Abstract
Recombinase mediated cassette exchange (RMCE) is a process in which site-specific recombinases exchange one gene cassette flanked by a pair of incompatible target sites for another cassette flanked by an identical pair of sites. Typically one cassette is present in the host genome, whereas the other gene cassette is introduced into the host cell by chemical or biological means. We show here that the frequency of cassette exchange is dependent on the relative and absolute quantities of the transgene cassette and the recombinase. We were able to successfully modify genomic targets not only by electroporation or chemically mediated gene transfer but also by using an adenovirus vector carrying both the transgene cassette to be inserted and the recombinase coding region. RMCE proceeds efficiently in cells in which the adenovirus vector is able to replicate. In contrast, insufficient quantities of the transgene cassette are produced in cells in which the virus cannot replicate. Additional transfection of the transgene cassette significantly enhances the RMCE frequency. This demonstrates that an RMCE system in the context of a viral vector allows the site directed insertion of a transgene into a defined genomic site.
INTRODUCTION
Foreign DNA can be transported into eukaryotic cells by physical, chemical or biological methods (e.g. microinjection, liposomes, electroporation, gene-gun or viral vectors) (1). Stable integration of the foreign DNA will only occur in a small proportion of the cells that have taken up the DNA. Integration of the foreign DNA is mediated by cellular DNA repair enzymes and occurs at random sites of the host genome. If the foreign DNA contains sequences that are identical to the host genome, it can be inserted by homologous recombination in a small fraction of the transduced cells. Retroviral vectors integrate their DNA into the host genome efficiently and murine leukemia virus (MLV) and lentivirus-based vectors are excellent tools for both animal transgenesis and gene therapy (2,3). However, retroviral vectors suffer from some limitations. In the context of animal transgenesis, the limited packaging capacity of retroviral vectors dictates the use of small transgene expression cassettes, which often lack the regulatory elements required for the tissue-specific and abundant expression of the transgene (4). The integration at random sites often places the transgene into an environment that is not supportive of its expression. MLV-based vectors are additionally silenced by epigenetic mechanisms. In the context of gene therapy, retroviral vectors also carry the risk of insertional mutagenesis (2,5). If a transgene or a therapeutic gene would be inserted into the host genome at a pre-selected site, abundant and continuous expression of the foreign DNA could be accomplished in the absence of genome mutagenesis. In principle targeting of a defined site in a eukaryotic genome can be achieved by using either DNA-base pairing or sequence-specific DNA–protein interactions (6). Site-specific recombinases can insert transgenes, introduced into the host cell by chemical, physical or biological means, into defined genomic sites. If the transgene cassette is flanked by a pair of incompatible recombinase target sites, it can be inserted into a genomic target which carries the same arrangement of two incompatible target sites without the introduction of excess plasmid sequences. This process is termed recombinase mediated cassette exchange (7–9). The molar excess of the newly introduced cassette leads to an effective exchange of the cassette in the genome (present in one copy) with the transfected transgene cassette (present as multiple copies per cell). The recombinase target sites themselves can be inserted into the host cell genome at random or by homologous recombination (3).
We show here that the site-specific recombinase Cre is able to mediate integration of a transgene into a predefined genomic site not only in the form of naked DNA but also in the context of a viral vector. However, the integration efficiency is highly dependent on the relative and absolute amounts of transgene DNA and recombinase protein present in the cell. In addition, in order to achieve suitable concentrations of the transgene cassette, the viral vector needs to be replication competent. The experiments shown in here demonstrate that the elements of a recombinase mediated cassette exchange approach can be delivered in the context of a viral vector (i.e. adenovirus) which is regularly used for gene therapy. Whereas adenovirus DNA remains episomal throughout the viral life cycle a transgene embedded in the viral DNA can be mobilized by Cre such that it integrates site-specifically into the host genome. This strategy will pave the way for a safer approach to gene therapy and also for the targeted delivery of transgenes to specific genomic loci in transgenic animals.
MATERIALS AND METHODS
Cells
Human embryonic kidney cells (HEK 293; ECACC No: 85120602) were cultivated in high glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% foetal calf serum, Penicillin/Streptomycin and 2 mM Glutamine. Baby hamster kidney cells (BHK 21; ECACC No: 85011433) were cultivated in minimum essential medium (MEM) supplemented with 10% foetal calf serum, Penicillin/Streptomycin, 2 mM Glutamine, 0.1 mM non-essential amino acids and 1.0 mM sodium pyruvate. HM1 mouse embryonic stem cells (10) and HC11 mouse mammary gland cells (11) were cultivated as described (12). The cell line HM1 RMCE2272-98 has been described before and contains an HPRT selection marker gene inserted into the murine β-casein gene by homologous recombination (13). Cell culture reagents were purchased from Invitrogen, foetal calf serum was purchased from Sigma.
HEK293 and HC11 cells were transfected using the Gene Juice Reagent (Novagen) according to the manufacturer’s recommendations. HM1 embryonic stem (ES) cells were electroporated using a Bio-Rad Gene Pulser using various conditions as indicated in Table 2. HC11 cells were electroporated in a 4 mm cuvette at 250 V and 950 µF. Protein transfections were carried out using the ProteoJuice reagent (Novagen) as suggested by the supplier.
Table 2.
Recombinase mediated cassette exchange efficiency is dependent on electroporation conditions
| Electroporation conditions | Number of colonies | Site-specific integration (%) |
|---|---|---|
| 250 V/960 μF (+Cre) | 221 ± 17 | 100 (12 of 12) |
| 250 V/960 μF (+Cre) | 73 ± 8 | 92 (11 of 12) |
| 250 V/960 μF (+Cre) | 154 ± 7 | 83 (10 of 12) |
| 250 V/960 μF (+Cre) | 68 ± 6 | 92 (11 of 12) |
| 800 V/3 μF (+Cre) | 53 ± 5 | 0 (0 of 12) |
| 800 V/3 μF (+Cre) | 21 ± 2 | 0 (0 of 12) |
| 800 V/3 μF (+Cre) | 46 ± 3 | 0 (0 of 12) |
| 800 V/3 μF (+Cre) | 61 ± 7 | 0 (0 of 12) |
| 800 V/3 μF (−Cre) | 57 ± 3 | nd |
| 800 V/3 μF (−Cre) | 62 ± 6 | nd |
Colony numbers are average values from four cell culture dishes (±SD). Site-specific integration was assessed by PCR as shown in Figure 1. nd: not determined.
All transfections were carried out using identical amounts of plasmid DNA. In control reactions the plasmids pCMV-bgal (Clontech) or pBluescript (Stratagene) were co-transfected.
DNA constructs
The plasmid pBK2272-HPRT was derived from the plasmid pPGK-HPRT (14) from which the 2.9-kb EcoRI insert was excised and inserted into the plasmid pBK-CMV-2272, which carries a pair of incompatible lox sites (loxP and lox2272) at the fringes of its multiple cloning site. pB2272-neo is based on the plasmid pB2272 (13) and carries the same pair of incompatible lox sites at the fringes of its multiple cloning site. The PGK-neo expression cassette is inserted as an EcoRI/HindIII 1.8-kb fragment. The plasmid pB-lox1/2-hyg/luc2 was described previously (15).
The plasmids used for the generation of adenovirus vectors are based on the Stratagene AdEasy system and the system was used as recommended by the supplier. The plasmid pShuttle was used as basis for the generation of vectors carrying (i) a Cre-expression cassette derived from the plasmid pMC1-Cre (16), (ii) a PGK-neo expression cassette derived from pB2272-neo and (iii) in case of the plasmid pShuttle-G5 two copies of the chicken β-globin insulator element (derived as a 2.5-kb EcoRI/BamHI fragment from the plasmid pJC13-1, a gift of Gary Felsenfeld; NIH, Bethesda). Average virus titres were in the range of 106 pfu/ml. Infections were performed at an multiplicity of infection (MOI) of one unless stated otherwise.
The vector pB-bcas7-2272-hytk carries a PGK-hytk (hygromycin-phosphotransferase thymidine kinase fusion gene) expression cassette flanked by a pair of incompatible lox sites (lox2272 and loxP) embedded into the backbone of a mouse β-casein gene. The construct was derived from the plasmids pB-bcas6 and pB2272-hytk (13) by in vitro Cre mediated recombination. In order to generate stable cell clones an 8-kb NheI/SfiI fragment was excised from the vector and electroporated into HEK 293 cells (1 µg of DNA in 1 × 107 cells in a 0.2 cm cuvette at 110 V). Cells were selected in medium containing 100 μg/ml of Hygromycin B.
The plasmid pDSred-mito-2272neo was derived from the Clontech plasmid pDSred2-mito, which was cut with NheI and blunt ended using Escherichia coli DNA polymerase I. The PGK-neo cassette flanked by a tandem pair of lox2272 sites was isolated from pB2272-neo-2272 as a 1.9-kb SacI/KpnI fragment. The insert was also blunt-ended using E. coli DNA polymerase I.
PCR
polymerase chain reaction (PCR) amplifications were done using Taq polymerase from various suppliers. Oligonucleotides were purchased from MWG or Sigma-Genosys. Primer sequences, amplicon size and annealing temperatures are given in Table 1. Template DNA for PCR analyses was isolated as described (12). Real-time PCR amplifications were carried out using the Roche Light-Cycler with Roche reagents as described (17).
Table 1.
Primer combinations used for PCR analysis
| Name | Sequence | Annealing temperature (°C) | Amplicon length (bp) |
|---|---|---|---|
| bcas10 | 5′ GTA ACC ATA AAA CTT CTC CAG GGA CTT GG 3′ | ||
| bcas3 | 5′ AGA GGA TCC GTA AGA CGT CAC CTG CTC ACC 3′ | 55 | 1317 |
| bcas10 | 5′ GTA ACC ATA AAA CTT CTC CAG GGA CTT GG 3′ | ||
| bgalint.1 | 5′ TGT TGG TCA AAG TAA ACG ACA TGG TGA CT 3′ | 55 | 1203 |
| neoint.4 | 5′ GCG CAT CGC CTT CTA TCG CCT TCT TGA C 3′ | ||
| bcas3 | 5′ AGA GGA TCC GTA AGA CGT CAC CTG CTC ACC 3′ | 60 | 1023 |
| PGK1 | 5′ CGA GGC CCG GCA TTC TGC ACG C 3′ | ||
| CMVseq.1 | 5′ GGA CTT TCC AAA ATG TCG TA 3′ | 50 | 2941 |
| PGK5 | 5′ AAG CGC ATG CTC CAG ACT GCC TTG GGA AA 3′ | ||
| CMVseq.1 | 5′ GGA CTT TCC AAA ATG TCG TA 3′ | 51 | 421 |
| PGK5 | 5′ AAG CGC ATG CTC CAG ACT GCC TTG GGA AA 3′ | ||
| pBKpA | 5′ GCT ATT GCT TTA TTT GTA ACC ATT A 3′ | 52 | 622 |
| hytk1 | 5′ AGA GCT GCA TCA GGT CGG AGA CGC TGT CG 3′ | ||
| bcas3 | 5′ AGA GGA TCC GTA AGA CGT CAC CTG CTC ACC 3′ | 57 | 1147 |
| bcas6 | 5′ TAA GGG CCA GAG TAG ATC 3′ | ||
| hytk2 | 5′ TCC TGG ATT ACG ACC AAT CG 3′ | 48 | 215 |
| bcas6 | 5′ TAA GGG CCA GAG TAG ATC 3′ | ||
| PGK5 | 5′ AAG CGC ATG CTC CAG ACT GCC TTG GGA AA 3′ | 50 | 936 |
| neoint.4 | 5′ GCG CAT CGC CTT CTA TCG CCT TCT TGA C 3′ | ||
| pBKpA | 5′ GCT ATT GCT TTA TTT GTA ACC ATT A 3′ | 51 | 1074 |
| neoint.2 | 5′ CCA GTC ATA GCC GAA TAG CCT CTC CAC CC 3′ | ||
| pBKpA2 | 5′ TTC ACT GCA TTC TAG TTG TGG TTT GTC 3′ | 56 | 1106 |
| DSred-1R | 5′ AGC GCA TGA ACT CGG TGA TGA C 3′ | ||
| CMVseq.1 | 5′ GGA CTT TCC AAA ATG TCG TA 3′ | 59 | 529 |
Virus preparation
Adenovirus was prepared using the Stratage AdEasy system as recommended by the supplier. The virus titre was determined using a TCID50 protocol.
Immuno-histochemistry
Cells were seeded onto coverslips in a 24-well plate and transfected with plasmid DNA and/or protein. 48 h post-transfection, medium was removed and the cells were washed in PBS, fixed in 2% para-formaldehyde for 20 min and then washed again three times with PBS. Cells were blocked in 3% BSA in 1xTBST overnight at 4°C. An MBP-specific rabbit antiserum (New England Biolabs) was added at a 1 : 200 dilution in 1xTBST and incubated at room temperature for 2 h. The cells were washed three times for 10 min in 1× TBST. Then the second antiserum (goat-anti-rabbit FITC linked) was added at a dilution of 1:100 and the cells on the coverslips were incubated at room temperature for 2 h in the dark. Cells were then washed as before. The cover-slips were mounted in a mounting medium containing DAPI (4′,6-diamidino-2-phenylindole; Vector Labs) and photographed at a magnification of 100-fold on a Leica fluorescent microscope.
RESULTS
The Cre recombinase system can be used to insert genes at predefined sites in the mammalian genome which have been tagged with a lox target site. This has been demonstrated in a number of cell types and also (albeit at reduced efficiency) in fertilized mouse oocytes (7–9). We have utilized the Cre recombinase to insert genes into the murine β-casein gene with a view to expressing these genes in the milk of transgenic animals (4,18–20). Using recombinase mediated cassette exchange for this approach allows the target specific insertion of a transgene in a defined orientation without introducing any excess vector sequences. In order to express proteins effectively in the milk of transgenic animals, we have chosen the β-casein gene as a target site which is able to equip an inserted transgene with all required regulatory elements to allow for abundant expression of the transgene in the lactating mammary gland (3,18).
Electroporation conditions determine the frequency of site-specific recombination
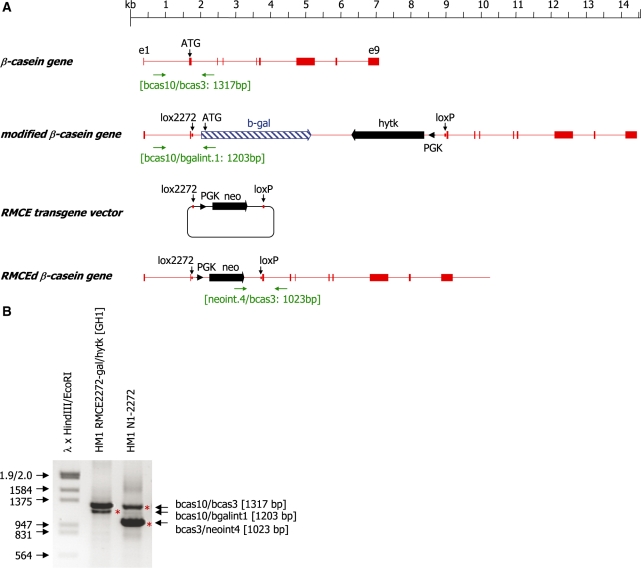
Electroporation is the method of choice for the transfer of DNA into embryonic stem cells. However, different electroporation conditions have been published for the successful DNA transfer into mouse ES cells (21,22). We therefore tried to delineate the parameters which would best support site-specific recombination in HM1 ES cells. We set out to analyse the precise electroporation conditions which provide the optimum recombination frequency. A pair of incompatible lox sites (loxP and lox2272) in conjunction with a β-galactosidase open reading frame and a hytk (hygromycin-phosphotransferase thymidine kinase fusion gene) selection marker expression cassette was inserted into the second exon of the β-casein gene in HM1 embryonic stem cells using sequential homologous and site-specific recombination (13). The ATG of the β-casein gene was deleted during that process, such that the ATG of β-galactosidase is the first translation start codon in a chimeric mRNA initiating at the first exon of the β-casein gene (Figure 1). The resulting cell line HM1 RMCE2272-gal/hytk (GH1) was then transfected with the plasmid pB2272-neo together with a two-fold excess of a Cre expression vector. The cells were selected in medium containing 200 µg/ml G418. Under these conditions cells which have incorporated the selection marker gene at random sites or by site-specific recombination will survive. We found that, surprisingly, the frequency of site-specific integration was heavily dependent on the electroporation conditions used (Table 2). At 250 V and 960 µF almost all of the colonies selected carried a site-specific insertion of the neo gene at the β-casein locus. In contrast, transfections carried out at 800 V and 3 µF, although generating the same number of resistant colonies, showed no site-specific insertion of the neo gene into the predefined β-casein target site (Table 2).
Figure 1.
(A) Schematic representation of the murine β-casein gene and its derivatives after homologous recombination and RMCE. Exons of the β-casein gene are indicated as solid boxes, the neomycin (neo) and hytk selection marker genes are indicated as solid arrows, respectively and the β-galactosidase gene (β-gal) is indicated as a hatched arrow. The PGK promoter elements directing expression of the selection marker genes are indicated as black arrowheads. The positions of the lox2272 and loxP sites and the translational start codon (ATG) are marked by vertical arrows. The primer binding sites (horizontal arrows) used for genotyping and the sizes of the expected PCR products are indicated. (B) PCR analysis of genomic DNA isolated from the cell clones HM1 RMCE2272-gal/hytk (GH1), and the cell clone HM1 N1-2272 derived from it. A 1317-bp band is detected in both samples and represents the unmodified β-casein allele. HM1 RMCE2272-gal/hytk (GH1) cells carry an insertion of a β-galactosidase open reading frame and a PGK-hytk expression cassette at one of the β-casein alleles as indicated by the occurrence of a 1203-bp PCR product. Cell clone HM1 N1-2272 was derived after an RMCE event which exchanged the β-gal and hytk genes for the neo selection marker gene. The correct modification is indicated by the generation of a 1023-bp PCR product and the concomitant loss of the 1203-bp band. Phage λ DNA digested with HindIII and EcoRI was used as molecular weight marker.
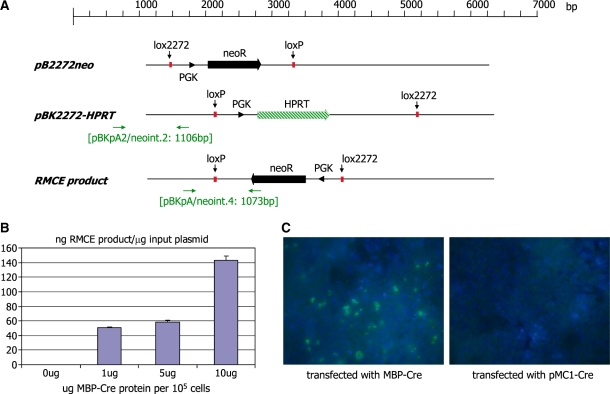
As we suspected that the amount of Cre protein expressed under these different electroporation conditions may be critical for the frequency of the RMCE reaction, we analysed the impact of the Cre protein concentration on the recombinase mediated cassette exchange (RMCE) frequency (Figure 2). HEK 293 cells were transfected with the plasmids pB2272-neo and pBK2272-HPRT. RMCE between the two plasmids can readily be detected and quantified by PCR (Figure 2A). Subsequently, varying amounts of MBP-Cre protein (23) were transfected into the cells using the Proteo-Juice reagent and the RMCE frequency was determined by real-time PCR. The results demonstrate that the RMCE frequency is indeed directly correlated with the concentration of Cre protein present in the cell. If 10 μg of MBP-Cre protein are tranfected per 105 cells, 14% of the transfected DNA undergo RMCE (Figure 2B). MBP-Cre protein can be detected by immuno-histochemistry in around 5–7% of the cells (Figure 2C). As transfection of DNA and protein will not have occurred necessarily in the same cells, it is possible that the maximum frequency of RMCE may be higher than 14%. Alternatively, as the RMCE reaction will reach an equilibrium of the different recombination products (which can be formed via either or both of the lox sites), the 14% RMCE product detected may represent the equilibrium state of the RMCE reaction.
Figure 2.
(A) Schematic representation of the plasmids pB2272-neo and pBK2272-HPRT. The neomycin resistance marker genes (neo) are indicated as solid arrows. The PGK promoter is indicated as an arrowhead. The HPRT selection marker gene is indicated as a striped arrow. The primer binding sites (horizontal arrows) used for genotyping and the sizes of the expected PCR products are indicated. (B) Quantitative PCR analysis of recombinase mediated cassette exchange in HEK 293 cells. Cells were transfected with equimolar amounts of the plasmids pBK2272-HPRT and pB2272-neo. Twenty four hours post-plasmid transfection the cells were transfected with different amounts of an MBP-Cre fusion protein using the Proteo-Juice reagent (Novagen). DNA was isolated from cells 36 h later and analysed by real-time PCR using the primer pair neoint.2/pBKpA2 (yielding a 1106-bp fragment) to quantify the concentration of the input plasmid pBK-2272HPRT and the primer pair neoint.4/pBKpA (yielding a 1073-bp fragment) to quantify the concentration of RMCE product generated. The concentrations are presented as pg RMCE product per ng input plasmid in correlation with the amount of MBP-Cre protein transfected into 1 × 105 cells. (C) Immunohistochemistry of cells transfected with the plasmids pB2272-neo and pBK2272-HPRT and the protein MBP-Cre (or the plasmid pMC1-Cre; right panel). The MBP section of the protein was detected using a 1 : 200 dilution of an MBP-specific rabbit antiserum and a goat-anti-rabbit FITC-linked secondary antiserum.
Chemical-mediated gene transfer supports RMCE
Most researchers have used electroporation to mediate DNA transfection in the context of RMCE experiments (8,24,25). We wanted to determine whether electroporation per se is a significant stimulus for the efficient integration of DNA via RMCE or whether (in our hands) a chemical (rather than a physical) method for DNA transfer would work as well as electroporation. We therefore used a chemical DNA transfer methods (Gene Juice, Novagen) to mediate site-specific insertion of the transgene cassette.
As HM1 ES cells do not display high transfection frequencies with the Gene Juice reagent, this experiment was carried out in a derivative of the mouse mammary gland cell line HC11 cells termed HC11-bcas-F9 (18). The cells carry the same modification as the HM1 ES cells used above and were transduced with the plasmids pB2272-neo and pMC1-Cre using the Gene Juice reagent and, in parallel, by electroporation. Seventeen percent of the selected cell colonies carried the correct site-specific integration of the marker gene into the β-casein gene when the cells were transfected using Gene Juice. Electroporation with the same construct lead to 28% of the cells carrying the correct integration. The absolute number of resistant cell colonies was similar for both techniques (Table 3), however, addition of Cre to the transfection mixture decreased the number of resulting selection resistant colonies when the cells were transfected with Gene Juice and increased the number of resistant colonies when the cells were electroporated. This suggests that the transfection method can indeed have an influence on the relative frequency of random integration and recombinase mediated integration. Gene Juice mediated transfections may present the DNA in a way that is more amenable to random insertion than DNA brought into the cell by electroporation. This in turn may be dependent on the different biochemical environments the DNA is exposed to in the two DNA transfer methods. However, these results also demonstrate that transfection methods other than electroporation can be used successfully to bring about recombinase mediated cassette exchange.
Table 3.
Recombinase mediated cassette exchange efficiency after liposome transfection and electroporation in two experiments conducted in parallel
| Selection | pMC1-Cre | Colonies Gene Juice transfection (1.5 pg DNA per cell) | Colonies electroporation (2 pg DNA per cell) |
|---|---|---|---|
| G418 | (−) | 178, 131 | 78, 40 |
| G418 | (+) | 42, 38 (3 of 17 positive, 18%) | 178, 155 (7 of 25 positive, 28%) |
| G418/Ganc. | (−) | 133, 146 | 54, 78 |
| G418/Ganc. | (+) | 71, 87 | 208, 218 |
pMC1-Cre was co-transfected with the plasmid pB2272-neo in a 3 : 1 ratio.
The recombinase expression cassette and the transgene cassette can be delivered in one plasmid
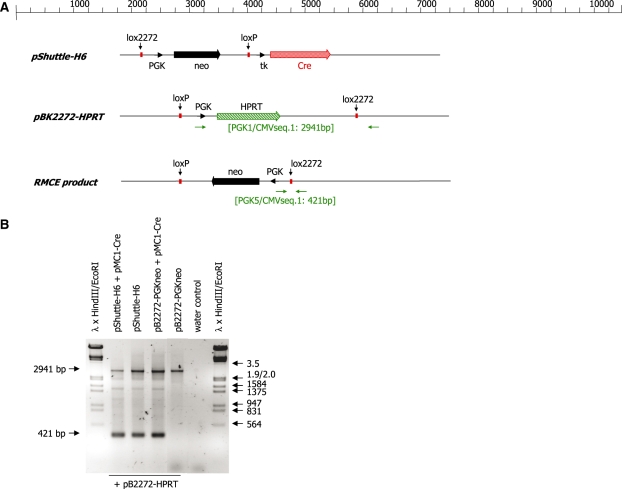
We then wanted to analyse the efficiency with which the recombinase and the transgene cassette mediate RMCE reactions in the context of a biological gene transfer vehicle i.e. a viral vector. Transfections aimed at achieving recombinase mediated cassette exchange typically utilize an excess of Cre expression plasmid over the plasmid carrying the transgene construct (13,26–28). In the experiments described above a 2- or 3-fold excess of Cre expression construct was used. In the context of a viral vector carrying both the transgene and the recombinase gene, the genes would be present at an equimolar ratio. In order to test whether such a construct would support site-specific recombination of a transgene, a plasmid carrying the neomycin resistance marker cassette derived from pB2272-neo (flanked by a pair of incompatible lox sites) was joined to a Cre expression cassette in the context of the plasmid pShuttle (Stratagene) (Figure 3A). First, the construct was co-transfected into BHK cells together with an acceptor plasmid (pBK2272-HPRT) carrying the same pair of lox sites. As shown in Figure 3B the plasmid (termed pShuttle-H6) supported site-specific recombination with the acceptor plasmid in the presence or absence of any additional Cre expression vector (see lanes pShuttle-H6 and pShuttle-H6 + pMC1-Cre) as demonstrated by the occurrence of the indicative 421-bp PCR product in a transient transfection.
Figure 3.
(A) Schematic representation of the plasmids pShuttle-H6 and pBK2272-HPRT. The neomycin resistance marker genes (neo) are indicated as solid arrows. The Cre open reading frame is indicated as a shaded arrow. The PGK and the tk promoter are indicated as arrowheads. The HPRT selection marker gene is indicated as a striped arrow. The primer binding sites (horizontal arrows) used for genotyping and the sizes of the expected PCR products are indicated. (B) PCR analysis of DNA isolated from HEK 293 cells transfected with the indicated plasmids. The 2941-bp product is generated from the non-recombined pBK2272-HPRT plasmid. The 421-bp PCR product is indicative of a recombinase mediated cassette exchange between the PGKneo cassette (derived from plasmid pB2272-neo) and the PGK-HPRT cassette. Phage λ DNA digested with HindIII and EcoRI was used as molecular weight marker.
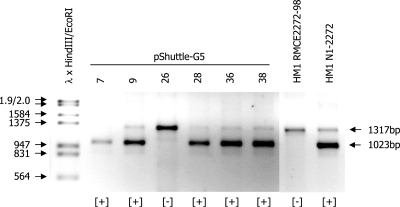
The construct pShuttle-H6 was then transfected into HM1-2272-98 cells (13). These derivatives of HM1 cells carry a modified β-casein locus incorporating an HPRT selection marker gene flanked by the loxP/lox2272 pair of Cre target sites. Interestingly, none of the G418 resistant cell clones derived from this transfection carried a site-specific insertion of the neo cassette at the β-casein gene even if electroporations were carried out under the conditions which had previously lead to a high efficiency of RMCE (cf. Table 1). In the plasmid pShuttle-H6 the neo expression cassette and the Cre expression cassette are placed in tandem. A corresponding plasmid in which the orientation of the neo expression cassette was reversed also failed to yield any G418 resistant cell clones which carried a site-specific insertion after transfection into HM1 RMCE2272-98 cells (data not shown). We have previously shown that expression cassettes in close vicinity can interfere with each others expression (15). Therefore the vector was modified by the insertion of two copies of the chicken β-globin insulator between the Cre and neo expression cassettes. The resulting vector (pShuttle-G5, Figure 5A) was again transfected into HM1-2272 98 cells and G418 resistant colonies were derived. This time the vast majority of stable cell clones (85%) had taken up the neo cassette by site-specific recombination (as shown for representative clones in Figure 4). This confirms that it is possible to provide the Cre expression cassette and the transgene which is to be integrated into the host genome in a single contiguous DNA segment. The results also demonstrate that vector design is a critical factor determining the efficiency of RMCE.
Figure 5.
(A) Schematic representation of the linear plasmids pShuttle-G5, pBK2272-HPRT and the expected recombination product. The neomycin resistance marker genes (neo) are indicated as solid arrows. The Cre open reading frame is indicated as a shaded arrow. The PGK and the tk promoter are indicated as arrowheads. The copies of the β-globin insulator element (INS) are indicated as vertically striped boxes. The positions of the lox2272 and loxP sites are marked by vertical arrows. (B) PCR analysis of DNA isolated from HEK 293 and BHK cells (as indicated) transfected/infected with pBK2272-HPRT plus the indicated plasmids and virus vectors. The 622-bp PCR product is generated from the non-recombined pBK2272-HPRT plasmid. The 421-bp PCR product (derived from the primer pair PGK5/CMVseq.1) is indicative of a recombinase mediated cassette exchange between the PGK-neo cassette and the PGK-HPRT cassette. Phage λ DNA digested with HindIII and EcoRI was used as molecular weight marker.
Figure 4.
PCR analysis using the primer combination bcas3, bcas10 and neoint.4 on DNA isolated from representative cell clones derived after electroporation of HM1 RMCE2272-98 cells with the plasmid pShuttle-G5 and selection of the transfected cells in medium containing 200 µg/ml of G418. The 1317-bp band is detected in all samples and represents the unmodified β-casein allele. Cell clones modified by an RMCE event, which has inserted the PGK-neo selection marker cassette, display an additional 1023-bp PCR product. Phage λ DNA digested with HindIII and EcoRI was used as molecular weight marker.
An adenovirus vector can mediate successful RMCE
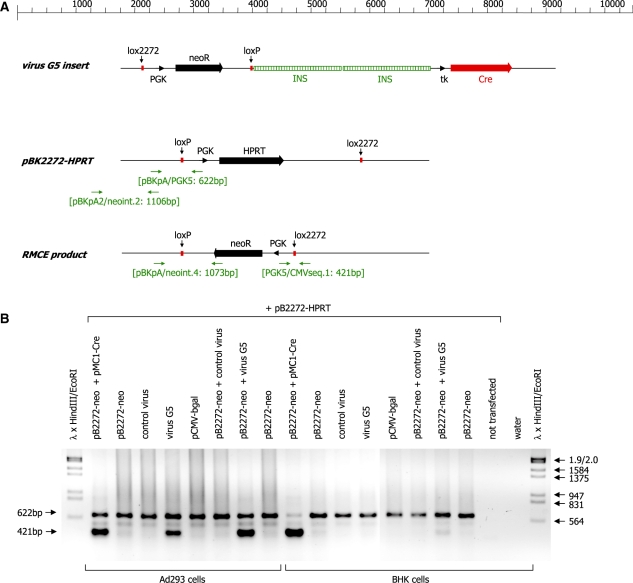
We therefore established an adenovirus vector based on the plasmid pShuttle-G5 and tested its ability to support site-specific recombination in HEK 293 and BHK cells. The G5 virus could be amplified to a typical titer of around 1 × 106 pfu/ml. This is significantly lower than the titer we obtained for the control virus encoding the β-galactosidase gene, which yields a titer in excess of 1 × 108 pfu/ml in our hands. We suspect that this is due to the fact that the size of the total insert is ∼7 kb, which is close to the maximum packaging capacity of the AdEasy vector system (7.5 kb), which carries deletions in the E1 and E3 genes. When infected with the β-galactosidase control adenovirus BHK and HEK 293 cells at an MOI of 10, essentially all cells become infected, demonstrating that both cell types are susceptible to adenovirus infection (data not shown).
HEK 293 cells carry segments of the adenovirus genome which are deleted from the recombinant virus and complement the viral vector to allow virus replication. BHK cells, in contrast, do not support viral replication. We analysed whether the transgene cassette present in the adenovirus vector can be mobilized such that it integrates site-specifically into an acceptor plasmid. HEK 293 and BHK cells were transiently transfected with the acceptor plasmid pBK2272-HPRT and subsequently infected with the adenovirus vector G5 or the control virus. As a positive control the cells were also transfected with the plasmid pMC1-Cre. In order to determine whether the availability of the mobilizable transgene cassette is a limiting factor, in a parallel reaction the infected cells were also transfected with the plasmid pB2272-neo, which corresponds exactly to the cassette present in the viral vector. DNA was isolated from the transfected and infected cells 24 h post-infection and analysed by PCR with the primer combination: CMVseq.1/PGK5/pBKpA. In the absence of recombination a PCR product of 622 bp is generated by the primer pair pBKpA/PGK5, whereas successful recombination is evidenced by the occurrence of a 421-bp product derived from primer pair PGK5/CMVseq.1 (Figure 5A).
As shown in Figure 5B transfection of HEK 293 cells with the acceptor plasmid pBK2272-HPRT, the donor plasmid pB2272-neo and the Cre expression plasmid pMC1-Cre leads to the generation of a 421-bp product indicative of a successful site-specific recombination. The same product is also generated when the cells are infected with the adenovirus G5 irrespective of whether the donor plasmid pB2272-neo was co-transfected. This indicates that in this assay format the G5 virus is capable of expressing sufficient amounts of Cre and to act as donor of the 2272-neo cassette. The PCR analysis shown in Figure 5B analyses the 3′ end of the recombination event (Figure 5A). Analysis of the 5′ end of the recombination event confirms these results (data not shown). BHK cells in contrast which do not allow replication of the adenovirus vector do only show the band indicative of an RMCE event after co-transfection of the plasmids pB2272-neo and pMC1-Cre. In addition, there is a faint band detectable in the cells which have been transfected with the plasmid pB2272-neo and infected with the G5 virus.
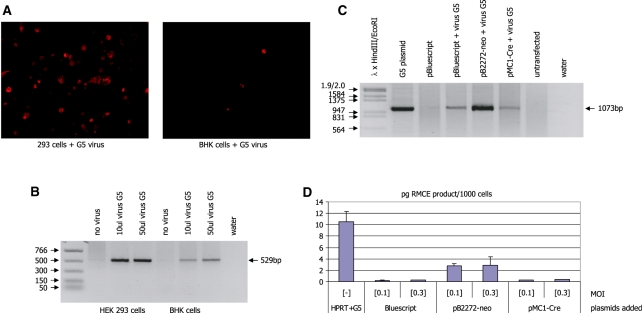
In order to assess whether the abundance of Cre or the neo-RMCE donor cassette is limiting for the RMCE reaction two further experiments were carried out. First, HEK 293 cells and BHK cells were transfected with the plasmid pDSred-mito-2272neo. This plasmid carries a PGKneo cassette flanked by two tandem lox2272 sites inserted between the CMV promoter and the open reading frame of the red fluorescent protein DSred-mito gene. The neo-cassette prevents activation of the DSred-mito gene. Cre removes the neo-cassette and the ensuing gene expression can be detected as red fluorescent staining in the mitochondria of the cells. Infection of the G5 virus was able to induce DSred-mito gene expression both in HEK 293 and BHK cells even though expression was much more abundant in HEK 293 where virus replication is possible (Figure 6A). This is confirmed by PCR using the primer pair DSred-1R/CMVseq.1 which yields a 529-bp product indicative of a site-specific recombination event in both cell types. The product can be detected in HEK 293 and BHK cells (albeit at a lower intensity) indicating that there is sufficient Cre protein present in the BHK cells to support recombination.
Figure 6.
(A) Red fluorescent staining in HEK 293 and BHK cells transfected with the plasmid pDSred-mito-2272-PGKneo and infected with the G5 adenovirus. Red staining of the mitochondria indicates the presence of the plasmid and the virus-derived Cre protein in the same cell. (B) PCR analysis of DNA derived from the cells in A using the primer combination DSred1R/CMVseq.1. Recombination between the two identical lox2272 sites in the plasmid, which is a prerequisite for the activation of the red fluorescent protein is indicated by the occurrence of a 529-bp band (marked by the arrow). (C) PCR analysis of DNA isolated from HEK293 cells transfected with the indicated plasmids and infected with adenovirus G5 at an MOI of 0.1. RMCE is detected using the primer pair pBKpA/neoint.4 which yields an indicative 1073 bp product (marked by the arrow). (D) Real-time PCR analysis of DNA isolated from HEK293 cells transfected with the indicated plasmids and infected with adenovirus G5 at an MOI of 0.1 or 0.3 (as indicated). RMCE is detected using the primer pair PGK5/CMVseq.1 (as in Figure 5) which yields an indicative 421-bp product. The amount of PCR product is shown as pg per 1000 cells.
Subsequently, we used a lower concentration of the G5 virus (MOIs of 0.1 and 0.3) in HEK 293 cells to assess whether under these conditions additional transfection of a Cre-expression plasmid (pMC1-Cre) or the RMCE plasmid pB2272-neo would augment RMCE. As shown in Figure 6C the addition of extra mobilizable transgene cassette DNA significantly enhances RMCE frequency (lane pB2272-neo + virus G5; Figure 6C). No significant difference is observed if the pMC1-Cre plasmid is transfected in addition to infection with the virus G5 (Figure 6C). Real-time PCR analysis confirms that there is a 14-fold increase in RMCE product when the plasmid pB2272-neo containing the transgene cassette is transfected in addition to the G5 infection in HEK 293 cells (Figure 6D). In contrast, transfection of the plasmid pMC1-Cre in addition to the virus infection only augments the occurrence of the RMCE product by 1.4-fold.
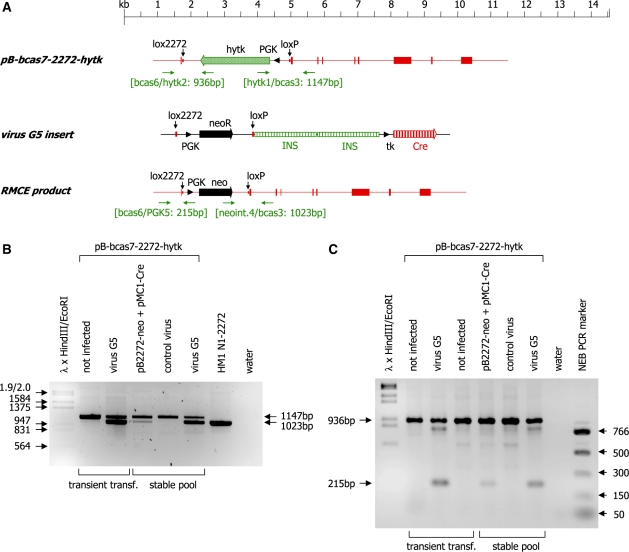
We then assessed whether the adenoviral vector would also be able to support integration of the neo-selection marker cassette from the virus into a target site embedded into the genome. We therefore generated a plasmid carrying a modified β-casein gene (pB-cas7-2272-hytk), which can serve as a target for RMCE, and transfected it stably into HEK 293 cells (Figure 7A). The stably transfected cells were subsequently infected with the virus G5 or the control virus. Genomic DNA was isolated 24 h post-infection and analysed by PCR. The primer combination bcas3/hytk1/neoint.4 was used to detect recombination at the 3′ end of the integrated cassette. A 1147-bp PCR product (amplified by the primer pair bcas3/hytk1) is detected in the unmodified β-casein gene, whereas a 1028-bp product (amplified by the primer pair bcas3/neoint.4) is indicative of a β-casein gene modified by RMCE (Figure 7B). The primer combination bcas6/hytk2/PGK5 was used to assess recombination at the 5′ end of the integrated cassette. A 936-bp PCR product (amplified by the primer pair bcas6/hytk2) is detected in the unmodified β-casein gene, whereas a 215-bp product (amplified by the primer pair bcas6/PGK5) is indicative of a β-casein gene modified by RMCE (Figure 7C). Recombination was readily detected in HEK 293 cells transiently or stably transfected with the pB-cas7-2272-hytk plasmid indicating that the G5 virus is able to serve as both transgene donor and source of Cre expression, mediating stable insertion of a transgene cassette into a predefined genomic site in HEK 293 cells. Comparison of the relative intensities of the PCR products representing the stably integrated β-casein gene and the RMCE product demonstrates that infection with the G5 virus is significantly more efficient in mediating RMCE than transfection of the plasmids pB2272-neo and pMC1-Cre.
Figure 7.
(A) Schematic representation of the plasmid pB-bcas7-2272-hytk, the insert of the virus vector G5 and the resulting RMCE product. Exons of the β-casein gene are indicated as shaded boxes, the neomycin and hytk selection marker genes are indicated as solid and shaded arrows, respectively and the Cre ORF (Cre) is indicated as a vertically striped arrow. The PGK promoter elements directing expression of the selection marker genes are indicated as black arrowheads. The positions of the lox2272 and loxP sites are marked by vertical arrows. The primer binding sites (horizontal arrows) used for genotyping and the sizes of the expected PCR products are indicated. (B) PCR analysis of DNA isolated from HEK 293 cells transiently or stably transfected with pB-bcas7-2272-hytk. The cells were either co-transfected with the indicated plasmids or infected with the indicated virus vectors 24 h post-transfection. The 3′ end of the RMCE reaction was analysed using the primer combination bcas3/neoint.4/hytk1. Successful recombination at the loxP site is indicated by the presence of the 1023-bp PCR product. Non-recombined DNA yields a PCR product of 1147 bp. Phage λ DNA digested with HindIII and EcoRI was used as molecular weight marker. (C) The 5′ end of the RMCE reaction was analysed using the primer combination bcas6/PGK5/hytk2. Successful recombination at the lox2272 site is indicated by the presence of the 215-bp PCR product. Non-recombined DNA yields a PCR product of 936 bp. Phage λ DNA digested with HindIII and EcoRI and the NEB PCR marker (New England Biolabs) were used as molecular weight markers.
DISCUSSION
The goal for genome modification in gene therapy and transgenesis is that genome alterations can be introduced rapidly and accurately. Cre recombinase has proved a useful tool for genome modifications due to its high activity in mammalian cells (29) and its high degree of DNA sequence specificity. We show here that Cre-mediated cassette exchange is highly dependent on electroporation conditions and vector design. Our results confirm our expectations that a single contiguous segment of DNA which carries both, the Cre expression cassette and a mobilizable transgene cassette, is able to mediate stable integration of the transgene into a genomic target. However, the presence of an insulator element between the two genes is essential for efficient recombination to take place. The requirement for the insulator element may suggest that transcription of the gene cassettes can interfere with Cre mediated recombination. Alternatively, transcription of the neo cassette may reduce expression of the neighboring Cre recombinase gene thus resulting in a diminished level of site-specific recombination. We and others have demonstrated that adjacent transcription cassettes can silence each other (15,30).
An adenovirus vector which acts both as transgene donor and source of Cre expression is able to mediate site-specific transgene integration into extra-chromosomal and chromosomal target sites. However, the vector is only efficient in cells which allow adenovirus vector replication. This is largely due to the limiting amounts of transgene cassette that can be generated in an infected cell which does not support virus replication. This can be concluded from the finding that augmenting the transgene cassette by transfection significantly increases the RMCE frequency, whereas augmenting the Cre-expression cassette does not. The limiting amounts of transgene cassette may also be responsible for the different RMCE frequencies obtained with different electroporation conditions. The conditions which strongly favor random integration over RMCE may not allow sufficient quantities of transgene DNA to enter the cell, whereas the conditions which strongly favor RMCE over random integration do.
E1 gene deleted adenovirus vectors do not allow a significant degree of DNA replication in cells in which the E1 gene product is not supplied exogenously. E1 encodes the earliest viral gene product and is essential for the early steps of virus DNA replication. This may limit the total amount of viral DNA that is produced in an infected cell. Therefore alternative adenovirus vector designs may be used to enhance the concentration of viral DNA in the infected cell.
Adenovirus vectors have been used successfully to mediate integration of transgene cassettes using transposable elements (e.g. sleeping beauty and L1) demonstrating the usefulness of combining viral vectors with enzymes mediating gene integration (albeit non-site-specific) to improve transgene expression (31,32).
It has been shown that the specificity of site-specific recombinases can be modulated such that they recognize sites existing in the human and animal genomes (33,34). We show in here that an adenovirus vector containing a Cre expression cassette and a transgene cassette can mediate site-specific gene integration into a genomic target site. In combination with Cre mutants of altered site-specificity and viral vectors whose inability to multiply in host cells is restricted at the step of packaging rather than DNA replication this approach may lead to vectors which allow site-directed insertion of genes into mammalian genomes. This approach can therefore be applied to the insertion of transgenes encoding recombinant proteins in animals and may also have implications for similar approaches in gene therapy. This system can also be applied to any other recombinase system including the ΦC31 recombinase (33,35,36) and the λ integrase (37), which mediate the site-specific integration of transgenes into the host genome.
FUNDING
The Scottish Executive Environment and Rural Affairs Department (ROAME 31190); the BBSRC (Gene Technologies underpinning Health Care project, grant no 12599); the Hannah Development Fund and the Genomia Seed Fund. Funding for open access charge: Rowett Institute of Nutrition and Health, University of Aberdeen, core funding.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to Dr Colin Wilde for helpful discussions. They thank Dr Gary Felsenfeld, NIH, Bethesda for the chicken β-globin insulator plasmid.
REFERENCES
- 1.Sorrell DA, Kolb AF. Targeted modification of mammalian genomes. Biotechnol. Adv. 2005;23:431–469. doi: 10.1016/j.biotechadv.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Sandrin V, Russell SJ, Cosset FL. Targeting retroviral and lentiviral vectors. Curr. Top. Microbiol. Immunol. 2003;281:137–178. doi: 10.1007/978-3-642-19012-4_4. [DOI] [PubMed] [Google Scholar]
- 3.Coates CJ, Kaminski JM, Summers JB, Segal DJ, Miller AD, Kolb AF. Site-directed genome modification: derivatives of DNA-modifying enzymes as targeting tools. Trends Biotechnol. 2005;23:407–419. doi: 10.1016/j.tibtech.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Kolb AF. Genome engineering using site-specific recombinases. Cloning Stem Cells. 2002;4:65–80. doi: 10.1089/153623002753632066. [DOI] [PubMed] [Google Scholar]
- 5.Sadelain M. Insertional oncogenesis in gene therapy: how much of a risk? Gene Ther. 2004;11:569–573. doi: 10.1038/sj.gt.3302243. [DOI] [PubMed] [Google Scholar]
- 6.Kolb AF, Coates CJ, Kaminski JM, Summers JB, Miller AD, Segal DJ. Site-directed genome modification: nucleic acid and protein modules for targeted integration and gene correction. Trends Biotechnol. 2005;23:399–406. doi: 10.1016/j.tibtech.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Waterhouse P, Griffiths AD, Johnson KS, Winter G. Combinatorial infection and in vivo recombination: a strategy for making large phage antibody repertoires. Nucleic Acids Res. 1993;21:2265–2266. doi: 10.1093/nar/21.9.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouhassira EE, Westerman K, Leboulch P. Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood. 1997;90:3332–3344. [PubMed] [Google Scholar]
- 9.Shmerling D, Danzer CP, Mao X, Boisclair J, Haffner M, Lemaistre M, Schuler V, Kaeslin E, Korn R, Burki K, et al. Strong and ubiquitous expression of transgenes targeted into the beta-actin locus by Cre/lox cassette replacement. Genesis. 2005;42:229–235. doi: 10.1002/gene.20135. [DOI] [PubMed] [Google Scholar]
- 10.Magin TM, McWhirJ, Melton DW. A new mouse embryonic stem cell line with good germ line contribution and gene targeting frequency. Nucleic Acids Res. 1992;20:3795–3796. doi: 10.1093/nar/20.14.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ball RK, Friis RR, Schoenenberger CA, Doppler W, Groner B. Prolactin regulation of beta-casein gene expression and of a cytosolic 120-kd protein in a cloned mouse mammary epithelial cell line. EMBO J. 1988;7:2089–2095. doi: 10.1002/j.1460-2075.1988.tb03048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolb AF, Ansell R, McWhir J, Siddell SG. Insertion of a foreign gene into the beta-casein locus by Cre-mediated site-specific recombination. Gene. 1999;227:21–31. doi: 10.1016/s0378-1119(98)00607-6. [DOI] [PubMed] [Google Scholar]
- 13.Kolb AF. Selection-marker-free modification of the murine beta-casein gene using a lox2272 site. Anal. Biochem. 2001;290:260–271. doi: 10.1006/abio.2000.4984. [DOI] [PubMed] [Google Scholar]
- 14.Kolb AF, Ansell R, McWhir J, Siddell SG. Insertion of a foreign gene into the beta-casein locus by Cre-mediated site-specific recombination. Gene. 1999;227:21–31. doi: 10.1016/s0378-1119(98)00607-6. [DOI] [PubMed] [Google Scholar]
- 15.Kolb AF, Siddell SG. Genomic targeting of a bicistronic DNA fragment by Cre-mediated site-specific recombination. Gene. 1997;203:209–216. doi: 10.1016/s0378-1119(97)00515-5. [DOI] [PubMed] [Google Scholar]
- 16.Gu H, Zou YR, Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 17.Flint DJ, Boutinaud M, Whitelaw CB, Allan GJ, Kolb AF. Prolactin inhibits cell loss and decreases matrix metalloproteinase expression in the involuting mouse mammary gland but fails to prevent cell loss in the mammary glands of mice expressing IGFBP-5 as a mammary transgene. J. Mol. Endocrinol. 2006;36:435–448. doi: 10.1677/jme.1.01873. [DOI] [PubMed] [Google Scholar]
- 18.Robinson C, Kolb AF. Analysis of mammary specific gene locus regulation in differentiated cells derived by somatic cell fusion. Exp. Cell Res. 2009;315:508–522. doi: 10.1016/j.yexcr.2008.10.029. [DOI] [PubMed] [Google Scholar]
- 19.Kolb AF. Engineering immunity in the mammary gland. J. Mammary Gland Biol. Neoplasia. 2002;7:123–134. doi: 10.1023/A:1020395701887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolb AF. The prospects of modifying the antimicrobial properties of milk. Biotechnol. Adv. 2001;19:299–316. doi: 10.1016/S0734-9750(01)00069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao L, Zabel C, Herrmann M, Nolden T, Mertes F, Magnol L, Chabert C, Hartl D, Herault Y, Delabar JM, et al. Proteomic shifts in embryonic stem cells with gene dose modifications suggest the presence of balancer proteins in protein regulatory networks. PLoS ONE. 2007;2:e1218. doi: 10.1371/journal.pone.0001218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dennie MT, Patricia AL. Electroporation of Murine Embryonic Stem Cells: A Step-by-Step Guide. Stem Cells. 2004;22:243–249. doi: 10.1634/stemcells.22-3-243. [DOI] [PubMed] [Google Scholar]
- 23.Kolb AF, Siddell SG. Genomic targeting with an MBP-Cre fusion protein. Gene. 1996;183:53–60. doi: 10.1016/s0378-1119(96)00470-2. [DOI] [PubMed] [Google Scholar]
- 24.Araki K, Araki M, Yamamura K.-I. Site-directed integration of the cre gene mediated by Cre recombinase using a combination of mutant lox sites. Nucleic Acids Res. 2002;30:e103. doi: 10.1093/nar/gnf102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soukharev S, Miller JL, Sauer B. Segmental genomic replacement in embryonic stem cells by double lox targeting. Nucleic Acids Res. 1999;27:e21. doi: 10.1093/nar/27.18.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Araki K, Araki M, Miyazaki J, Vassalli P. Site-specific recombination of a transgene in fertilized eggs by transient expression of Cre recombinase. Proc. Natl Acad. Sci. USA. 1995;92:160–164. doi: 10.1073/pnas.92.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Araki K, Araki M, Yamamura K. Targeted integration of DNA using mutant lox sites in embryonic stem cells. Nucleic Acids Res. 1997;25:868–872. doi: 10.1093/nar/25.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Araki K, Imaizumi T, Okuyama K, Oike Y, Yamamura K. Efficiency of recombination by Cre transient expression in embryonic stem cells: comparison of various promoters. J. Biochem. (Tokyo) 1997;122:977–982. doi: 10.1093/oxfordjournals.jbchem.a021860. [DOI] [PubMed] [Google Scholar]
- 29.Andreas S, Schwenk F, Kuter-Luks B, Faust N, Kuhn R. Enhanced efficiency through nuclear localization signal fusion on phage PhiC31-integrase: activity comparison with Cre and FLPe recombinase in mammalian cells. Nucleic Acids Res. 2002;30:2299–2306. doi: 10.1093/nar/30.11.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eszterhas SK, Bouhassira EE, Martin DI, Fiering S. Transcriptional interference by independently regulated genes occurs in any relative arrangement of the genes and is influenced by chromosomal integration position. Mol. Cell. Biol. 2002;22:469–479. doi: 10.1128/MCB.22.2.469-479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yant SR, Ehrhardt A, Mikkelsen JG, Meuse L, Pham T, Kay MA. Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat. Biotechnol. 2002;20:999–1005. doi: 10.1038/nbt738. [DOI] [PubMed] [Google Scholar]
- 32.Soifer HS, Kasahara N. Retrotransposon-adenovirus hybrid vectors: efficient delivery and stable integration of transgenes via a two-stage mechanism. Curr. Gene Ther. 2004;4:373–384. doi: 10.2174/1566523043346084. [DOI] [PubMed] [Google Scholar]
- 33.Sclimenti CR, Thyagarajan B, Calos MP. Directed evolution of a recombinase for improved genomic integration at a native human sequence. Nucleic Acids Res. 2001;29:5044–5051. doi: 10.1093/nar/29.24.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buchholz F, Stewart AF. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat. Biotechnol. 2001;19:1047–1052. doi: 10.1038/nbt1101-1047. [DOI] [PubMed] [Google Scholar]
- 35.Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl Acad. Sci. USA. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorpe HM, Wilson SE, Smith MC. Control of directionality in the site-specific recombination system of the Streptomyces phage phiC31. Mol. Microbiol. 2000;38:232–241. doi: 10.1046/j.1365-2958.2000.02142.x. [DOI] [PubMed] [Google Scholar]
- 37.Christ N, Droge P. Genetic manipulation of mouse embryonic stem cells by mutant lambda integrase. Genesis. 2002;32:203–208. doi: 10.1002/gene.10031. [DOI] [PubMed] [Google Scholar]