairs. Fluorescence spectra and kinetic time courses were acquired on a FL900 Edinburgh Instruments Spectrometer (Edinburgh Analytical Instruments Ltd). A constant reaction temperature of 22°C was maintained. Excitation and emission spectral bandwidths of 5 nm were used. Fluorescence emission and excitation spectra were taken before the kinetic experiments in order to verify the maximum absorption and emission wavelengths of YOYO-1 of 491 and 509 nm, respectively, which were used for the collection of all kinetic time traces. A 100-μl fluorescence microcuvette with 3-mm pathlengths was used; and 63 μl of the YOYO-1, DNA, magnesium and BfAddAB mixture was incubated for 1 min in the cuvette prior to the start of each experiment. After 60 s of fluorescence signal acquisition, BfAddAB activity was triggered by adding 7 μl of a 10 × concentrated Mg2+-ATP solution, which resulted in a drop of fluorescence intensity. All experiments were carried out in triplicate. The linear region of the fluorescence intensity decay was determined and the mean and standard deviation of the slope values calculated and plotted (in arbitrary units) against the Mg2+-ATP concentration. The fluorescence intensity change reached an endpoint and this was assumed to indicate complete unwinding of the DNA allowing a conversion of fluorescence intensity change into amount of YOYO displaced and base pairs of DNA unwound/digested. Control experiments without adding Mg2+-ATP confirmed a Mg2+-ATP -dependent BfAddAB activity. There was no significant drop in fluorescence intensity over the time course of this control experiment.
The ATPase activity of BfAddAB was monitored via the ATP-coupled consumption of NADH and its subsequent decrease in photometric absorption at 340 nm using an enzyme system consisting of pyruvate kinase and lactate dehydrogenase. The buffer was supplemented with 250 μM NADH, 1 mM phosphoenol pyruvate and the enzymes pyruvate kinase and lactic dehydrogenase (7 and 10 enzyme units/ml, respectively, Sigma–Aldrich). Kinetic absorption time traces were acquired on a Cary 50 UV-Visible Spectrometer (Varian Inc.). A constant reaction temperature of 22°C was used. The change in absorption was converted to a change in NADH and ATP concentration after subtraction of a background hydrolysis rate found in the absence of DNA. Omission of DNA or the presence of a 45 base single-stranded oligonucleotide rather than pUC18 DNA gave the same background hydrolysis rate which was subtracted from the rates observed with double stranded DNA.
RESULTS
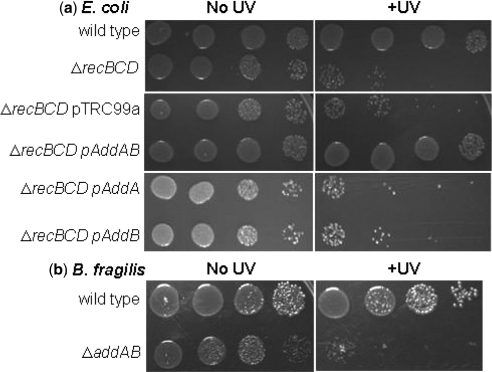
Since the overall homologies between the B. fragilis, L. lactis and B. subtilis AddAB proteins are low, we used two approaches to examine the capabilities of these proteins to initiate repair of DNA damage by HR. First, we determined if genes BF2191 and BF2192 could rescue the recombination deficiency of an E. coli strain containing a deletion of recBCD. An E. coli ΔrecBCD strain is significantly more sensitive to ultraviolet radiation than the parent (Figure 1a). Expression of a plasmid-borne copy of B. fragilis addAB in the E. coli ΔrecBCD strain, however, was capable of rescuing the UV-sensitive phenotype of these cells. Expression of either addA or addB alone did not rescue the UV-sensitivity of the ΔrecBCD strain (Figure 1a). Secondly, we used our recently described allelic replacement technique to delete the addAB genes in B. fragilis and replace them with an erythromycin resistance cassette (25). The resultant B. fragilis ΔaddAB strain had greatly reduced viability and was more sensitive to UV irradiation compared to the wild type strain (Figure 1b); a phenotype similar to that displayed by a recBCD deletion in E. coli. Together, these data support the bioinformatic assignment of the two proteins as AddA and AddB homologues and demonstrate their functionality for initiating repair of DSBs following DNA damage.
Figure 1.
Genetic characterization of B. fragilis addAB. (a) Serial dilutions of E. coli cultures were spotted onto LB plates and irradiated with ultraviolet light (+UV). The viability of the ΔrecBCD strain is greatly reduced upon exposure, compared to the wild type, but can be rescued by expression of addAB. Expression of either addA or addB, or the presence of the plasmid vector pTRC99a, did not affect sensitivity to ultraviolet light. (b) Serial dilutions of B. fragilis cultures spotted onto BHI-S plates. Deletion of addAB from B. fragilis increased sensitivity of the bacterium to UV compared to the wild-type strain.
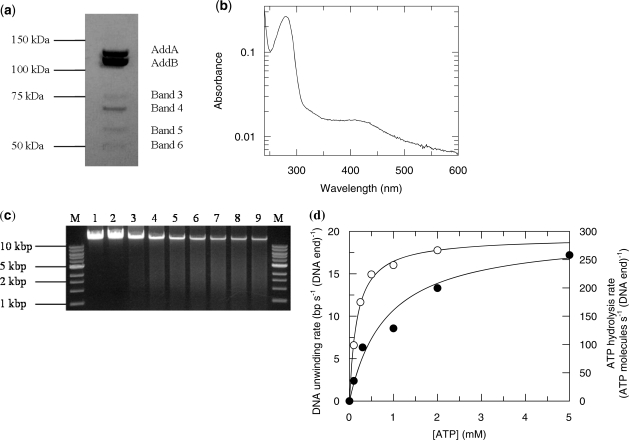
Over-expressed B. fragilis BfAddAB was purified to near homogeneity with only a few minor contaminants (Figure 2a). Mass spectrometry of peptides, produced by in-gel trypsinolysis, clearly identified the primary bands as AddA and AddB. The identified peptides covered nearly 100% of the sequence of the two proteins. The minor bands, labelled 3, 4, 5 and 6 in Figure 2b were all clearly identified as fragments of BfAddAB. The purified complex showed absorbance around 410 nm (Figure 2b), which is consistent with the presence of conserved cysteine residues in the C-terminus of AddB forming the essential Fe-S cluster previously identified in BsAddAB from B. subtilis (16). The exonuclease activity of the proteins was tested by the ability to degrade lambda DNA (Figure 2c). Incubation of BfAddAB with naked DNA resulted in nearly complete digestion of the substrate in 15 min, but only if Mg2+ and ATP were present (Figure 2c, lanes 3–9). Omission of either Mg2+ or ATP prevented degradation of the substrate (Figure 2c, lanes 1 and 2). BfAddAB did not degrade supercoiled circular plasmid DNA and its exonuclease activity was inhibited by the presence of Ca2+ (data not shown).
Figure 2.
Properties of B. fragilis BfAddAB. (a) Purified BfAddAB protein showing the presence of minor contaminants. Bands 3, 4, 5 and 6 were identified via MALDI-TOF MS as proteolytic fragments. The migration of molecular weight markers is indicated. (b) UV–vis absorption spectrum of BfAddAB showing, in addition to the usual aromatic amino acid absorbance around 280 nm, an absorbance peak around 410 nm attributed to the Fe-S cluster in AddB. (c) DNA agarose gel electrophoresis of BfAddAB-mediated DNA unwinding. The lanes contain, respectively, DNA ladder serving as a DNA size marker (M), lambda DNA incubated with 2 mM Mg2+ only (1), lambda DNA incubated with 1 mM ATP only (2) and lambda DNA reacted with 1 mM ATP and 2 mM Mg2+ for 1, 2, 3, 5, 7, 10 and 15 min (3–9). The BfAddAB unwinding reaction was stopped by adding EDTA to 10 mM. The reaction was carried out at 22°C using 170 nM BfAddAB. (d) Michaelis–Menten plots of bulk measurements of YOYO-1 displacement activity converted to base pairs translocated per AddAB-DNA end complex per second (open circle, left-hand scale, pUC18 dsDNA end and enzyme concentrations were 1.2 nM and 40 nM, respectively) and ATPase activity in ATP molecules hydrolysed per AddAB-DNA end complex per second (closed circle, right-hand scale, the pUC18 dsDNA end and enzyme concentrations were 2.5 nM and 40 nM, respectively).
The exonuclease activity of BfAddAB was further characterized by monitoring the displacement of YOYO-1 iodide molecules from linearized pUC18 plasmid DNA. In this assay and the following ATPase assay, the BfAddAB was in >10-fold excess over the concentration of DNA ends. Thus, we assumed that every DNA end had an enzyme attached and hence we could calculate a catalytic rate constant, kcat, in terms of the concentration of complexes of BfAddAB bound to a DNA end. The measured effect relies on the drastically reduced fluorescence quantum yield of the dye from 80% to <1% when it is displaced from double-stranded DNA. The drop in fluorescence intensity was assumed to be proportional to the amount of YOYO-1 displaced and therefore to the number of base pairs digested. Analysis of the data (Figure 2d) gave a Michaelis–Menten constant, KM, of 0.17 ± 0.02 mM and a catalytic rate constant, kcat, of 26.2 ± 3.1 bp (DNA end)−1 s−1.
The ATPase activity of BfAddAB was also assessed with a coupled enzyme assay. Every ATP molecule consumed is linked to the reduction of one NADH molecule concomitant with a reduction in absorption at 340 nm. The plot of these data versus [ATP] (Figure 2d) was fitted to the Michaelis–Menten equation giving a KM of 0.84 ± 0.17 mM and a kcat of 295 ± 19 ATP molecules (DNA end)−1 s−1.
The single-molecule TIRF instrument was tested using stretched lambda DNA and a sample of purified RecBCD enzyme (data not shown; RecBCD was a kind gift from Mr Jakob Zipprich, Edinburgh). Shortening of the DNA in the presence of magnesium and 1 mM ATP was observed. A wide range of rates was observed and the average unwinding rate was 130 ± 102 bp s−1 at 22°C. This rate is slower than the 500 bp s−1 observed by others near this temperature (7) but we attribute this to the age of our preparation of RecBCD (>2 years old).
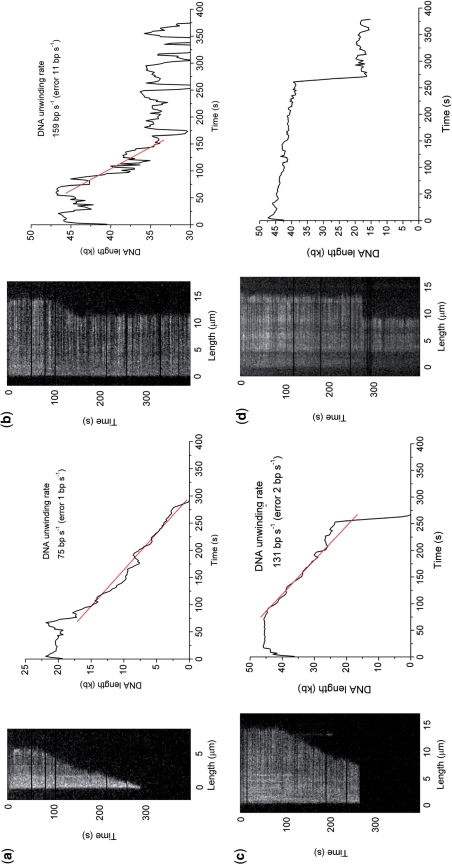
Figure 3 shows typical kymographs for BfAddAB-mediated degradation of single DNA molecules derived from movies (Supplementary Movies). Given the sequence similarity and the similarity in function demonstrated above between BfAddAB and BsAddAB (17) and the similarity between AddAB and RecBCD (15), we assert that the shortening of the DNA observed in Figure 3 is due to a DNA unwinding helicase activity of BfAddAB displacing the YOYO-1 dye. By measuring the length of DNA during each reaction, the plot of length against time allows the derivation of unwinding rate (number of DNA base pairs unwound from the duplex per unit time) and processivity (number of base pairs uninterruptedly unwound by one BfAddAB enzyme). Most kymographs showed a processive unwinding of the DNA (Figure 3a) that could be prematurely terminated by either enzyme dissociation (Figure 3b), or photo-damage (Figure 3c). In Figure 3c, initial shortening of the DNA occurred but at ∼120 s, the DNA end transiently became stuck on the surface and then broke off remaining as a bright spot until ∼200 s when it bleached. After the end broke off from the surface, progressive shortening of the DNA restarted due to the binding of a new molecule of BfAddAB. At ∼280 s the surface tether destabilized and the DNA molecule escaped from the field of view. Figure 3d shows a control kymograph obtained under exactly the same conditions, but without Mg2+-ATP. After ∼280 s, photo-damage truncated the DNA molecule. No gradual shortening of the DNA was observed in the control measurements.
Figure 3.
Kymographs of BfAddAB unwinding of YOYO-1-stained lambda DNA at 2 mM Mg2+-ATP. Adjacent to each kymograph is a plot of DNA molecule length (kb) with time (seconds) smoothed using a five-point moving average. The BfAddAB enzyme rates were determined by linear least squares fitting of the slope. To calculate the processivity of each individual event, the DNA molecule length was determined before and after unwinding by BfAddAB. DNA in these experiments had a staining ratio of 1:20 dye:bp. (a) A truncated DNA molecule whose BfAddAB-mediated unwinding commenced at 80 s and proceeded to completion. (b) A DNA molecule whose unwinding stalled presumably due to dissociation of the BfAddAB. (c) DNA unwinding was stopped prematurely because of tether instabilities which results in loss of the active BfAddAB–DNA complex from the field of view. Note that at ∼120 s the right-hand DNA end became stuck to the surface and broke off. (d) Control image. The DNA sample was incubated with BfAddAB but without Mg2+-ATP. No unwinding was ever observed, only instantaneous truncations such as that at ∼280 s due to photodamage.
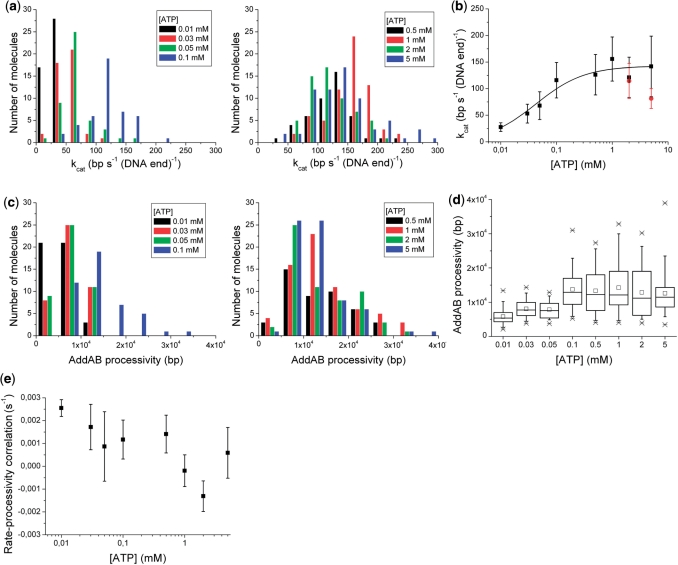
The BfAddAB unwinding rates and processivities were measured for single molecules at eight different [ATP] ranging from 0.01 to 5 mM, at a constant shear rate of 510 s−1 at 22°C. The distribution of enzyme unwinding rates varied with [ATP], ranged from around 25 to 250 bp (DNA end)−1 s−1 and was approximately symmetrical with a shape like a Gaussian distribution (Figure 4a). The mean and standard deviation of the unwinding rate were calculated for all single-molecule events at each [ATP]. A Michaelis–Menten analysis for the mean unwinding rate (Figure 4b), gave a Michaelis constant of 0.043 ± 0.010 mM for ATP and a maximum DNA unwinding rate of 143 ± 7 bp (DNA end)−1 s−1. The processivity data showed a skewed distribution rather than a symmetrical distribution at all [ATP] (Figure 4c). Processivities of up to 40 000 bp were reached by individual enzymes. The mean value, however, was around 14 000 bp for high [ATP] with large variation between molecules (Figure 4d).
Figure 4.
[ATP]-dependent BfAddAB kinetic measurements. (a) A histogram of unwinding rates for each [ATP]. Unwinding rates for each single molecule were grouped in to bins of 25 bp (DNA end)−1 s−1. (b) Michaelis–Menten plot of mean BfAddAB rates dependent on [ATP]. The fit yielded a KM and a kcat of 0.043 ± 0.010 mM and 143 ± 7 bp (DNA end)−1 s−1, respectively. The black data were obtained with [Mg2+] of 2 mM for [ATP] ≤1 mM, and 3 and 6 mM Mg2+ for 2 and 5 mM ATP, respectively. The red data were obtained with 2 mM Mg2+ (these data are not shown in Figure 4a). The error bars represent one standard deviation. (c) A histogram showing the processivity of BfAddAB molecules for eight different [ATP]. The processivity data were grouped in bins of 5000 bp. (d) Box plot of ATP dependence of BfAddAB processivity (number of molecules analysed for 0.01, 0.03, 0.05, 0.1, 0.5, 1, 2 and 5 mM ATP: 45, 44, 45, 45, 46, 68, 59 and 69, respectively). Cross mark indicates smallest and largest values, open square is mean, error bars are 95% confidence limits, the large box encompasses the middle 50% of data values and the line across this box is the median value of the data. (e) Correlation of BfAddAB rate with BfAddAB processivity as a function of [ATP]. At each [ATP], a straight line was fitted to a plot of the rate versus processivity for all of the single molecules (Supplementary Figure S1). The slopes of these lines are plotted here versus [ATP]. These data suggest there is a weak positive linear correlation between BfAddAB rate and BfAddAB processivity at low [ATP] concentrations and no correlation at higher concentrations.
Supplementary Figure S1 shows the correlation of unwinding rate versus processivity for each BfAddAB molecule at each [ATP]. Although there is considerable scatter, there is a tendency at low [ATP] for a slow BfAddAB molecule to also exhibit a low processivity. Figure 4e shows this correlation of the unwinding rate and processivity as a function of [ATP]. A positive correlation between unwinding rate and processivity was observed at the lower ATP concentrations of 0.01, 0.03, 0.1 and 0.5 mM. For example, at 0.01 mM ATP, a BfAddAB complex with an unwinding rate of 50 bp (DNA end)−1 s−1 would unwind approximately three times the amount of DNA compared to a complex with an unwinding rate of 25 bp (DNA end)−1 s−1. At higher [ATP] there was no apparent correlation between unwinding rate and processivity.
Once shortening of the DNA had started, we observed no pausing during the translocation; in other words, the unwinding process once halted did not restart during the observation period. The distribution of these halting sites on the lambda DNA was random (data not shown). Pauses of short duration would not be visible in our experiment as the camera takes one 0.2-s exposure every 2 s and we would estimate that pauses of <10 s would not be noticeable. It has been noted that RecBCD pauses for of order 10 s at its chi site (8) and that BsAddAB pauses for minutes at its chi site (17). While we currently do not know the sequence of the analagous chi site recognized by B. fragilis BfAddAB, the absence of long pauses at defined positions on the DNA during our single molecule observations could indicate that if BfAddAB has the same pausing time as the BsAddAB then either a chi site is absent from the DNA substrate or is inefficiently recognized by BfAddAB.
Shear rates of 306, 510 and 714 s−1 at 1 mM ATP gave processivities of 13 600, 14 200 15 000 bp, respectively (Supplementary Figure S2), which when extrapolated to zero shear gives a processivity of 12 600 bp on average. However, given the standard deviations of ±50% in processivity this shear rate dependence was not significant. Concomitantly, unwinding rates fell from 164 ± 59 to 156 ± 42 to 143 ± 33 bp (DNA end)−1 s−1 as the shear rate increased (Supplementary Figure S2), which when extrapolated to zero shear gives an unwinding rate of 182 bp (DNA ends)−1 s−1, although again the dependence on shear rate is small compared to experimental error. During unwinding, BfAddAB needs to displace the YOYO-1 from the DNA. The unwinding rates increased by 14% and 41% when the staining ratio decreased from 1:20 to 1:50 dye:bp for shear rates of 306 s−1 and 714 s−1, respectively, but these changes were within the standard deviation of the data (Supplementary Figure S3). Processivity showed no dependence on the amount of dye at 714 s−1 and an increase of 38% at 306 s−1 when the staining ratio decreased from 1:20 to 1:50 dye:bp but again the change was within the standard deviation of the data (Supplementary Figure S3). Thus shear rate and amount of YOYO-1 may have a small influence on BfAddAB activity.
DISCUSSION
Despite the fact that E. coli has provided the paradigm for DSB repair in bacteria, the RecBCD complex is predominantly present in the γ-proteobacteria and some members of the δ- and β-proteobacteria, but absent from most other bacterial phyla (15). In the many bacteria that do not encode RecBCD, pre-synaptic processing of DSBs is performed by the less-well studied AddAB system. Our results provide the first report on the enzymatic properties, translocation rates and processivity of the AddAB helicase from the Gram-negative bacterium B. fragilis.
The single-molecule experiments were backed up with data obtained from classical bulk experiments. Clearly, there are discrepancies between our single-molecule and bulk experiments and we attribute this to the fact that our single-molecule experiments select only active BfAddAB bound to DNA ends ignoring all others which are not bound. This unbound fraction will include those defective in some way in binding or enzymatic activity. These defective AddAB may still contribute to bulk assays. For instance, the maximum unwinding rate was determined to be 143 ± 7 bp (DNA end)−1 s−1 and 26.2 ± 3.1 bp (DNA end)−1 s−1 for the single-molecule and bulk YOYO-1 iodide displacement assay, respectively. Similarly, the KM values obtained from the YOYO-1 iodide displacement assay and the ATPase assay were larger than from the single-molecule assay.
Whilst bearing this caveat in mind, we compare the maximum unwinding rate with the ATP hydrolysis rate. The obtained kcat value from the coupled ATPase assay is 295 ± 19 ATP molecules (DNA end)−1 s−1. This value is approximately twice the maximum rate of unwinding and would suggest that two ATP molecules are hydrolysed per DNA base pair unwound. RecBCD which uses two helicase motors was shown to consume 2–3 ATP molecules per DNA base pair unwound (28,29). Interestingly, Yeeles et al. (19) found that the BsAddAB enzyme has a DNA unwinding and a dsDNA-dependent ATPase activity of 550 bp s−1 (DNA end)−1 and 170 ATP s−1 (DNA end)−1, respectively, which implies that <1 ATP molecule is needed to unwind three DNA base pairs for this enzyme. Therefore the coupling between ATP hydrolysis and DNA unwinding observed with BfAddAB is comparable to that observed with the other recombination enzymes.
The single-molecule data demonstrate that the B. fragilis BfAddAB complex is highly processive and can unwind an average of 14 000 bp per event, with some enzymes unwinding up to 40 000 bp, and can translocate along DNA at rates up to 250 bp s−1 from a single DNA end at 22°C. The rate of movement of BfAddAB along DNA is comparable with the reported value from single molecule experiments for another SF1 helicase, UvrD, that has a maximum translocation rate of 275 bp s−1 at 25°C (30). The major difference between BfAddAB and UvrD is the processivity, since UvrD can only unwind an average of 255 bp per event. The high processivity of BfAddAB is surprising since it only contains a single motor protein, AddA. In contrast, the RecBCD complex contains two helicase motors that contribute to the high processivity of the enzyme at a rate of ∼500–1500 bp s−1, with an average of 30 000 bp being unwound at 37°C (7,8,31). When the RecD motor protein is disabled, however, the RecBCDK29Q complex moves along DNA at a reduced rate of 800 bp s−1 (37°C) and can only unwind an average of 4900 bp (31). In this case, having a single 3′–5′ helicase motor is not sufficient to provide the same extent of DNA unwinding as observed with AddAB. Since both RecBCDK29Q and wild type B. fragilis BfAddAB translocate at lower rates than wild type RecBCD but yet are fully capable of DSB repair, in their respective genetic backgrounds, this suggests that translocation speed may not be a critical factor for DSB repair. The predominant feature of these two classes of helicases, however, is the ability to translocate along significant lengths of DNA in order to reach and interact with the appropriate recognition site.
Why is the single motor BfAddAB complex so processive? We speculate that one possible function of AddB, in addition to providing a nuclease, might be to stimulate the helicase function of AddA. Many SF1 proteins are inefficient helicases as monomers and will only unwind DNA efficiently when they are present as multimers, for example, UvrD, Rep and PcrA (32–34). In the case of Rep, the 2B domain within the protein is responsible for autoinhibition of helicase activity, but not translocation on single-stranded DNA (33,35). The analogous 2B domain in AddA, between motor motifs IV and V, contains an additional 135 aa compared to Rep. This extended region might form part of the dimer interface with AddB, enhancing the stability of the complex and stimulating the helicase activity of AddA. This would be similar to the structural role of domain 2B in RecB, which forms an extensive interface with the RecC component (36). The rate of unwinding might also be important for the processivity of the BfAddAB complex, since at low Mg2+-ATP concentrations we observed a correlation between the two components, such that a faster translocating complex was more likely to unwind more DNA. Chi sites are often a few thousands of bp apart hence to stand a chance of finding one, an enzyme like RecBCD or AddAB must move on average at least this distance to allow generation of the substrate required for RecA-mediated strand invasion (31,37). Although it has been suggested that a highly processive enzyme would be able to push other proteins, such as transcriptional regulators and nucleoid-associated proteins, out of its way in the search for a chi site (22,31,37), it is also true that a processive enzyme must possess a very low dissociation rate constant (of order 1 min−1) from DNA compared to a typical nanomolar binding affinity DNA-binding protein with a dissociation rate constant of order <1 s−1 (assuming a diffusion limited association rate) (38). In such a situation, the processive enzyme merely needs to wait for the blocking proteins to dissociate rather than having to actively force them out of its way. By remaining bound to DNA for long periods of time, RecBCD and AddAB ensure that they can find their chi sites even in the presence of many other proteins bound to the damaged DNA molecule.
It is not clear why some bacteria have evolved to use a twin motor helicase for resecting DSBs, while many others use the single motor AddAB system. It has been suggested that the Mycobacterium AdnAB represents an ancestral system with two motors and two nucleases, with AddAB resulting from the loss of one motor but maintaining two nucleases and RecBCD then representing a more recent acquisition of a second motor in the form of RecD (39). This argument is supported by the many different variants of RecD that are encoded in diverse bacterial genomes, which are not associated with RecBCD and do not appear to be involved in DSB repair (40). The widespread distribution of AddAB within bacteria might reflect the shorter chi recognition sequences used by the single motor system, since this could enhance the spread of addAB genes by horizontal gene transfer because the probability of finding a correct chi site in a new genetic background would be increased. Interestingly, it has been suggested that genes encoding AddAB may have been frequently transferred between bacteria and even into the archaea (15).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Biotechnology and Biological Sciences Research Council RASOR grant (BB/C511599/1) and BB/D001870/1 (to D.T.F.D.); a studentship (to FP); Wellcome Trust (GR080463MA to D.T.F.D., GW.B., Malcolm Walkinshaw and James Naismith); University of Edinburgh Development Trust (ID 3072 to M.R.); and the Marie-Curie Network ‘From FLIM to FLIN’ (MRTN-CT-2005-019481 to M.R.). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Andrew Garry (COSMIC) for technical assistance, Drs Jochen Arlt and Cristina Flors (COSMIC and Chemistry) for optics advice, Prof. Eric Greene (Columbia University) for advice on DNA stretching, Dr Dale Wigley and Dr Robert Court (Cancer Research UK) for supplying plasmids and instructions for preparing RecBCD, Mr Jakob Zipprich for preparing RecBCD, and Dr John White, Dr Gareth Roberts, and Mr Laurie Cooper (Chemistry) for assistance with protein preparation.
REFERENCES
- 1.Cox MM, et al. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 2.Pennington JM, Rosenberg SM. Spontaneous DNA breakage in single living Escherichia coli cells. Nat. Genet. 2007;39:797–802. doi: 10.1038/ng2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dillingham MS, Spies M, Kowalczykowski SC. RecBCD enzyme is a bipolar DNA helicase. Nature. 2003;423:893–897. doi: 10.1038/nature01673. [DOI] [PubMed] [Google Scholar]
- 4.Taylor AF, Smith GR. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–893. doi: 10.1038/nature01674. [DOI] [PubMed] [Google Scholar]
- 5.Chédin F, Kowalczykowski SC. A novel family of regulated helicases/nucleases from Gram-positive bacteria: insights into the initiation of DNA recombination. Mol. Microbiol. 2002;43:823–834. doi: 10.1046/j.1365-2958.2002.02785.x. [DOI] [PubMed] [Google Scholar]
- 6.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bianco PR, et al. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 8.Handa N, Bianco PR, Baskin RJ, Kowalczykowski SC. Direct visualization of RecBCD movement reveals cotranslocation of the RecD motor after chi recognition. Mol. Cell. 2005;17:745–750. doi: 10.1016/j.molcel.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 9.Spies M, Amitani I, Baskin RJ, Kowalczykowski SC. RecBCD enzyme switches lead motor subunits in response to chi recognition. Cell. 2007;131:694–705. doi: 10.1016/j.cell.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spies M, Kowalczykowski SC. The RecA binding locus of RecBCD is a general domain for recruitment of DNA strand exchange proteins. Mol. Cell. 2006;21:573–580. doi: 10.1016/j.molcel.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Zuñiga-Castillo J, Romero D, Martínez-Salazar JM. The recombination genes addAB are not restricted to gram-positive bacteria: genetic analysis of the recombination initiation enzymes RecF and AddAB in Rhizobium etli. J. Bacteriol. 2004;186:7905–7913. doi: 10.1128/JB.186.23.7905-7913.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rocha EP, Cornet E, Michel B. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS. Genet. 2005;1:e15. doi: 10.1371/journal.pgen.0010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mertens K, Lantsheer L, Ennis DG, Samuel JE. Constitutive SOS expression and damage-inducible AddAB-mediated recombinational repair systems for Coxiella burnetii as potential adaptations for survival within macrophages. Mol. Microbiol. 2008;69:1411–1426. doi: 10.1111/j.1365-2958.2008.06373.x. [DOI] [PubMed] [Google Scholar]
- 14.Amundsen SK, et al. Helicobacter pylori AddAB helicase-nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Mol. Microbiol. 2008;69:994–1007. doi: 10.1111/j.1365-2958.2008.06336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cromie GA. Phylogenetic ubiquity and shuffling of the bacterial RecBCD and AddAB recombination complexes. J. Bacteriol. 2009;191:5076–5084. doi: 10.1128/JB.00254-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinha KM, Unciuleac MC, Glickman MS, Shuman S. AdnAB: a new DSB-resecting motor-nuclease from mycobacteria. Genes Dev. 2009;23:1423–1437. doi: 10.1101/gad.1805709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chédin F, Handa N, Dillingham MS, Kowalczykowski SC. The AddAB helicase/nuclease forms a stable complex with its cognate chi sequence during translocation. J. Biol. Chem. 2006;281:18610–1861. doi: 10.1074/jbc.M600882200. [DOI] [PubMed] [Google Scholar]
- 18.Yeeles JT, Dillingham MS. A dual-nuclease mechanism for DNA break processing by AddAB-type helicase-nucleases. J. Mol. Biol. 2007;371:66–78. doi: 10.1016/j.jmb.2007.05.053. [DOI] [PubMed] [Google Scholar]
- 19.Yeeles JT, Cammack R, Dillingham MS. An iron-sulfur cluster is essential for the binding of broken DNA by AddAB-type helicase-nucleases. J. Biol. Chem. 2009;284:7746–7755. doi: 10.1074/jbc.M808526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patrick S. In: Molecular Medical Microbiology. Sussman M, editor. London: Academic Press; 2002. pp. 1921–1948. [Google Scholar]
- 21.Cerdeño-Tárraga AM, et al. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 22.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 23.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 24.Visnapuu ML, Duzdevich D, Greene EC. The importance of surfaces in single-molecule bioscience. Mol. Biosyst. 2008;5:394–403. doi: 10.1039/b800444g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patrick S, Houston S, Thacker Z, Blakely GW. Mutational analysis of genes implicated in LPS and capsular polysaccharide biosynthesis in the opportunistic pathogen Bacteroides fragilis. Microbiology. 2009;155:1039–1049. doi: 10.1099/mic.0.025361-0. [DOI] [PubMed] [Google Scholar]
- 26.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 27.Busscher HJ, van der Mei HC. Microbial adhesion in flow displacement systems. Clin. Microbiol. Rev. 2006;19:127–141. doi: 10.1128/CMR.19.1.127-141.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roman LJ, Kowalczykowski SC. Characterization of the adenosinetriphosphatase activity of the Escherichia coli RecBCD enzyme: relationship of ATP hydrolysis to the unwinding of duplex DNA. Biochemistry. 1989;28:2873–2881. doi: 10.1021/bi00433a019. [DOI] [PubMed] [Google Scholar]
- 29.Korangy F, Julin DA. Efficiency of ATP hydrolysis and DNA unwinding by the RecBC enzyme from Escherichia coli. Biochemistry. 1994;33:9552–9560. doi: 10.1021/bi00198a022. [DOI] [PubMed] [Google Scholar]
- 30.Dessinges MN, Lionnet T, Xi XG, Bensimon D, Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc. Natl Acad. Sci. USA. 2004;101:6439–6444. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dillingham MS, Webb MR, Kowalczykowski SC. Bipolar DNA translocation contributes to highly processive DNA unwinding by RecBCD enzyme. J. Biol. Chem. 2005;280:37069–37077. doi: 10.1074/jbc.M505520200. [DOI] [PubMed] [Google Scholar]
- 32.Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J. Mol. Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Brendza KM, et al. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc. Natl Acad. Sci. USA. 2005;102:10076–10081. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niedziela-Majka A, Chesnik MA, Tomko EJ, Lohman TM. Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro. J. Biol. Chem. 2007;282:27076–27085. doi: 10.1074/jbc.M704399200. [DOI] [PubMed] [Google Scholar]
- 35.Cheng W, et al. The 2B domain of the Escherichia coli Rep protein is not required for DNA helicase activity. Proc. Natl Acad. Sci. USA. 2002;99:16006–6011. doi: 10.1073/pnas.242479399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 37.Roman LJ, Eggleston AK, Kowalczykowski SC. Processivity of the DNA helicase activity of Escherichia coli recBCD enzyme. J. Biol. Chem. 1992;267:4207–4214. [PubMed] [Google Scholar]
- 38.Halford SE. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem. Soc. Trans. 2009;37:343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 39.Unciuleac MC, Shuman S. Characterization of the mycobacterial AdnAB DNA motor provides insights to the evolution of bacterial motor-nuclease machines. J. Biol. Chem. 2010;285:2632–2641. doi: 10.1074/jbc.M109.076133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montague M, Barnes C, Smith HO, Chuang RY, Vashee S. The evolution of RecD outside of the RecBCD complex. J. Mol. Evol. 2009;69:360–371. doi: 10.1007/s00239-009-9290-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.