Abstract
Differences between male and female mammals are initiated by embryonic differentiation of the gonad into either a testis or an ovary. However, this may not be the sole determinant. There are reports that embryonic sex differentiation might precede and be independent of gonadal differentiation, but there is little molecular biological evidence for this. To test for sex differences in early-stage embryos, we separated male and female blastocysts using newly developed non-invasive sexing methods for transgenic mice expressing green fluorescent protein and compared the gene-expression patterns. From this screening, we found that the Fthl17 (ferritin, heavy polypeptide-like 17) family of genes was predominantly expressed in female blastocysts. This comprises seven genes that cluster on the X chromosome. Expression analysis based on DNA polymorphisms revealed that these genes are imprinted and expressed from the paternal X chromosome as early as the two-cell stage. Thus, by the time zygotic genome activation starts there are already differences in gene expression between male and female mouse embryos. This discovery will be important for the study of early sex differentiation, as clearly these differences arise before gonadal differentiation.
INTRODUCTION
In eutherian mammals, gender is determined genetically at the time of syngamy and females (XX) have twice as many X chromosomes as males (XY). However, soon after fertilization in females, one of the X chromosomes which is derived from father becomes inactivated and, after implantation, one of the X chromosomes becomes inactivated randomly in the embryo proper. This equalizes the dosage of X-linked genes between sexes (1–3). This is called ‘X chromosome inactivation’ and demonstrates that differences in sex chromosome constitution between sexes start to be compensated prior to embryonic implantation. Contrary to X inactivation, the presence of the Y chromosome leads to fundamental differences between males and females. To date, it has been understood that, after implantation, expression of the Y-linked Sry gene determines the sex of the gonads (4) and that sex hormones secreted from the differentiated gonads influence the fetus and allow various sexual characteristics to become apparent (5).
However, there are some reports that suggest that this differentiation of gonads is not the sole determinant of all gender differences. For instance, in several mammalian species, male embryos develop faster than female embryos prior to implantation (6). Moreover, preimplantation male and female embryos show differences in glucose metabolism and pentose phosphate pathway activity (7,8) and female rat neurons harvested and cultured prior to gonadal differentiation develop more tyrosine hydroxylase or prolactin-immunoreactive neurons (9). These early sex differences may have some effects on sexual differentiation thereafter (10). In spite of these observations, little molecular biological evidence about early sex differences has been established so far.
In searching for genetic clues on the nature of sex differentiation before gonadal differentiation, we compared the gene-expression patterns of male and female blastocysts. We have already developed a method to sex blastocysts using a transgenic mouse line in which the X chromosome is tagged with an enhanced green fluorescent protein (EGFP) transgene (11–13). We then compared gene-expression patterns between sexed blastocysts using DNA microarrays. We have reported previously that two Y-linked genes (Dby and Eif2s3y) are expressed specifically in male embryos, and that two X-linked genes (Xist and Rhox5/Pem) are imprinted and expressed predominantly in female blastocysts (14). These genes were thought to be candidates possibly involved in sex differentiation. In detail, Dby and Eif2s3y encode an RNA helicase and a translation-initiation factor, respectively, and are necessary for spermatogenesis, but there is no report that they are involved in sex differentiation (15,16). Another gene, Xist, is well known as a non-coding RNA responsible for X chromosome inactivation that compensates for dosage differences between sexes (17). Rhox5 is a homeobox gene (18) and we expected that Rhox5 would contribute to differentiation between male and female embryos. However, targeted disruption was shown to reduce sperm production, but no other abnormalities have been reported from gene-inactivation experiments (18,19). Thus, so far there is no gene positively identified to be involved in early sex differences and later sex differentiation.
In previous reports (14), we showed that there are sex-linked differences in gene expression at the blastocyst stage. However, the arrays we used (Agilent Mouse Development G4120A) mainly cover postimplantation stages and do not identify all the known genes. We suspected there might be undiscovered genes showing sex differences. In this report, to carry out more comprehensive gene-expression analysis, we used arrays capable of analyzing all the known mouse genes and compared male and female embryonic gene expression at the blastocyst stage. From this screening, we found imprinted genes involved in sex-linked differential expression and determined the time of onset of differences in the expression of these genes.
MATERIALS AND METHODS
Animals
The handling and surgical manipulation of all experimental animals were carried out in accordance with the guidelines of the Committee on the Use of Live Animals in Teaching and Research of Tokyo Medical and Dental University. The B6C3F1 TgN (act EGFP) Osb CX-38 (G38) transgenic mouse strain described in our previous paper (12) was used to distinguish between male and female embryos.
Blastocyst collection and RNA extraction
B6C3F1 strain female mice at 8 weeks of age were superovulated with 5 IU of pregnant mare serum gonadotropin followed by 5 IU of human chorionic gonadotropin (hCG) 48 h later and were mated with XGFPY male mice. Four-cell stage embryos were collected from the oviducts 55 h after the hCG injection, placed in potassium simplex optimization medium (KSOM) and incubated in a humidified atmosphere of 5% carbon dioxide (CO2) in air at 37°C for an additional 38 h. Mid-stage blastocysts were collected, and male (EGFP-negative) and female (EGFP-positive) embryos were separated by observing green fluorescence under a dissecting microscope. To verify differential gene expression in non-transgenic embryos in vivo, wild-type C57BL/6 blastocysts were obtained from the uteri of superovulated C57BL/6 females that had been mated with wild-type C57BL/6 males 92 h after hCG injection. Genomic DNA and RNA were extracted from individual blastocysts. Male and female blastocysts were pooled separately following the determination of sex by polymerase chain reaction (PCR) using Ube1x primers as follows: 5′-TGGTCTGGACCCAAACGCTGTCCACA-3′ and 5′-GGCAGCAGCCATCACATAATCCAGATG-3′ (20). For each experiment, 30–50 wild-type blastocysts were sexed and pooled samples were used for expression analysis.
Comparative expression analysis using DNA microarrays
Total RNAs were extracted from over 150 male or female sexed blastocysts (as determined above). Half of the isolated total RNA from each sex was labeled with Cy-3 and Cy-5 using a Low Input Fluorescent Linear Amplification kit (Agilent) following the manufacturer’s instructions. The Custom Oligo Microarray consisting of two 22 k slides (GEO accession numbers GPL8326 and GPL8329) was used in this study. To improve accuracy, samples were collected from three independent preparations and all experiments were run in triplicate. The hybridization experiments were duplicated in a reciprocal labeling manner to reduce dye-integration bias and six hybridizations were carried out for the entire analysis. Combining plural array results and statistical analyses were carried out using Luminator software. The probe sequence of this array set was designed based mainly on the representative transcripts of the FANTOM2 database (36 176 transcripts; http://fantom2.gsc.riken.jp/), the expressed sequence tag data obtained from a cDNA library of mouse primordial germ cells (4689 transcripts; Abe et al., unpublished data) and manually selected sequences of interest (357 transcripts). The sequences of all probes were Basic Local Alignment Search Tool (BLAST) searched against NCBI GenBank (Build 37) and the top-hit accession number was considered as the unique ID of the probe. Any alternative transcripts were regarded as independent and the number of distinct transcripts in our microarray set was then counted.
Reverse transcription PCR of candidate genes for sexually differential expression
Reverse transcription was carried out with the pooled total RNA extracted from 100 blastocysts. One percent of the resulting cDNA samples was amplified by PCR. Primer sets were as follows: 5′-AAGTGTGACGTTGACATCCG-3′ and 5′-GATCCACATCTGCTGGAAGG-3′ for β-actin, 5′-CTCATCCTCATGTCTTCTCCG-3′ and 5′-GATTCCAGATAGACAGGCTGG-3′ for Xist and 5′-AGAGATGAGCAAGGTGCACA-3′ and 5′-CGAACCTAGAGCCCTGGAG-3′ for Rhox5, see Supplementary Table S1 for primers to amplify Fthl17 (ferritin, heavy polypeptide-like 17) gene. All PCR reactions were replicated at least once. The same PCR conditions were used to examine the wild-type blastocysts, except for the amount of starting materials (30–50 blastocysts).
Verification of imprinting
Two reciprocal sets of F1 hybrid blastocysts, (C57BL/6 × JF1) F1 and (JF1 × C57BL/6) F1, were produced by in vitro fertilization. In each experiment at least 30 blastocysts were sexed by PCR as described above and pooled according to sex. The pooled RNA was treated with DNase to eliminate genomic DNA contamination, and reverse transcription was performed using Superscript III reverse transcriptase (Invitrogen). One-third of the resulting cDNA samples derived from pooled RNAs were amplified by PCR using the above primers. For PCR amplification of each Fthl17 family gene, r–Taq DNA polymerase (TOYOBO) that does not have 3′→5′ exonuclease activity was used. In the allelic expression analysis of Fthl17-L6, pooled 30 blastocysts were used for each reverse transcription (RT)–PCR reaction, and the Sty I restriction enzyme was used for detecting single nucleotide polymorphisms (SNPs). In the RT–PCR experiments, no genomic DNA contamination was detected.
In situ hybridization
Whole mount in situ hybridization was performed as described (21). The probes encompass the whole length of the Fthl17 sequence (NM_031261).
Bisulfite sequencing
The bisulfite sequence reaction was performed as described (22), with a smaller quantify of starting samples (600 male blastocysts and 600 female blastocysts). See Supplementary Table S2 for primers to amplify the CpG loci.
RESULTS AND DISCUSSION
DNA microarray
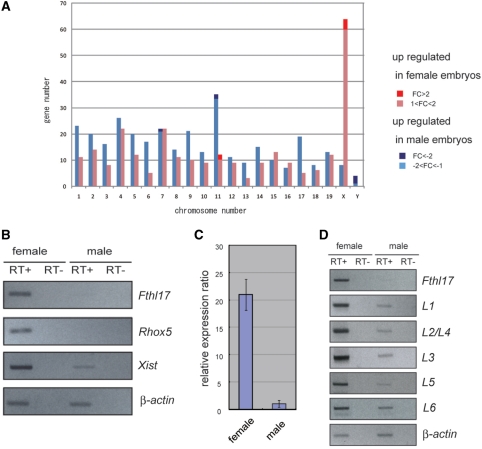
To compare gene-expression patterns between male and female blastocysts, we used a DNA microarray (22 K-1 and 22 K-2) that represents all known 41 222 mouse transcripts, whose sequences were mainly obtained from the Riken FANTOM2 Database (see ‘Materials and Methods’ section). Using six hybridization experiments and statistical analysis, 413 transcripts were found to be expressed to a higher degree in female blastocysts (P < 0.001) and 460 transcripts were expressed to a higher degree in males (P < 0.001). Mapping these transcripts revealed that many were concentrated on the X chromosome (Figure 1A). Although most of the differences were less than 2-fold, there were a few differentially expressed transcripts showing greater variations (Table 1 for the 22 K-1 array and Table 2 for the 22K-2 array). The candidate 19 probes contained the male-specific genes Dby and Eif2s3y, and the female-predominant genes Xist and Rhox5 that we reported previously, confirming the validity of the experiments. We were interested in discovering other differentially expressed genes and carried out RT–PCR to confirm the expression levels of other candidate genes. The results showed that the Fthl17 gene was expressed predominantly in female embryos (Figure 1B). Furthermore, the real-time PCR analysis revealed that in female embryos the Fthl17 expression level was 21 times greater than that seen in males (Figure 1C). The same results were obtained with wild-type blastocysts not expressing EGFP recovered from the uterus (Supplementary Figure S1), ruling out the possibility that the differential expression was caused by the artificial conditions. Originally, Fthl17 was reported to be expressed specifically in spermatogonia (testicular stem cells) (23) as a gene expected to be a metal ion-binding protein (data from the NCBI database). In addition, we found here that it was expressed at the blastocyst stage and showed predominantly female expression.
Figure 1.
Sex-linked differential gene expression at the blastocyst stage. (A) The chromosomal distribution of differentially expressed genes. Red and blue bars show genes that were significantly upregulated in female and male embryos, respectively (P < 0.001). Dark red and blue bars correspond to the genes showing >2-fold changes. (B) RT–PCR analysis of the differentially expressed genes. Genes showing >2-fold expression differences were selected and RT–PCR was carried out using female (GFP-positive) and male (GFP-negative) samples. (C) Real-time PCR measurements of Fthl17 gene expression in female and male samples. (D) RT–PCR analysis of each Fthl17 family gene using specific RT–PCR primers (see Supplementary Figure S2 and Supplementary Table S1 for primer sequences).
Table 1.
List of the differentially expressed genes according to embryo sex (22 K-1)
| Gene name | Accession number | Fold change | P | Male intensity | Female intensity | Map position |
|---|---|---|---|---|---|---|
| 2310065F04Rik | AK010044 | 8.4 | 0 | 1199 | 10 013 | chr11 |
| Flotillin 2 | AK030199 | 2.1 | 0 | 2193 | 4518 | chr11 |
| EST | AK029640 | 2 | 1.03 (10−19 | 3387 | 6802 | Not mapped uniquely |
| 1810057C19Rik | NM_026433 | −2.5 | 3.87 (10−5 | 564 | 240 | chr11 |
| Dby | AK020213 | −3 | 1.46 (10−22 | 5292 | 1744 | chrY |
| EG330552 | AK086712 | −3.5 | 4.18 × 10−4 | 461 | 132 | chr7 |
| H2-M10.1 | NM_013544 | −6.8 | 7.09 × 10−7 | 258 | 69 | Not mapped uniquely |
| Dby | BC021453 | −7.2 | 2.25 (10−29 | 724 | 99 | chrY |
| Eif2s3y | NM_012011 | −35.8 | 0 | 2418 | 68 | chrY |
Table 2.
List of the differentially expressed genes according to embryo sex (22K-2)
| Gene name | Accession number | Fold change | P | Male intensity | Female intensity | Map position |
|---|---|---|---|---|---|---|
| Xist | NR_001463 | 23.6 | 0 | 529 | 12 475 | chrX |
| Xist | NR_001463 | 22.4 | 0 | 554 | 12 467 | chrX |
| Fthl17 | AF285569.1 | 6.5 | 0 | 1453 | 9556 | chrX |
| Rhox5/Pem | NM_008818 | 4.6 | 0 | 394 | 1845 | chrX |
| Rhox5/Pem | NM_008818 | 4.3 | 0 | 425 | 1869 | chrX |
| Bex1 | NM_009052 | 3.3 | 9.28 × 10−7 | 353 | 1265 | chrX |
| Bex1 | NM_009052 | 2.3 | 9.82 × 10−5 | 379 | 940 | chrX |
| EST | U306415-1 | 2.1 | 7.22 × 10−8 | 246 613 | 529 132 | Not mapped uniquely |
| EST | U306415-1 | 2.1 | 1.07 × 10−6 | 246 644 | 523 398 | Not mapped uniquely |
| B230343A10Rik | B230343A10 | −2.3 | 2.44 × 10−5 | 210 | 92 | chr11 |
All genes showing >2-fold (200% P < 0.001) change and <–2.0-fold (50%) change in expression level are listed. Some probes were redundant, but the results of all probes are listed.
Identification of the Fthl17 gene family
Next, to characterize this gene in more detail, we carried out database searching in GenBank (Build 37.1) and found that the Fthl17 gene (NM_031261) has seven highly homologous nucleotide sequences covering approximately 150 kb on the X chromosome. Therefore, we named these genes Fthl17-L1 through Fthl17-L6 and have called this group the ‘Fthl17 gene family’ (Supplementary Figure S2). The expression level of each gene in this family was examined using specific RT–PCR primers amplifying individual transcriptional units. All seven genes were transcribed successfully and showed predominantly female expression (Figure 1D).
Verification of imprinting of the Fthl17 gene family
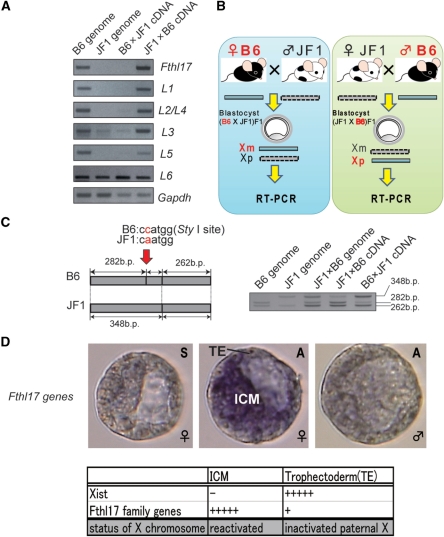
How does this family of genes show predominant expression in females? Because both the X-linked genes Rhox5 and Xist are imprinted and expressed from the paternal X chromosome, they show a female predominant expression. Therefore, we examined whether the Fthl17 genes were also imprinted. First, to distinguish paternal from maternal expression, we searched for DNA polymorphisms between Mus musculus musculus and the Japanese wild mouse, M. mus. molossinus. Southern blot analysis revealed that this family of genes was conserved in M. mus. molossinus (JF1 strain) as well as in M. mus. musculus (B6, C3H and 129sv strains; Supplementary Figure S3). Therefore, we tried to detect the B6 and JF1 alleles by PCR using specific primers for this gene family. However, the B6 allele was detected, but the JF1 alleles were not, with the exception of Fthl17-L6 (Figure 2A). This was probably caused by DNA polymorphisms in the specific PCR primer sequences. We then used the B6 allele to examine whether these genes (Fthl17 and Fthl17-L1-L5) were active when they were derived paternally. As shown in Figure 2B, the active alleles of the Fthl17 genes were examined in F1 blastocysts derived from mating between C57BL/6 females and JF1 males (B6 × JF1) and the reciprocal cross of JF1 females with C57BL/6 males (JF1 × B6). The RT–PCR products clearly showed that these genes (Fthl17 and Fthl17-L1-L5) were active only when they were derived from the males (Figure 2A). Furthermore, we also examined allelic expression of Fthl17-L6 gene using a SNP discovered between B6 and JF1 alleles (Figure 2C). There was predominant expression from the paternal allele. All of these results indicate that the Fthl17 gene family is imprinted and predominantly expressed from the paternal X chromosome.
Figure 2.
Verification of imprinting in the Fthl17 gene family. (A) Genomic PCR and RT–PCR amplification for Fthl17. Left two samples: PCR products of genomic DNA (B6 genome, JF1 genome). Right two samples: RT–PCR products of B6 (C57/BL/6) × JF1 (JF1/Ms) F1 and JF1 × B6 F1 blastocyst samples. (B) Scheme for verification of imprinting in the Fthl17 gene family using intersubspecific hybrid F1 mouse embryos. The active alleles of the Fthl17 genes were examined in F1 blastocysts derived from mating between C57BL/6 females and JF1 males (B6 × JF1) and the reciprocal cross of JF1 females with C57/BL/6 males (JF1 × B6). (C) Allelic expression analysis was carried out using a SNP in the Fthl17-L6 gene. (D) Whole mount in situ hybridization in female and male blastocysts. ‘S’ indicates the sense strand probe and ‘A’ indicates the antisense strand probe.
Expression of Fthl17 genes in blastocysts
To examine the relationship between Fthl17 expression pattern and the pattern of X chromosome inactivation, whole mount in situ hybridization of the Fthl17 gene family was carried out on blastocysts. The genes were barely detected in male embryos. However, in female embryos, strong signals were detected in both the inner cell mass (ICM) and trophectoderm (TE) with stronger signals in the ICM than in the TE (Figure 2D). This is markedly different from the expression pattern of Xist, which is a marker of X inactivation. As reported, Xist was not expressed in the ICM but only in the TE. We conclude that the Fthl17 family of genes was mainly expressed from a reactivated paternal X chromosome (Xp) in the ICM and was slightly expressed from an inactivated paternal X chromosome in the TE, indicating escape from imprinted inactivation at this stage.
Expression of Fthl17 genes before and after the blastocyst stage
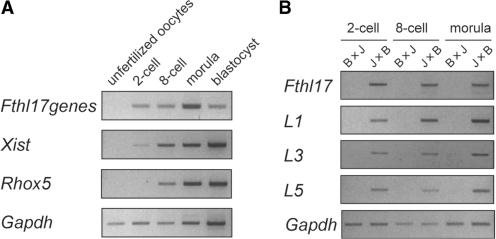
To determine the time of onset of differences in sex-linked gene expression, we examined the expression levels of Fthl17 genes before the blastocyst stage. Interestingly, expression started at the two-cell stage, which corresponds to the onset of zygotic genome activation (Figure 3A). Furthermore, allelic expression analysis also revealed that four highly expressed genes, Fthl17, Fthl17-L1, -L3 and -L5, were imprinted and expressed from the paternal allele as early as the two-cell stage (Figure 3B). Because of the low-level expression of Fthl17-L6, allelic expression analysis using the SNP between B6 and JF1 could not be done. However, the above results demonstrate that, at least in M. mus. musculus (C57/BL6), the Fthl17 family of genes, exactly similar to Xist, is expressed from inactivated Xp chromosomes during preimplantation stages and thus comprise one of the earliest sets of imprinted genes that is expressed. It is known that the fertilized oocyte stores maternally derived mRNA and that the zygotic genome remains largely inactive. After fertilization, de novo transcription of embryonic genes starts mainly after the two-cell stage. Our results clearly demonstrate that epigenetic gene regulation controls the differential expression of male and female embryos from this very early stage of embryonic development. As for postimplantation stages, we could not detect Fthl17 family gene expression in embryos at day 9.5 and 12.5, so no information on the imprinting status was obtained (data not shown).
Figure 3.
(A) Expression of Fthl17 in preimplantation stages (unfertilized oocytes, two-cell, eight-cell, morula and blastocyst). The RT–PCR primers for Fthl17 were designed to detect all genes in this family. (B) Allelic expression analysis of the Fthl17 gene family (Fthl17, Fthl17-L1, -L3 and -L5) at preimplantation stages (two-cell, eight-cell and morula).
Effect of X inactivation and genomic imprinting on X-linked gene expression
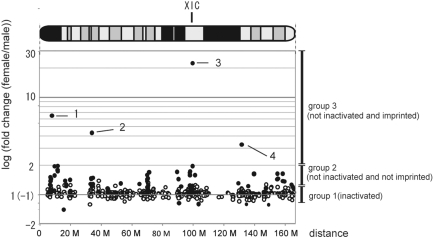
We were also interested in the fact that the differentially expressed 873 transcripts (P < 0.001) were concentrated on the X chromosome (Figure 1A). This suggests that at preimplantation stages this chromosome is not completely inactivated. To understand the positional effect of X inactivation, we plotted the female versus male signal ratio of 1021 distinct X-linked transcripts as a fold change (FC) on a physical map of the X chromosome (Figure 4). All 585 expressed transcripts mapped on chromosome X with a signal intensity >500 (including genes showing no statistically significant expression differences) could be divided into three groups. Group 1 included 512 transcripts showing no marked differences between male and female embryos (−1.2 < FC < + 1.2) indicating that at the blastocyst stage most, but not all, transcripts had undergone X chromosome inactivation. Group 2 contained 63 transcripts that showed greater expression in female than in male embryos; however, their FC values showed <2-fold difference (1.2 < FC < 2.0). These genes probably had not been inactivated at this stage. As far as we know, only six mouse genes are reported to escape X inactivation after implantation (24). Thus, inactivation during preimplantation stages is not considered as strict as at postimplantation stages, possibly because of the reactivation of the X chromosome in the ICM or incomplete inactivation in the TE. At the morula stage there is reported to be a gradient of gene silencing, with the lowest degree of silencing near the chromosome ends and the highest degree of silencing near Xic (1). However, in our experiments the blastocysts did not show such a gradient. Instead, the silencing tended to cover the entire chromosome. Our observation is consistent with a recent report by Patrat et al. (25) on the inactivation of 18 X-linked genes in blastocysts (Supplementary Figure S4). Finally, group 3 contained four transcripts with >2-fold difference between male and female embryos. This group of genes had not been inactivated, but were probably imprinted and predominantly expressed in the female embryos. We concentrated on these and identified three imprinted genes: the Fthl17 group, Rhox5 and Xist. These data demonstrate that genomic imprinting could be utilized for marking differences in gene expression between sexes.
Figure 4.
Genomic imprinting of X-linked genes. Fold change (FC) values of transcripts mapped on the X chromosome. Changes in expression of the transcripts mapped on the X chromosome are plotted along the physical map positions. A set of 586 transcripts showing significant expression levels (greater than 500 units of signal intensity) was selected from 1021 distinct transcripts mapped uniquely on the X chromosome. Black spots correspond to transcripts judged to be differentially expressed between male and female blastocysts at a statistically significant level (P < 0.001) and white spots correspond to others not showing statistically significant differences. Besides groups 1–3 (see ‘Results and Discussion’ section), there were six transcripts showing FC values in the range –1.2 to –1.5, which could be explained by the margin of error in microarray analysis. Spot 1, Fthl17; spot 2, Rhox5; spot 3, Xist; spot 4, Bex1.
Genomic imprinting on X-linked genes
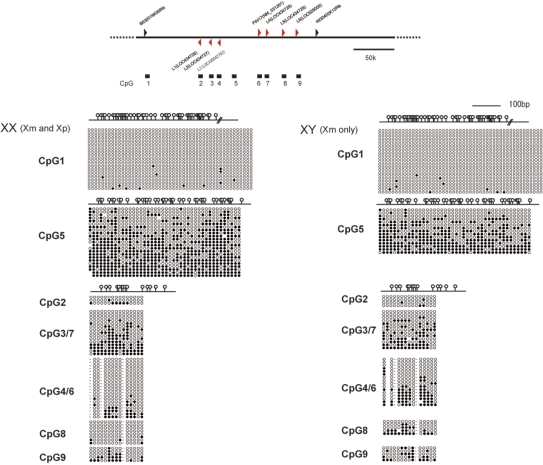
So far, six imprinted genes (our three identified genes and Xlr-3b, -4b and -4c) have been identified on the X chromosome (26,27). However, the mechanism of imprinting on the X chromosome is unknown. For the autosomal chromosomes, differentially methylated regions (DMRs) have been discovered that control imprinted expression. To investigate whether the Fthl17 family of genes is controlled by DMRs, we searched for their sequences for 300 kb around the Fthl17 gene. From this we selected nine areas corresponding to CpG islands and CpG-rich regions and carried out bisulfite sequencing using male and female blastocyst genomic DNA samples. As far as we examined, there was no DMR signal in the Fthl17 locus (Figure 5). However, a control DMR was detected in the H19-imprinted region, indicating that the bisulfite sequencing was working well (Supplementary Figure S5). Although parental allele could not be distinguished using SNPs, these results suggest that imprinting of the Fthl17 family of genes is maintained by a mechanism independent from standard DNA methylation. In addition, it was reported that Xist genes are also not differentially methylated before implantation (28). Therefore, imprinting on X chromosomes in preimplantation embryos could be controlled in general by methylation-independent mechanisms. In general, there are different organizations of the chromatin of the two parental genomes during the first cell divisions (29,30), but it is not fully understood how long this differential chromatin organization remains in later stages. It would be interesting to examine the relationship of this differential chromatin organization and imprinting on the X chromosome in preimplantation-stage embryos. Further searching for DMRs would be necessary in the Fthl17 region, and the precise mechanisms of imprinting remain to be elucidated.
Figure 5.
Methylation analysis of Fthl17 genomic sequences at the blastocyst stage. The upper panel indicates the position of CpG islands and a CpG-rich region in sequences spanning 150 kb either side of Fthl17 (NM_031261). The lower panel indicates the results of bisulfite sequence analysis of male and female blastocyst genomes. Filled ovals indicate methylated CpGs and open ovals indicate unmethylated CpGs. Female samples contained both the paternal (Xp) and maternal (Xm) alleles, whereas male samples contained only the Xm alleles. CpG-2, −3, −4, −6, −7, −8 and −9 are located inside the Fthl17 family of genes.
The role of sex-linked differentially expressed genes in preimplantation embryos
It appears likely that sex-linked differentially expressed genes in preimplantation embryos serve as factors producing sex differences, which are independent of later gonadal differentiation. These factors could involve differences in growth rates and metabolic factors between the sexes. Concerning the growth rate, it has been hypothesized that the paternally derived X chromosome and/or genes expressed from both X chromosomes retard development prior to gonadal sex differentiation (31). Although the growth effects mediated by the X chromosome appear to be complicated, the paternally expressed Fthl17 family of genes (group 3 described above) and X-linked genes that are not inactivated (group 2) could serve as good candidates for the cause of sex differences in the rate of growth. Furthermore, it has been pointed out that these sex differences arising prior to implantation might affect later development and eventually influence the male-to-female ratio at birth (10). Therefore, analyzing these early sex-linked differences will help us to understand other aspects of early embryonic development that may have effects at later stages.
CONCLUSIONS
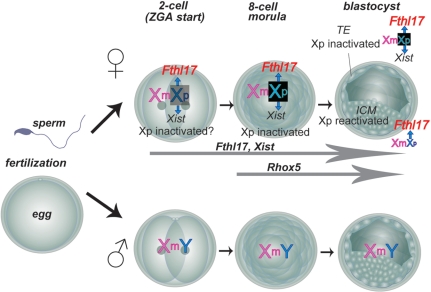
From our screening of genes that are differentially expressed between sexes in preimplantation mouse embryos, we have identified three imprinted genes. Rhox5, a homeobox gene, is predominantly expressed from the eight-cell stage (14). In addition, Xist has been reported to transcribe a non-coding RNA and to be expressed from the two-cell stage (32). Here we identified for the first time the Fthl17 family of genes having a metal ion-binding motif, which is also expressed from the two-cell stage. This confirms that there are already differences in gene expression between sexes at the time of zygotic genome activation (Figure 6). Previously there was scarcely any clue to the control of sex differentiation prior to gonadal differentiation. These X-linked imprinted genes—Fthl17 and Rhox5—offer clues to deciphering sex differentiation controlled by epigenetic gene regulation. Functional analysis of the Fthl17 family of genes using gene-manipulated mice will provide a novel area of investigation on the onset of sexual differentiation before gonadal differentiation.
Figure 6.
Summary of Fthl17 gene family expression in preimplantation stage embryos. The Fthl17 genes are imprinted and expressed from the paternal X chromosome as early as the two-cell stage. A significant difference in gene expression between male and female embryos appears at the time of zygotic genome activation (ZGA).
ACCESSION NUMBERS
The microarray data was deposited in the NCBI database. The Gene Expression Omnibus (GEO) accession number is GSE15611.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Program for Improvement of Research Environment for Young Researchers from Special Coordination Funds for Promoting Science and Technology (SCF) commissioned by MEXT of Japan; a Grant-in-Aid for Scientific Research from The Ministry of Education, Culture, Sports, Science and Technology; the 21st Century COE program from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Mochida Memorial Foundation for Medical and Pharmaceutical Research. Funding for open access charge: Program for Improvement of Research Environment for Young Researchers from Special Coordination Funds for Promoting Science and Technology (SCF) commissioned by MEXT of Japan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank T. Kohda and D. Endo for critical reading of the manuscript.
REFERENCES
- 1.Huynh KD, Lee JT. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature. 2003;426:857–862. doi: 10.1038/nature02222. [DOI] [PubMed] [Google Scholar]
- 2.Mak W, Nesterova TB, de Napoles M, Appanah R, Yamanaka S, Otte AP, Brockdorff N. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303:666–669. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- 3.Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 4.Goodfellow PN, Lovell-Badge R. SRY and sex determination in mammals. Annu. Rev. Genet. 1993;27:71–92. doi: 10.1146/annurev.ge.27.120193.000443. [DOI] [PubMed] [Google Scholar]
- 5.Jost A. Hormonal factors in the sex differentiation of the mammalian foetus. Philos. Trans. Roy. Soc. Lond. B. Biol. Sci. 1970;259:119–130. doi: 10.1098/rstb.1970.0052. [DOI] [PubMed] [Google Scholar]
- 6.Erickson RP. Does sex determination start at conception? Bioessays. 1997;19:1027–1032. doi: 10.1002/bies.950191113. [DOI] [PubMed] [Google Scholar]
- 7.Ray PF, Conaghan J, Winston RM, Handyside AH. Increased number of cells and metabolic activity in male human preimplantation embryos following in vitro fertilization. J. Reprod. Fertil. 1995;104:165–171. doi: 10.1530/jrf.0.1040165. [DOI] [PubMed] [Google Scholar]
- 8.Tiffin GJ, Rieger D, Betteridge KJ, Yadav BR, King WA. Glucose and glutamine metabolism in pre-attachment cattle embryos in relation to sex and stage of development. J. Reprod. Fertil. 1991;93:125–132. doi: 10.1530/jrf.0.0930125. [DOI] [PubMed] [Google Scholar]
- 9.Beyer C, Eusterschulte B, Pilgrim C, Reisert I. Sex steroids do not alter sex differences in tyrosine hydroxylase activity of dopaminergic neurons in vitro. Cell Tissue Res. 1992;270:547–552. doi: 10.1007/BF00645057. [DOI] [PubMed] [Google Scholar]
- 10.Gutierrez-Adan A, Perez-Crespo M, Fernandez-Gonzalez R, Ramirez MA, Moreira P, Pintado B, Lonergan P, Rizos D. Developmental consequences of sexual dimorphism during pre-implantation embryonic development. Reprod. Domest. Anim. 2006;41(Suppl. 2):54–62. doi: 10.1111/j.1439-0531.2006.00769.x. [DOI] [PubMed] [Google Scholar]
- 11.Hadjantonakis AK, Gertsenstein M, Ikawa M, Okabe M, Nagy A. Non-invasive sexing of preimplantation stage mammalian embryos. Nat. Genet. 1998;19:220–222. doi: 10.1038/893. [DOI] [PubMed] [Google Scholar]
- 12.Nakanishi T, Kuroiwa A, Yamada S, Isotani A, Yamashita A, Tairaka A, Hayashi T, Takagi T, Ikawa M, Matsuda Y, et al. FISH analysis of 142 EGFP transgene integration sites into the mouse genome. Genomics. 2002;80:564–574. doi: 10.1006/geno.2002.7008. [DOI] [PubMed] [Google Scholar]
- 13.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS. Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi S, Isotani A, Mise N, Yamamoto M, Fujihara Y, Kaseda K, Nakanishi T, Ikawa M, Hamada H, Abe K, et al. Comparison of gene expression in male and female mouse blastocysts revealed imprinting of the X-linked gene, Rhox5/Pem, at preimplantation stages. Curr. Biol. 2006;16:166–172. doi: 10.1016/j.cub.2005.11.071. [DOI] [PubMed] [Google Scholar]
- 15.Foresta C, Ferlin A, Moro E. Deletion and expression analysis of AZFa genes on the human Y chromosome revealed a major role for DBY in male infertility. Hum. Mol. Genet. 2000;9:1161–1169. doi: 10.1093/hmg/9.8.1161. [DOI] [PubMed] [Google Scholar]
- 16.Mazeyrat S, Saut N, Grigoriev V, Mahadevaiah SK, Ojarikre OA, Rattigan A, Bishop C, Eicher EM, Mitchell MJ, Burgoyne PS. A Y-encoded subunit of the translation initiation factor Eif2 is essential for mouse spermatogenesis. Nat. Genet. 2001;29:49–53. doi: 10.1038/ng717. [DOI] [PubMed] [Google Scholar]
- 17.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes. Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 18.Maclean J.A., 2nd, Chen MA, Wayne CM, Bruce SR, Rao M, Meistrich ML, Macleod C, Wilkinson MF. Rhox: a new homeobox gene cluster. Cell. 2005;120:369–382. doi: 10.1016/j.cell.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 19.Pitman JL, Lin TP, Kleeman JE, Erickson GF, MacLeod CL. Normal reproductive and macrophage function in Pem homeobox gene-deficient mice. Dev. Biol. 1998;202:196–214. doi: 10.1006/dbio.1998.8978. [DOI] [PubMed] [Google Scholar]
- 20.Chuma S, Nakatsuji N. Autonomous transition into meiosis of mouse fetal germ cells in vitro and its inhibition by gp130-mediated signaling. Dev. Biol. 2001;229:468–479. doi: 10.1006/dbio.2000.9989. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto M, Saijoh Y, Perea-Gomez A, Shawlot W, Behringer RR, Ang SL, Hamada H, Meno C. Nodal antagonists regulate formation of the anteroposterior axis of the mouse embryo. Nature. 2004;428:387–392. doi: 10.1038/nature02418. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi S, Uemura H, Kohda T, Nagai T, Chinen Y, Naritomi K, Kinoshita EI, Ohashi H, Imaizumi K, Tsukahara M, et al. No evidence of PEG1/MEST gene mutations in Silver-Russell syndrome patients. Am. J. Med. Genet. 2001;104:225–231. [PubMed] [Google Scholar]
- 23.Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 24.Disteche CM, Filippova GN, Tsuchiya KD. Escape from X inactivation. Cytogenet. Genome Res. 2002;99:36–43. doi: 10.1159/000071572. [DOI] [PubMed] [Google Scholar]
- 25.Patrat C, Okamoto I, Diabangouaya P, Vialon V, Le Baccon P, Chow J, Heard E. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc. Natl Acad. Sci. USA. 2009;106:5198–5203. doi: 10.1073/pnas.0810683106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies W, Isles A, Smith R, Karunadasa D, Burrmann D, Humby T, Ojarikre O, Biggin C, Skuse D, Burgoyne P, et al. Xlr3b is a new imprinted candidate for X-linked parent-of-origin effects on cognitive function in mice. Nat. Genet. 2005;37:625–629. doi: 10.1038/ng1577. [DOI] [PubMed] [Google Scholar]
- 27.Raefski AS, O'Neill MJ. Identification of a cluster of X-linked imprinted genes in mice. Nat. Genet. 2005;37:620–624. doi: 10.1038/ng1567. [DOI] [PubMed] [Google Scholar]
- 28.McDonald LE, Paterson CA, Kay GF. Bisulfite genomic sequencing-derived methylation profile of the Xist gene throughout early mouse development. Genomics. 1998;54:379–386. doi: 10.1006/geno.1998.5570. [DOI] [PubMed] [Google Scholar]
- 29.McLay DW, Clarke HJ. Remodelling the paternal chromatin at fertilization in mammals. Reproduction. 2003;125:625–633. doi: 10.1530/rep.0.1250625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Heijden GW, Dieker JW, Derijck AA, Muller S, Berden JH, Braat DD, van der Vlag J, de Boer P. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech. Dev. 2005;122:1008–1022. doi: 10.1016/j.mod.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 31.Burgoyne PS, Thornhill AR, Boudrean SK, Darling SM, Bishop CE, Evans EP. The genetic basis of XX-XY differences present before gonadal sex differentiation in the mouse. Philos. Trans. Roy Soc. Lond. B. Biol. Sci. 1995;350:253–260. doi: 10.1098/rstb.1995.0159. discussion 260–251. [DOI] [PubMed] [Google Scholar]
- 32.Kay GF, Barton SC, Surani MA, Rastan S. Imprinting and X chromosome counting mechanisms determine Xist expression in early mouse development. Cell. 1994;77:639–650. doi: 10.1016/0092-8674(94)90049-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.