Abstract
The transcriptional response of Streptococcus pneumoniae was examined after exposure to the GyrB-inhibitor novobiocin. Topoisomer distributions of an internal plasmid confirmed DNA relaxation and recovery of the native level of supercoiling at low novobiocin concentrations. This was due to the up-regulation of DNA gyrase and the down-regulation of topoisomerases I and IV. In addition, >13% of the genome exhibited relaxation-dependent transcription. The majority of the responsive genes (>68%) fell into 15 physical clusters (14.6–85.6 kb) that underwent coordinated regulation, independently of operon organization. These genomic clusters correlated with AT content and codon composition, showing the chromosome to be organized into topology-reacting gene clusters that respond to DNA supercoiling. In particular, down-regulated clusters were flanked by 11–40 kb AT-rich zones that might have a putative structural function. This is the first case where genes responding to changes in the level of supercoiling in a coordinated manner were found organized as functional clusters. Such an organization revealed DNA supercoiling as a general feature that controls gene expression superimposed on other kinds of more specific regulatory mechanisms.
INTRODUCTION
The Gram-positive bacterium Streptococcus pneumoniae, the pneumococcus, is an opportunistic pathogen of the commensal flora of human nasopharinx. Under specific circumstances, it can migrate to other niches (ear, lung, bloodstream, cerebrospinal fluid) causing diverse pathologies. In spite of the development of vaccines and chemotherapy, the pneumococcus continues to be a serious cause of illness and death and the main ethyological agent of community-acquired pneumonia. Pneumococcal resistance to some anti-microbial drugs has spread worldwide in the last two decades (1). The new fluoroquinolones, which act on type II DNA topoisomerases, have been explicitly recommended for the treatment of community-acquired pneumonia in adults (2). Type II DNA topoisomerases pass one DNA double helix through another by introducing a transient double-stranded break with the energy provided by ATP hydrolysis. They are tetrameric proteins that consist of two different subunits: GyrA2GyrB2 for DNA gyrase (gyrase) and ParC2ParE2 for topoisomerase IV (topo IV), ParC and ParE being homologous to GyrA and GyrB, respectively. Gyrase introduces negative supercoils in the DNA and is involved in replication, recombination and transcription, while topo IV acts mainly in the segregation of sister chromosomes after DNA replication. Fluoroquinolones inhibit gyrase and topo IV affecting mainly chromosome replication. However, they also stabilize a reaction intermediate in which the enzymes are covalently linked to the DNA originating double-stranded breaks that lead to cell death (3). Genetic and biochemical studies have shown that in Gram-positive bacteria, including S. pneumoniae (4,5), topo IV is the primary target for most fluoroquinolones and gyrase is a secondary target. The opposite is true for Gram-negative bacteria. On the other hand, novobiocin (Nov) is a chemically unrelated drug that primarily targets GyrB and is not used for treatment due to its toxicity.
In the Gram-negative bacteria, Escherichia coli DNA supercoiling is maintained homeostatically by the opposing activities of topo IV and DNA topoisomerase I (topo I), which relax DNA, and by gyrase, which introduces negative supercoils. In these bacteria, transcription of the topA gen (topo I) increases when negative supercoiling increases (6), and that of gyrA and gyrB increases after DNA relaxation (7–9). Likewise, gyrase expression activation in response to relaxation has also been observed in Streptomyces and Mycobacterium (10,11). In many bacteria, the DNA supercoiling level is affected by diverse environmental conditions, replication, growth stage and the infection process (12–14). Changes in DNA supercoiling have an effect on global genome transcription, as shown in E. coli (15,16) and Haemophilus influenzae (17). These changes, associated with transcriptional alterations, would be crucial when S. pneumoniae infects diverse niches in the human host. In addition, expression of gyrase and topo I genes in E. coli is subjected to global regulation by nucleoid-associated proteins such as FIS, CRP, IHF and HU (14). These proteins seem to control supercoiling architecture (18). These regulatory mechanisms, however, do not necessarily apply to all bacteria. S. pneumoniae, for instance, lacks proteins homologous to CRP and IHF.
The present study was designed to learn about the transcriptional response to DNA relaxation in the Gram-positive pathogen S. pneumoniae. Nov, an inhibitor of S. pneumoniae GyrB (19), was used to relax DNA. Changes in DNA topology were monitored by analysing the topoisomer distribution of a replicating plasmid. Global transcription response was analysed using microarray technology after exposing cells to two Nov concentrations. The results were validated and implemented by studying transcription of all DNA topoisomerase genes at five different Nov concentrations and at three intervals after adding the drug.
MATERIALS AND METHODS
Bacterial strains, growth and transformation of bacteria
Streptococcus pneumoniae was grown in a casein hydrolysate-based medium with 0.3% sucrose (AGCH) as energy source and transformed with chromosomal or pLS1 plasmid as described previously (20). Minimal inhibitory concentrations (MICs) were determined in the same medium. Strains used were R6 (Nov MIC = 1 µg/ml) and R6GyrB (Nov MIC = 16 µg/ml). R6GyrB was obtained by transformation with a PCR product from strain 755 obtained with oligonucleotides GyrBUP 5′-GCGCAAGCTTAGACATCAAGTGTACTATAGACAGC-3′ and GyrBRTR 5′-GCGGCAGCACTATCTCCATC-3′ and selection in Nov at 5 µg/ml.
Microscopy
To visualize cells, 4 ml of each culture were collected, centrifuged (4420g, 15 min, 4°C), washed in 10 mM phosphate buffer (pH 7.2) and fixed in 1 ml of paraformaldehyde 2% and glutaraldehyde 0.05% in PBS 1× for 4 h at 4°C. Fixed cells were washed with PBS and stored at 4°C until use. Cells were mounted onto 0.8% agarose pads and observed using an Olympus BX61 microscope equipped with a 100× phase-contrast objective and filter sets for fluorescence. To observe nucleoids, fixed cells were treated with DAPI (4-6-diamidino-2-phenylindole, a stain for nucleic acids that fluoresces blue under UV excitation when bound to double-stranded DNA) solution (1 vol of DAPI 2% in 1 vol of vectashield-mounting medium for fluorescence) before mounting them onto the pads.
Analysis of the topology of covalently closed circles
Plasmid DNA isolation from S. pneumoniae cultures grown on AGCH medium containing 1µg/ml of tetracycline (for selection of pLS1) and various Nov concentrations was performed as described before (21). Intact circular DNA molecules were analysed in neutral/neutral two-dimensional agarose gels. The first dimension was run at 1.5 V/cm in a 0.4% agarose (Seakem; FMC Bioproducts) gel in 1× Tris–borate–EDTA (TBE) buffer for 17−19 h at room temperature. The second dimension was run at 7.5 V/cm in 1% agarose gel in 1× TBE buffer for 7−9 h at 4°C. Chloroquine (Sigma) was added to the TBE buffer in both, the agarose and the running buffer. After electrophoresis gels were subjected to Southern hybridization. A 240-bp PCR fragment obtained from pLS1 DNA with 5′-biotinylated forward primer 5′-GTGCCGAGTGCCAAAATCAA-3′ and reverse 5′-TTTCAAGTACCGATTCACTTAATG-3′ was used as a probe on two-dimensional agarose gels transferred to nylon membrane (Inmobylon NY+, Millipore). Chemiluminiscent detection of DNA was performed with the Phototope®-Star kit (New England Biolabs). Images were captured in a VersaDoc MP400 system and analysed with the Quantity One program (BioRad). DNA linking number (Lk) was calculated by quantifying the amount of every topoisomer. DNA supercoiling density (σ) was calculated from the equation σ = ΔLk/Lk0. Linking number differences (ΔLk) were determined using the equation Lk = Lk − Lk0, in which Lk0 = N/10.5, where N is the size of the molecule in base pair and 10.5 the number of base pairs per one complete turn in B-DNA.
RNA extraction and real time RT–PCR experiments
Synthesis of cDNAs from 5 µg of total RNA was performed as previously described (22). The cDNAs obtained were subjected to quantitative real-time PCR (qPCR, Chromo 4, BioRad) in 20 µl reactions containing 2 µl of cDNA, 0.3 µM of each specific primer, and 10 µl of LightCycler FastStart Universal A SYBR Green Master (Roche). Amplification was achieved with 42 cycles of a tree-segment program: denaturation (30 s at 94°C), annealing (30 s at 45–56°C), and elongation (30 s at 68°C). To normalize the three independent cDNA replicate samples, values were divided by those obtained of the amplification of internal fragments of rpoB (22) and 16S rDNA (5′-GGTGAGTAACGCGTAGGTAA-3′ and 5′-ACGATCCGAAAACCTTCTTC-3′, forward and reverse primers, respectively).
Microarray data normalization and analysis
High density arrays A6701-00-01 from Roche NimbleGen were used. Arrays include two copies of 17 oligonucleotide probes (average size of 50 nt) for each 2037 protein coding genes from S. pneumoniae R6. Double-stranded cDNAs were obtained from total RNA with the SuperScriptTM Double-Stranded cDNA Synthesis Kit (Invitrogen). Labelling of double-stranded cDNAs with Cy3 and microarray hybridization was performed following the NimbleGen arrays user’s guide at the Functional Genomics Core Facility, Institut de Recerca Biomèdica, Barcelona. Microarrays were scanned with an GenePix 4000B scanner at 5 µm resolution and raw data were extracted and RMA normalized (23) using NimbleScan v2.4 (NimbleGen). RMA, a probe-level summarization method, identifies probes that are outliers in the overall behaviour of the expression measured for a given gene and reduces its contribution in the reported gene expression level by means of quantile normalization and background correction. Quantile normalization assumes that the expression distributions for all arrays under consideration are the same and attempts to adjust each array so that they are all equivalent. Then, the median of each group of 17 probes in each block of the array are considered. After this normalization, Partek Genomics Suite 6.4 was used to do a principal component analysis (PCA) and test for significance for differential gene expression using ANOVA. Each microarray experiment was carried out in triplicate with cDNA prepared from three independent cultures treated with Nov at each concentration. All microarray data are available at the Array Express (EBI, UK) database via accession numbers E-MTAB-140 and E-MTAB-141.
Synonymous codon-usage and statistical analysis
Correspondence analyses of general/relative synonymous codon usage and residue usage were performed using the CodonW server at http://mobyle.pasteur.fr/cgi-bin/portal.py?form=codonw. Briefly, this method allows for visualizing genes according to their codon/ residue usage frequencies in a multi-dimensional space by identifying the major trends as those axes which account for the largest fractions of the variation among genes. Functional classification of COGs was carried out according to COG classification (24). Gene COG evidences were downloaded from S. pneumoniae R6 page at NCBI. COG codes associated to two functional classes were not considered for the statistics. Significance of the functional bias was estimated by chi-squared analysis.
RESULTS
DNA topoisomer distribution varied according to the treatment with novobiocin
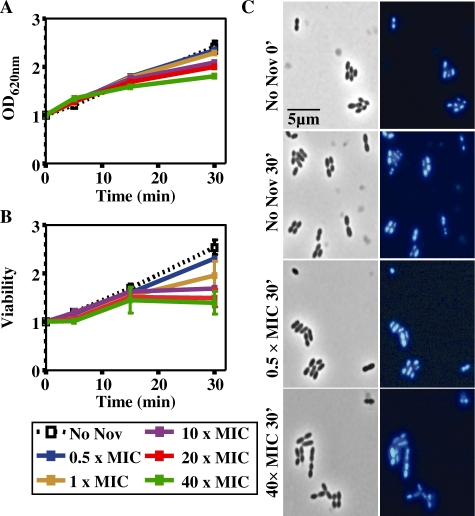
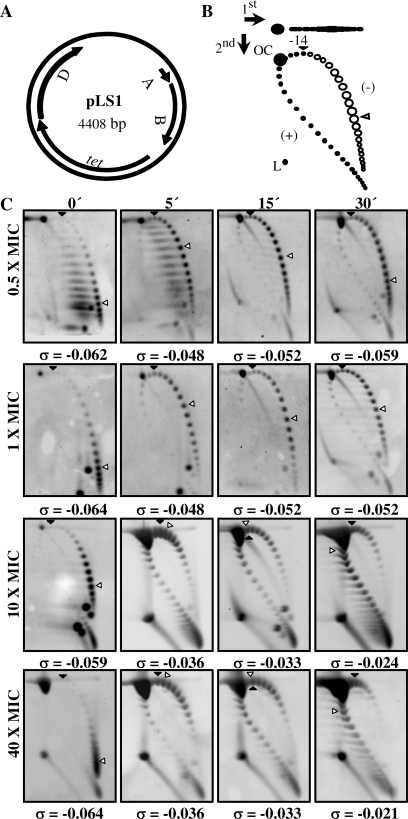
The effect of Nov was tested in the wild-type strain R6 (Nov MIC = 1 µg/ml) carrying plasmid pLS1, at concentrations ranging from 0.5× to 40× MIC. Cell growth (Figure 1A) and division (Figure 1B) were affected in a Nov concentration-dependent manner. However, even after 30 min of treatment at 40× MIC, cells did not show appreciable changes in nucleoid size or number of nucleoids per cell (Figure 1C), suggesting that they were still physiologically active. Alterations in supercoiling were detected by analyzing the topoisomer distributions of pLS1. This plasmid is appropriate to study gyrase activity, given that it replicates by a rolling circle mechanism (25) and all their genes are transcribed in the same direction (Figure 2A), avoiding problems of transcription interference during replication (26). First, pLS1 linking number (ΔLk) was quantified using two-dimensional agarose gel electrophoresis with 2 µg/ml chloroquine in the second dimension (21). This technique, which differs from the classical two-dimensional chloroquine gels, makes it possible to separate DNA molecules by mass and shape allowing the distinction of monomers, dimers and catenanes. No dimers or catenates were observed in pLS1, given their mode of replication. Under the conditions used, the induced ΔLk of monomers was −14. In the autoradiograms, topoisomers appeared distributed in a bubble-shaped arc, with negative supercoiled molecules located to the right and positive supercoiled ones to the left (Figure 2). At 0.5× MIC Nov, supercoiling densities (σ) were −0.062, −0.048, −0.052 and −0.059 at 0, 5, 15 and 30 min, respectively, indicating that after an initial relaxation, plasmids were able to recover the initial σ-value. While partial recovery (from −0.048 to −0.052) was also observed for 1× MIC Nov, this effect was not detected at higher concentrations of the drug. Note that regardless of the broad distribution of topoisomers observed at 0.5× and 1× MIC doses, the vast majority still showed negative supercoiling. The reverse was found using higher concentrations of the drug. Although direct extrapolation from what is observed in small plasmids to the bacterial chromosome is not reliable, the results obtained indicated that supercoiling did change in a significant manner in the presence of high concentrations of Nov.
Figure 1.
Growth kinetics of S. pneumoniae R6 after Nov addition. (A) Exponentially growing cultures in AGCH at OD620 nm = 0.4 were treated with the indicated Nov concentrations. Relative values of two independent replicates are indicated. (B) Relative values (mean ± SEM) are indicated, being the initial one 1.53 × 108 colony forming U/ml. (C) Left column, phase contrast images; right column, DAPI staining.
Figure 2.
pLS1 topoisomers distribution after novobiocin treatment. (A) Map of pLS1 (GenBank accession number M29725) showing genes location and direction of transcription: replication (A, repA and B, repB), truncated mobilization protein (D, orfD) and a tetracycline pump (tet). (B) Cartoon illustrating pLS1 topoisomer distribution after two-dimensional electrophoresis in agarose gels run in the presence of 1 and 2 µg/ml chloroquine in the first and second dimensions, respectively. Arrows at the top left corner indicate running direction during first and second dimensions, respectively. OC, open circle, L, linear forms. Negative supercoiled topoisomers are in white and positive supercoiled in black. It was previously determined that 2 µg/ml chloroquine introduces 14 positive supercoils. A black arrowhead indicates the topoisomer that migrated with ΔLk = 0 in the second dimension. Therefore, this topoisomer migrated with a ΔWr = −14 during the first dimension. An empty arrowhead indicates the most abundant topoisomer. (C) pLS1 topoisomer distribution after different Nov treatments (indicated to the left). Cultures were grown as described in ‘Material and Methods’ section. Samples were taken before the addition of the drug (time 0 min) and at the times indicated on top. The corresponding supercoiling density (σ) value is indicated below each autoradiogram.
Topoisomerase mRNAs levels varied in parallel to DNA relaxation
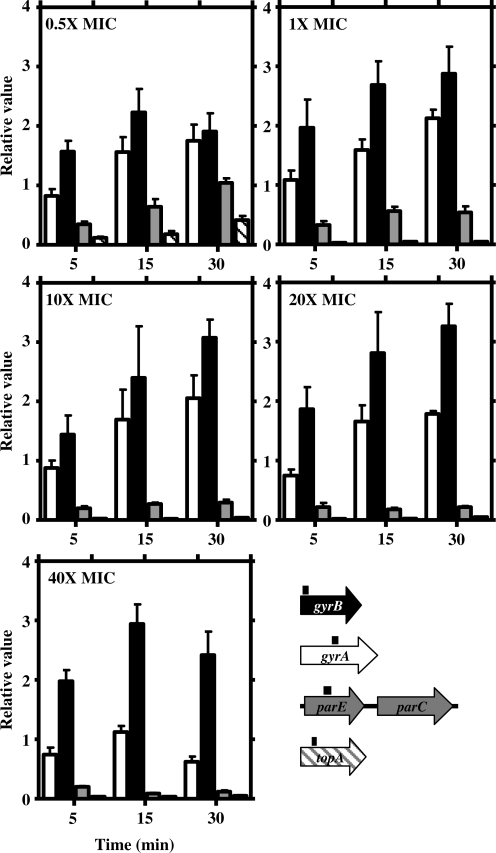
The relative expression of gyrase, topo IV and topo I genes were quantified using quantitative real-time PCR (qRT–PCR). Since parE and parC (topo IV) are transcribed from a single promoter (22), only parE was tested. In every case, the oligonucleotides used amplified regions of 118–205 bp close to the N-terminus (positions 371–240), except for gyrA, in which case amplification of a 158 bp region (positions 1431–1589) was used as the primers chosen to amplify this region showed a low identity to the corresponding regions of parC (22% versus 40% in the whole gene).
Relaxation by Nov caused an up-regulation of gyrase genes (gyrA and gyrB) and a down-regulation of topo I (topA) and topo IV (parE) genes (Figure 3). However, the up-regulation of gyrA was neither observed for 40× MIC nor for 5 min points, with just modest increases (1.6- to 2.1-fold) for the rest of the conditions used. Up-regulation of gyrB was dependent on the length of the treatment. Values (mean ± SD) for all Nov concentrations were 1.8 ± 0.2 (5 min) and 2.8 ± 0.3 (15 and 30 min). Down-regulation of both parE and topA augmented with increased Nov concentrations for all times except for 0.5 × MIC. At this concentration, decreases of 2.9 (parE) and 8.6 (topA) were observed at 5 min. Recovery at 15 min and 30 min was observed (1.6-fold for parE and 5.5-fold for topA), and total recovery at 30 min for parE and partial recovery for topA. At higher Nov concentrations (10 × MIC to 40 × MIC), down-regulation was observed for both parE (3.8- to 11.1-fold) and topA (20- to 50-fold).
Figure 3.
Transcriptional response to relaxation measured by qRT–PCR of DNA topoisomerases genes. Cultures of R6 and R6GyrB were grown as described in Figure 1. Total RNA was isolated, cDNA was synthesized and subjected to qRT–PCR. Data were normalized both to time 0 min and to that obtained from the amplification of rpoB and 16SrDNA genes. Relative values (mean of three independent replicates ±SD) are represented. A culture of R6GyrB (as R6, GyrBS127L, Nov MIC = 32 µg/ml) was treated with 16 µg/ml (0.5× MIC) and cultures of R6 (Nov MIC = 1 µg/ml) with 1, 10, 20 and 40 µg/ml (1× to 40× MIC). Genes used for the qRT–PCR with fragments amplified represented by black boxes above the corresponding gene are showed in different colours.
Transcriptional response to DNA relaxation occurred in clusters
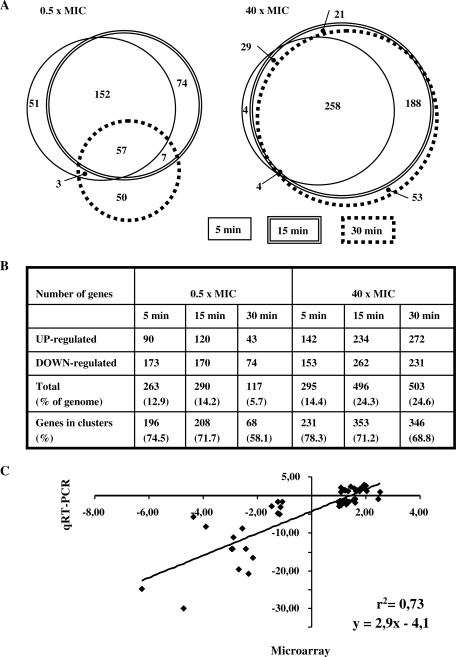
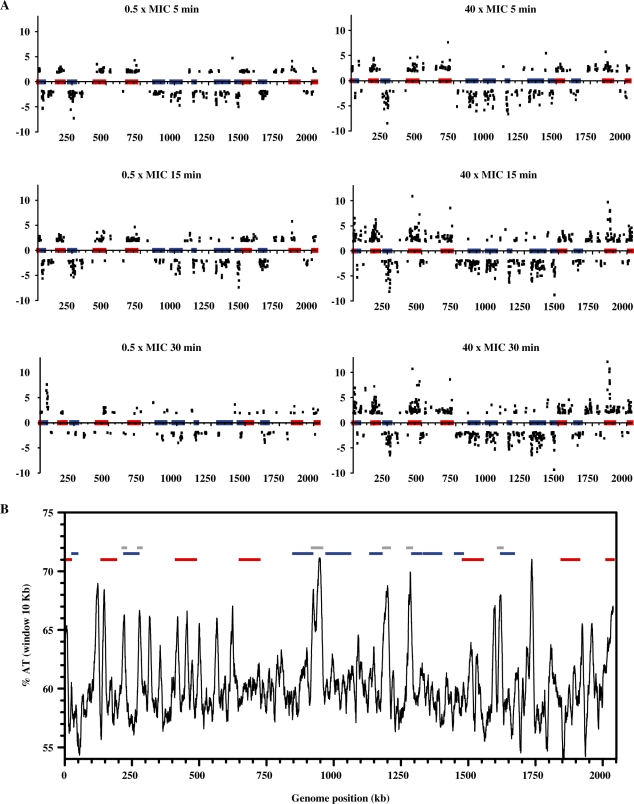
Two conditions, 0.5× and 40× MIC, were used for gene expression analysis on microarrays. Cultures of the wild-type strain R6 (Nov MIC = 1 µg/ml) were treated with Nov at 40 µg/ml and R6GyrB cells (as R6, GyrBS127L, Nov MIC = 32 µg/ml) were treated at 16 µg/ml. Gene expression variations were measured as the differences among values at time 0 and at 5, 15 and 30 min of Nov exposure. The fold variation values considered were only those ≥2, with P < 0.01. The whole transcriptomic response to relaxation is shown in Supplementary Table S1. For these experiments, S. pneumoniae R6 cultures were grown in triplicate as described in the ‘Materials and Methods’ section. Triplicate hybridisation datasets were normalized and analysed as described in the ‘Materials and Methods’ section. We detected variations of up to 12.1-fold for genes with increased expression levels and up to 9.3-fold for genes with decreased transcript levels. Similar figures were also observed for the transcriptional response to DNA relaxation in other bacteria (15,17). At low Nov concentrations (0.5× MIC), a similar response was observed at 5 and 15 min (Figure 4A). The total number of responsive genes was 263 and 290, respectively, accounting for 12.9% and 14.2% of the genome (Figure 4B). A majority of them (209 genes, >70%) responded similarly at both times, being 51 and 74 specific for 5 and 15 min, respectively (Figure 4A). The response was attenuated after 30 min at 0.5× MIC, the number of responsive genes being reduced >2-fold (117 genes, 5.7% of the genome) and only 57 of them were common with those of 5 and 15 min, respectively (Figure 4A). The number of genes affected by relaxation at the highest Nov concentration (40× MIC) at 5 min was similar to those found at 5 and 15 min for 0.5× MIC, 295 genes (14.4% of the genome). For longer times, the number of responsive genes increased considerably, being 496 and 503 (about 24% of the genome) at 15 and 30 min, respectively. Among them, 258 were common to all times at 40× MIC (Figure 4A).
Figure 4.
Gene expression analysis and validation of microarray data by qRT–PCR. (A) Venn diagram with three circles, each one corresponding to one time interval in each condition, showing the differentially expressed genes in six conditions. The area is proportional to the number of genes. A complete list of the genes can be found in Supplementary Table S1. (B) Classification of the responsive genes in the six conditions. (C) Correlation of the changes in RNA transcript levels using data from microarray hybridizations and qRT–PCR. Values can be found in Supplemental Table S2.
These transcriptome data from microarrays were validated by qRT–PCR. The reverse transcription products of 14 genes (Supplementary Table S2) were measured with samples taken at 0.5× and 40× MIC (5 and 15 min). Ratios of microarray and qRT–PCR showed a good correlation (r2 = 0.73, P < 10−4) and a slope of 2.9, indicating that qRT–PCR (as a single-gene technique) was more sensitive than microarray hybridization (Figure 4C).
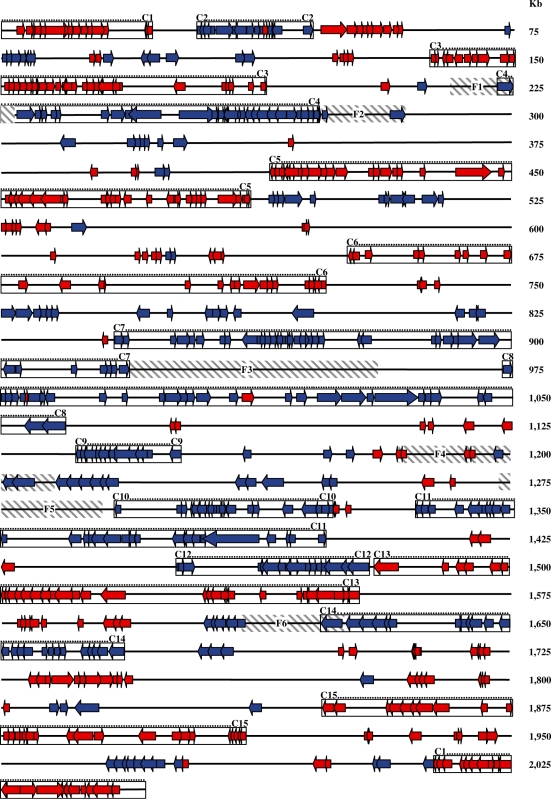
Surprisingly, a majority of the responsive genes were located contiguously in the chromosome, forming clusters that showed coordinated up- or down-regulation (Figures 5A and 6; Supplementary Table S1). There were three exceptions at 40× MIC (15 and 30 min): spr0039 in the down-regulated cluster 2 that was up-regulated; spr0994 and spr1029 in the down-regulated cluster 8 were also up-regulated (Figure 6; Supplementary Table S1). Clusters were evident at 0.5× MIC (5 and 15 min) and at 40× MIC (5, 15 and 30 min), while at 30 min at 0.5× MIC, genes became dispersed (Figure 5A). Considering the transcriptomic results of all experimental points except 30 min at 0.5× MIC, a total of 15 clusters were determined by inspection (Figures 5A and 6; Supplementary Table S1). The minimal criterion for cluster assignment was that clusters should contain at least six successive genes in a group of at least 14 showing the same kind of response (up- or down-regulation). Gaps allowed between responsive genes were no bigger than 12 kb for genes within the cluster and smaller than 8 kb for genes located within the boundaries of the cluster. The size of the clusters varied from 14.64 to 85.62 kb (average ± SD: 51.76 ± 21.77) and contained 15–43 responsive genes (average ± SD: 28 ± 9). Intercluster regions varied from 2.07 to 179.85 kb, and contained between 0 and 36 responsive genes (Table 1). Relative gene density in clusters (number of responsive genes per kilobase divided by total number of genes per kilobase) was higher (0.35–0.88 average ± SD: 0.57 ± 0.16) than in the intercluster regions (0.00–0.35, average ± SD: 0.13 ± 0.10). Even with this conservative definition of a cluster, included genes accounted for >68% of the responsive genes. In addition, a total of 136 genes were common to 0.5× MIC (5 and 15 min) and to 40× MIC (5, 15 and 30 min). A total of 124 (91.2%) were located in clusters.
Figure 5.
Localization of relaxation responsive genes in the S. pneumoniae chromosome. (A) Cultures were grown as described in Figure 1 and treated with different Nov concentrations. Samples were taken before the addition of the drug (time 0 min), and at indicated times, total RNA was isolated, double-stranded cDNA was synthesized, labelled with Cy3 and hybridized on high-density arrays. The relative fold variation of each gene is represented against the 3′ location of each open reading frame in the S. pneumoniae R6 chromosome (bases 1 to 2 038 615). Boxes indicate the transcriptional clusters: up-regulated in red, down-regulated in blue. (B) Analysis of percentage of AT content calculated at intervals of 100 bp along the whole R6 genome applying a window of 10 kb. Boxes indicate transcriptional clusters defined in (A). Flanking regions are shown in grey.
Figure 6.
Representation of the R6 genome showing responsive genes at five conditions and clusters location. Conditions considered were 0.5× MIC (5 and 15 min) and 40× MIC (5, 15 and 30 min). Genes are showed as arrows which head indicates the direction of transcription (as provided in http://cmr.jcvi.org/tigr-scripts/CMR/GenomePage.cgi?database=ntsp02). Red, up-regulated genes; blue, down-regulated genes. Values can be found in Supplementary Table S1. Boxes shown the clusters defined in Table 1. Flanking regions are shown in grey stripped boxes.
Table 1.
Characteristics of the transcriptional clusters
| Regiona | Genes included |
Size (kb) |
Responsive genesb |
|||||
|---|---|---|---|---|---|---|---|---|
| Interval (spr number) | Total number | Interval | Total | Total | Relative densityc | |||
| Cluster 1 | 2022 | 2046 | 25 | 2 014 115 | 2 038 403 | 24 288 | 21 | 0.84 |
| Cluster 1 | 1 | 19 | 19 | 1 | 21 851 | 21 850 | 15 | 0.79 |
| IC | 20 | 27 | 8 | 21 852 | 29 601 | 7749 | 0 | 0.00 |
| Cluster 2 | 28 | 44 | 17 | 29 602 | 45 417 | 15 815 | 15 | 0.88 |
| IC | 45 | 128 | 84 | 45 418 | 137 884 | 92 466 | 29 | 0.35 |
| Cluster 3 | 129 | 181 | 53 | 137 885 | 189 483 | 51 598 | 38 | 0.72 |
| IC | 182 | 231 | 50 | 189 484 | 222 573 | 33 089 | 2 | 0.04 |
| Cluster 4 | 232 | 272 | 41 | 222 574 | 273 257 | 50 683 | 33 | 0.80 |
| IC | 273 | 415 | 143 | 273 258 | 414 804 | 141 546 | 15 | 0.10 |
| Cluster 5 | 416 | 484 | 69 | 414 805 | 486 548 | 71 743 | 43 | 0.62 |
| IC | 485 | 643 | 159 | 486 549 | 651 794 | 165 245 | 30 | 0.19 |
| Cluster 6 | 644 | 721 | 78 | 651 795 | 722 781 | 70 986 | 27 | 0.35 |
| IC | 722 | 855 | 134 | 722 782 | 849 965 | 127 183 | 20 | 0.15 |
| Cluster 7 | 856 | 932 | 77 | 849 966 | 918 890 | 68 924 | 36 | 0.47 |
| IC | 933 | 988 | 56 | 918 891 | 973 795 | 54 904 | 0 | 0.00 |
| Cluster 8 | 989 | 1061 | 73 | 973 796 | 1 059 415 | 85 619 | 31 | 0.42 |
| IC | 1062 | 1131 | 70 | 1 059 416 | 1 136 390 | 76 974 | 7 | 0.10 |
| Cluster 9 | 1132 | 1149 | 18 | 1 136 391 | 1 151 027 | 14 636 | 15 | 0.83 |
| IC | 1150 | 1299 | 150 | 1 151 028 | 1 292 605 | 141 577 | 24 | 0.16 |
| Cluster 10 | 1300 | 1336 | 37 | 1 292 606 | 1 324 004 | 31 398 | 19 | 0.51 |
| IC | 1337 | 1352 | 16 | 1 324 005 | 1 336 102 | 12 097 | 2 | 0.13 |
| Cluster 11 | 1353 | 1411 | 59 | 1 336 103 | 1 397 930 | 61 827 | 31 | 0.53 |
| IC | 1412 | 1469 | 58 | 1 397 931 | 1 451 431 | 53 500 | 3 | 0.05 |
| Cluster 12 | 1470 | 1498 | 29 | 1 451 432 | 1 478 397 | 26 965 | 15 | 0.52 |
| IC | 1499 | 1501 | 3 | 1 478 398 | 1 480 465 | 2067 | 0 | 0.00 |
| Cluster 13 | 1502 | 1570 | 69 | 1 480 466 | 1 552 472 | 72 006 | 33 | 0.48 |
| IC | 1571 | 1651 | 81 | 1 552 473 | 1 623 065 | 70 592 | 16 | 0.20 |
| Cluster 14 | 1652 | 1698 | 47 | 1 623 066 | 1 667 915 | 44 849 | 27 | 0.57 |
| IC | 1699 | 1877 | 179 | 1 667 946 | 1 847 797 | 179 851 | 36 | 0.20 |
| Cluster 15 | 1878 | 1929 | 52 | 1 847 798 | 1 911 030 | 63 232 | 30 | 0.58 |
| IC | 1930 | 2021 | 92 | 1 911 031 | 2 014 114 | 103 083 | 22 | 0.24 |
aIC, intercluster.
bResponsive genes included were those showing significant fold variations (≥2 and P < 0.01) at 0.5× MIC 5 and 15 min and 40× MIC 5, 15 and 30 min.
cRelative gene density of responsive genes is defined as the number of responsive genes per kilobase divided by the total cell density (total number of genes per kilobase) in the region.
Clusters and flanking regions showed different codon composition and functional preferences
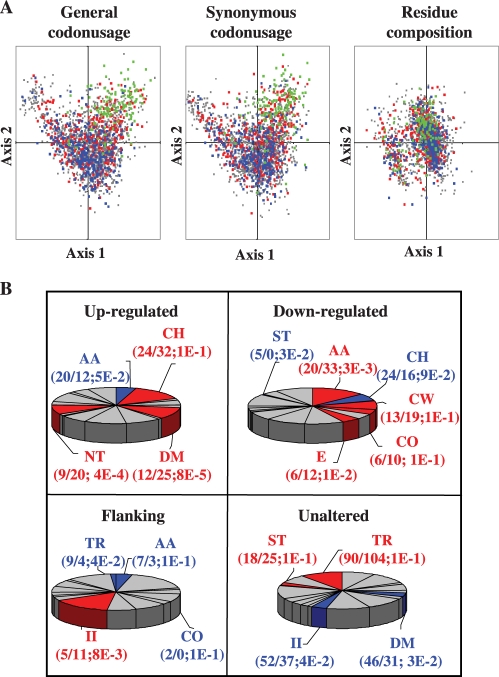
Measurement of the AT content along the chromosome revealed a number of noticeable peaks mainly located at regions flanking the down-regulated clusters although, secondarily, also in internal regions of the up-regulated clusters and intercluster zones. We selected the sharpest peaks, which corresponded to regions with the highest AT content. These regions contained at least 10 contiguous genes (11–40 genes) with >62% AT content (a condition only fulfilled by 29% of the genes in the genome) and an average AT content higher than 65%. Regions with two or more contiguous gene exceptions, AT content lower than 62%, or where these exceptions amounted to >20% of the genes in the region were discarted. The six regions that satisfied these criteria contained a total of 117 genes (Figure 5B; Table 2). These were termed flanking regions since they were located in intercluster regions overlapping or close to (<3 kb) down-regulated cluster boundaries. There was only one exception, flanking region 4, corresponding to intercluster region 9–10. When codon usage was analysed, although up-regulated genes were evenly dispersed in a codon correspondence analysis, down-regulated genes were concentrated at the bottom core of the correspondence analysis graph (Figure 7A). Genes of flanking regions grouped along the right horn which is inferred as an abnormal codon composition (27), are the consequence of both synonymous codon substitutions and residue usage frequency, although the latter seems less determinant. Flanking genes presented residue composition biased to those with a higher AT content in nearly all amino acid groups, such as Lys>Arg/His (basic charged), Asn > Gln (amides), Ile>Leu/Val (hydrophobic aliphatic) and Ser > Gly/Ala (small).
Table 2.
Characteristics of the six defined flanking regions including 117 genes
| Region |
Size (kb) |
Genes included |
%AT average |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Flanking peak | Relative location | Interval | Totala | Interval (spr number) | Total Numberb | In all region | Into genesc (P-valued) | ||
| 1 | Upstream 4 | 216.0 | 226.8 | 10.8 | 223 | 234 | 12 | 66.0 | 65.6 (2E-5) |
| 2 | Downstream 4 | 273.8 | 284.9 | 11.1 | 274 | 284 | 11 | 66.2 | 66.6 (1E-6) |
| 3 | Downstream 7 | 919.6 | 955.5 | 35.8 | 933 | 972 | 40 | 68.4 | 68.0 (4E-28) |
| 4 | Intercluster 9/10 | 1184.3 | 1207.2 | 22.9 | 1188 | 1210 | 23 | 67.3 | 66.1 (1E-10) |
| 5 | Upstream 10 | 1273.8 | 1289.6 | 15.8 | 1278 | 1293 | 16 | 67.9 | 66.9 (1E-9) |
| 6 | Upstream 14 | 1610.9 | 1626.3 | 15.4 | 1638 | 1653 | 15 | 67.5 | 67.9 (5E-11) |
aAverage size was 18.6 kb.
bAverage amount of genes was 19.5.
cData averaged by gene. %AT content into gene coding sequences was 66.9%.
dSignificance of altered AT-content of genes estimated by unpaired t-test between genes in the flanking region and the whole genome.
Figure 7.
Characteristics of S. pneumoniae R6 genes in relation to its transcriptional relaxation response. (A) Plot of the first two axes generated by correspondence analysis for codon usage, synonymous codon usage, and residue composition in 2236 S. pneumoniae R6 protein-coding genes. Up-regulated (red), down-regulated (blue), flanking (green) and unaltered (grey) gene values corresponding to the two most relevant axes derived from correspondence analysis for general codon, and residue composition are shown. (B) COG functional classes representation in domains. COG functional classes with a significantly increase (red), decrease (blue) or unaltered (grey) number of members. The ratio of the number of expected and observed members, together with the significance of the change through P-value assessed by chi-squared test (P ≤ 1E-1) are shown in parentheses below each functional class. AA, amino acid transport and metabolism; CH, carbohydrate transport and metabolism; CO, coenzyme transport and metabolism; CW, cell wall/membrane/envelope biogenesis; DM, defence mechanisms; E, energy production and conversion; II, inorganic ion transport and metabolism; NT, nucleotide transport and metabolism; ST, signal transduction; T, transcription; TR, translation, ribosome structure and biogenesis.
Functional preferences were analysed using the 24 functional classes of the COG (clusters of orthologous groups of proteins) system that unambiguously covers 74% of the R6 coding sequences (Figure 7B). This is a phylogenetic classification of proteins from complete genomes in which each COG includes proteins that are thought to be orthologous, i.e. connected through vertical evolutionary descent. Up-regulated genes were enriched in nucleotide management and defence systems, whereas down-regulated genes were enriched in amino acid and cell wall metabolism, along with energy production and conversion. Flanking genes were those with the lowest rate of COG annotation (only 61%). They were enriched in carbohydrate and inorganic ion transport/metabolism and especially in hypothetical proteins. Unaltered genes were enriched in genes involved in translation.
DISCUSSION
For the first time, we have determined the transcriptional response to DNA relaxation for a Gram-positive bacteria. All DNA topoisomerases of S. pneumoniae were involved in this response. The topoisomer distribution of plasmid pLS1 (Figure 2) showed a mean superhelical density of −0.062, a figure compatible with that reported for E. coli (28). At a subinhibitory Nov concentration (0.5 × MIC), after an initial relaxation (5 and 15 min), a cellular response allowed restoration of the native level of supercoiling (30 min). While a similar effect was observed for 1× MIC, increased degrees of relaxation with no restoration of supercoiling were observed at higher concentrations, compatible with the inhibition of cell division (Figure 1). Modest increases in superhelical densities of 23% (0.5 × MIC) and 25% (1× MIC) allowed full recovery of the supercoiling level at 0.5 × MIC and partial recovery at 1× MIC, in agreement with an estimate of the ±20% variation allowed for normal cell growth (29). Recovery of the supercoiling level at 0.5 × MIC coincides with non-evident effects in growth and division (Figure 1) and is interpreted as a consequence of a complete homeostatic response: transcription of gyrase genes increased to compensate for the induced relaxation, and transcription of topo I and topo IV genes decreased to prevent further relaxation. Although variations in transcription of DNA topoisomerase genes were evident by qRT–PCR (Figure 3), no increase for gyrA nor gyrB was observed in microarrays with the 2-fold variation considered as a threshold, due to the higher sensitivity of the qRT–PCR method. However, decreases using microarray technology were observed for the parE-parC operon (−2.45 to −2.98 at 15 and 30 min and 40× MIC) and for topA (−3.46 to −6.28 in all experimental conditions). The transcription increase of gyrase genes agrees with previous reports in E. coli (7). However, in S. pneumoniae, gyrA and gyrB seem to be independently regulated, given that both transcription levels and Nov concentration induction were different (Figure 3). Accordingly, the location of gyrA in the intercluster region 8–9 and that of gyrB in cluster 6 indicates they have different regulatory mechanisms. Likewise, while parE is located in the region between clusters 6 and 7, topA is located in cluster 9. These results suggest that several regulatory mechanisms regulate the homeostatic response to DNA relaxation. These include the organization of the chromosome in supercoiling domains and the existence of specific regulators for the DNA topoisomerase genes, such as the bending present in the gyrA promoter (30). Likewise, the lower level of transcription of topA compared to parE, and the location of the first in cluster 9 and of the latter in an intercluster region could be indicative of a differencial role in the regulation of DNA topology.
Recovery of supercoiling at 0.5× MIC was consistent with the down-regulation of parE and topA (Figure 3). At this Nov concentration, after an initial down-regulation of parE (2.9- and 1.6-fold at 5 min and 15 min) and topA (8.3- and 5.5-fold at 5 min and at 15 min), total recovery of parE and partial recovery of topA mRNA levels were observed at 30 min. Likewise, gene expression of the whole genome reflected the recovery of supercoiling at 0.5 × MIC. The general response at 30 min was attenuated, the number of responsive genes decreased from 263 and 290 at 5 and 15 min, to 117 at 30 min, respectively (Figure 4). Among these 117 genes, only half were common to times 5 and 15 min. However, at 40× MIC, in agreement with the observed inhibition of cell division (Figure 1) and with the continuous relaxation of the DNA (Figure 2), the number of responsive genes increased continuously from 295 at 5 min to about 500 at longer times (Figure 4). Altogether, these data indicated that under physiological conditions, ∼13% of the pneumococcal genome was involved in the cellular response allowing for recovery of the initial level of supercoiling from a moderate relaxation. This figure is consistent with the 15% response reported for inhibition of E. coli gyrase with a fluoroquinolone (16), but higher than the 7% reported by others (15). On the other hand, values as high as 37% were reported for a Nov treatment of H. influenzae (17).
The most outstanding feature in the genome response to DNA relaxation in pneumococcus was the clustering of responsive genes. Clusters include >68% of these genes, representing between 12.9% (at 0.5× MIC) and 24.6% (at 40× MIC) of the genome. Co-regulation clearly trespassed the operon limits. The direction of transcription of the responsive genes followed the predominant direction of transcription dictated by the location of the origin of replication with no preference for leading or lagging strands (Figure 6). The AT content along the genome correlated with clusters location, showing high values in up-regulated clusters, low values in down-regulated clusters, and clear peaks in at least one of the boundaries of down-regulated clusters (Figure 5). These results suggest that relaxation of DNA in high AT content regions favour the access of RNA polymerase to their promoters. On the contrary, a low AT content in down-regulated clusters obstructs the access of RNA polymerase. Accordingly, AT enrichment in both upstream and coding regions of up-regulated genes induced after relaxation was also reported in E. coli (15).
The AT content organizes the genome into topological domains of supercoiling (Figure 5). In this study, we detected 15 clusters of coordinately transcribed genes, with an average size of 51.76 kb. This size is about twice the 25-kb domain estimated in Salmonella enterica by measuring site-specific recombination between distant chromosomal sites (31) and about half the size (∼100 kb) of the domains detected in E. coli by UV-induced psoralen cross-linking (32). Different domain types showed different functional preferences. Up-regulated domains were enriched in nucleotide metabolism and defence functions, whereas down-regulated domains were supplemented in non-abundant housekeeping genes responsible for the maintenance of cellular life under normal conditions. This switch of functions after DNA relaxation suggests a mechanism for cells to feel a stress condition, whereas the highly relevant translation machinery, driven by abundant proteins, remains unchanged. Notably, the unaltered fraction contained most genes positioned in the left horn of the plot derived from the codon usage correspondence analysis, an area enriched in genes coding for abundant proteins involved in translation as ribosomal proteins and elongation factors (27), whose transcription efficiency seems to be protected from topological variations. The disparate codon and residue composition of the flanking regions suggest that selective pressure did not adapt this gene pool to the intrinsic codon usage bias of the pneumococcus, retaining instead a high AT content. Alterations in the synonymous codon usage have been correlated to protein abundance in S. pneumoniae (27). This study shows that DNA topology acts as a secondary force for codon selection. In addition, many flanking genes cannot be functionally annotated and for those that are, the excess of ABC-transporters suggests that these genes are redundant or dispensable. These features of the genes of the flanking regions lead to propose that these zones act as DNA with an architectural mission. Therefore, assuming that the chromosome has a stable length, the relaxation of AT-rich zones would cause a relative compression of proximal zones. The four-class chromosome architecture division into domains (up-regulated, down-regulated, flanking and unaltered) could work as a permanent background control of transcription. To our knowledge, this kind of organization remained unreported in bacteria, so the extent to how much this can be extensible to other prokaryotic species is open to fascinating future research.
In summary, this study shows functional (transcriptionally controlled) and physical (AT% content distribution, codon usage bias) evidence of chromosomal organization into domains of supercoiling. This domain organization makes supercoiling a general mechanism for controlling gene expression superimposed on other kinds of regulatory mechanisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Plan Nacional de I + D + I of Ministerio de Ciencia e Innovación (BIO2008-02154); Comunidad de Madrid to AG de la Campa (COMBACT-S-BIO-0260/2006); from Plan Nacional de I + D + I of Ministerio de Ciencia e Innovación (BFU2008-00408/BMC to J.B.S.) in part; Ciber Enfermedades Respiratorias is an initiative from Instituto de Salud Carlos III. AG de la Campa is an Investigador Científico from de CSIC. Juan de la Cierva contract provided by Spanish Ministerio de Educación y Cultura (to A.J.M.G.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Vicente for his help with microscopy.
REFERENCES
- 1.Jacobs MR, Felmingham D, Appelbaum PC, Grüneberg RN the Alexander Project Group. The Alexander project 1998–2000: susceptibility of pathogens isolated from community-acquired respiratory tract infection to commomnly used antimicrobial agents. J. Antimicrob. Chemother. 2003;52:229–246. doi: 10.1093/jac/dkg321. [DOI] [PubMed] [Google Scholar]
- 2.Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, Dowell SF, File T.M., Jr, Musher DM, Niederman MS, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin. Infect. Dis. 2007;44(Suppl 2):S27–S72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drlica K, Malik M, Kerns RJ, Zhao X. Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 2008;52:385–392. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muñoz R, de la Campa AG. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob. Agents Chemother. 1996;40:2252–2257. doi: 10.1128/aac.40.10.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernández-Moreira E, Balas D, González I, de la Campa AG. Fluoroquinolones inhibit preferentially Streptococcus pneumoniae DNA topoisomerase IV than DNA gyrase native proteins. Microb. Drug Resist. 2000;6:259–267. doi: 10.1089/mdr.2000.6.259. [DOI] [PubMed] [Google Scholar]
- 6.Tse-Dinh YC. Regulation of the Escherichia coli DNA topoisomerase I gene by DNA supercoiling. Nucleic Acids Res. 1985;13:4751–4763. doi: 10.1093/nar/13.13.4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menzel R, Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983;34:105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- 8.Menzel R, Gellert M. Modulation of transcription by DNA supercoiling: a deletion analysis of the Escherichia coli gyrA and gyrB promoters. Proc. Natl Acad. Sci. USA. 1987;84:4185–4189. doi: 10.1073/pnas.84.12.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menzel R, Gellert M. Fusions of the Escherichia coli gyrA and gyrB control regions to the galactokinase gene are inducible by coumermycin treatment. J. Bacteriol. 1987;169:1272–1278. doi: 10.1128/jb.169.3.1272-1278.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiara AS, Cundliffe E. Expression and analysis of two gyrB genes from the novobiocin producer, Streptomyces sphaeroides. Mol. Microbiol. 1993;8:495–506. doi: 10.1111/j.1365-2958.1993.tb01593.x. [DOI] [PubMed] [Google Scholar]
- 11.Unniraman S, Chatterji M, Nagaraja V. DNA gyrase genes in Mycobacterium tuberculosis: a single operon driven by multiple promoters. J. Bacteriol. 2002;184:5449–5456. doi: 10.1128/JB.184.19.5449-5456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorman CJ. DNA supercoiling and environmental regulation of gene expression in pathogenic bacteria. Infect. Immun. 1991;59:745–749. doi: 10.1128/iai.59.3.745-749.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rui S, Tse-Dhin YC. Topoisomerase function during bacterial responses to environmental challenge. Front. Biosci. 2003;8:d256–d263. doi: 10.2741/984. [DOI] [PubMed] [Google Scholar]
- 14.Travers A, Muskhelishvili G. DNA supercoiling – a global transcriptional regulator for enterobacterial growth? Nat. Rev. 2005;3:157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- 15.Peter BJ, Arsuaga J, Breier AM, Khodursky AB, Brown PO, Cozzarelli NR. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004;5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong KS, Xie Y, Hiasa H, Khodursky AB. Analysis of pleiotropic transcriptional profiles: a case study of DNA gyrase inhibition. PLoS Genet. 2006;2:e152. doi: 10.1371/journal.pgen.0020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gmuender H, Kuratli K, Di Padova K, Gray CP, Keck W, Evers S. Gene expression changes triggered by exposure of Haemophilus influenzae to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 2001;11:28–42. doi: 10.1101/gr.157701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vora T, Hottes AK, Tavazoie S. Protein occupancy landscape of a bacterial genome. Mol. Cell. 2009;35:247–253. doi: 10.1016/j.molcel.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muñoz R, Bustamante M, de la Campa AG. Ser-127-to-Leu substitution in the DNA gyrase B subunit of Streptococcus pneumoniae is implicated in novobiocin resistance. J. Bacteriol. 1995;177:4166–4170. doi: 10.1128/jb.177.14.4166-4170.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lacks SA, López P, Greenberg B, Espinosa M. Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pLS1. J. Mol. Biol. 1986;192:753–765. doi: 10.1016/0022-2836(86)90026-4. [DOI] [PubMed] [Google Scholar]
- 21.Martín-Parras L, Lucas I, Martínez-Robles ML, Hernández P, Krimer DB, Hyrien O, Schvartzman JB. Topological complexity of different populations of pBR322 as visualized by two-dimensional agarose gel electrophoresis. Nucleic Acids Res. 1998;26:3424–3432. doi: 10.1093/nar/26.14.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balsalobre L, de la Campa AG. Fitness of Streptococcus pneumoniae fluoroquinolone-resistant strains with topoisomerase IV recombinant genes. Antimicrob. Agents Chemother. 2008;52:822–830. doi: 10.1128/AAC.00731-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 24.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de la Campa AG, del Solar GH, Espinosa M. Initiation of replication of plasmid pLS1. The initiator protein RepB acts on two distant DNA regions. J. Mol. Biol. 1990;213:247–262. doi: 10.1016/S0022-2836(05)80188-3. [DOI] [PubMed] [Google Scholar]
- 26.Olavarrieta L, Hernández P, Krimer DB, Schvartzman JB. DNA knotting caused by head-on collision of transcription and replication. J. Mol. Biol. 2002;322:1–6. doi: 10.1016/s0022-2836(02)00740-4. [DOI] [PubMed] [Google Scholar]
- 27.Martín-Galiano AJ, Wells JM, de la Campa AG. Relationship between codon biased genes, microarray expression values and physiological characteristics of Streptococcus pneumoniae. Microbiology. 2004;150:2313–2325. doi: 10.1099/mic.0.27097-0. [DOI] [PubMed] [Google Scholar]
- 28.Deng S, Stein RA, Higgins NP. Organization of supercoil domains and their reorganization by transcription. Mol. Microbiol. 2005;57:1511–1521. doi: 10.1111/j.1365-2958.2005.04796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drlica K. Control of bacterial supercoiling. Mol. Microbiol. 1992;6:425–433. doi: 10.1111/j.1365-2958.1992.tb01486.x. [DOI] [PubMed] [Google Scholar]
- 30.Balas D, Fernández-Moreira E, de la Campa AG. Molecular characterization of the gene encoding the DNA gyrase A subunit of Streptococcus pneumoniae. J. Bacteriol. 1998;180:2854–2861. doi: 10.1128/jb.180.11.2854-2861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higgins NP, Yang X, Fu Q, Roth JR. Surveying a supercoil domain by using the gamma delta resolution system in Salmonella typhimurium. J. Bacteriol. 1996;178:2825–2835. doi: 10.1128/jb.178.10.2825-2835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinden RR, Pettijohn DE. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc. Natl Acad. Sci. USA. 1981;78:224–228. doi: 10.1073/pnas.78.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.