Abstract
To explore the higher order structure of transcribable chromatin in vivo, its local configuration was assessed through the accessibility of the chromatin to crosslinking with formaldehyde. The application of crosslinked and mildly sheared chromatin to sedimentation velocity centrifugation followed by size-fractionation of the DNA enabled us to biochemically distinguish between chromatin with heavily versus sparsely crosslinkable structures. The separated fractions showed a good correlation with gene expression profiles. Genes with poor crosslinking around the promoter region were actively transcribed, while transcripts were hardly detected from genes with extensive crosslinking in their promoter regions. For the inducible gene, Il2, the distribution of the promoter shifted in the gradient following T-cell receptor stimulation, consistent with a change in structure at this locus during activation. The kinetics of this switch preceded the chromatin change observed in a DNase I accessibility assay. Thus, this new chromatin fractionation technique has revealed a change in chromatin structure that has not been previously characterized.
INTRODUCTION
Ultracentrifugation employing a sucrose density gradient has been previously utilized for the analysis of chromatin structure (1). This technique separates molecules based on both their size and shape, with large dense structures migrating faster than small extended ones. When applied to enzymatically digested native chromatin, fragments containing loci such as the β-globin gene were found to sediment slower if they were isolated from tissues expressing the gene than from tissues that were not (2,3). This sedimentation difference was attributed to a change in the openness of the chromatin structure surrounding the gene which accompanies the onset of transcription. We were interested in applying this technique to do more fine structure analysis of chromatin changes around gene promoters. The technique as described, however, requires large pieces of chromatin to obtain significant shifts in velocity sedimentation. In order to obtain better resolution, we developed a significant modification that involves chemical crosslinking of the chromatin with formaldehyde followed by mild shearing. This traps a portion of the small chromatin fragments in faster sedimenting larger fractions, but only when the protein–DNA structure is dense enough to allow extensive crosslinking. The rest of the small fragments behave like short nucleosome arrays and sediment more slowly in the gradient. Subsequent reversal of the crosslinking by heat, followed by agarose gel electrophoresis of extracted DNA, allowed isolation of small pieces of DNA of comparable size (<1 kb) for a quantitative PCR analysis of promoter fragment distribution in the original gradient. This new chromatin fractionation assay has allowed us to detect dynamic changes in chromatin structure around the promoter of the interleukin 2 gene following T-cell activation.
MATERIALS AND METHODS
Preparation and activation of mouse T cells
All T-cell populations used in this report were derived from B10.A, TCR-5C.C7 transgenic, Rag-2 deficient mice (female, 5–8 weeks old) (4). Total splenocytes from eight mice, containing both naïve T cells and APC, were cultured for 3 days in 75 ml of complete medium (CM: RPMI1640 with 10% FCS, 2 mM glutamine, 50 µM 2-ME, and antibiotics) supplemented with 2 µM of the cytochrome c antigenic peptide (4). The activated T cells were expanded into 450 ml of fresh CM with 10 units/ml of recombinant murine IL-2 (Peprotech), and cultured for another 2 weeks of expansion and resting. For (re)activation of these cells, we used six-well plastic dishes (#3506, Costar) pre-coated with 10 µg/ml of anti-CD3ε Ab (clone 145-2C11, BD Biosciences) and a 1 : 2500 dilution of an ascites preparation of an anti-CD28 Ab (clone 37.51). Ten million cells in 1 ml CM were put into each well of a six-well plate pre-coated with both Abs.
RT–PCR
cDNA was synthesized from total RNA using Superscript III Reverse Transcriptase plus random primers (Invitrogen), and purified on a MinElute spin column (Qiagen) following treatment with RNase A plus RNase H and extraction with phenol/chloroform. The amount of cDNA was quantified with an OligoGreen Kit (Invitrogen). For the conventional PCR (Figure 3), 250 pg of cDNA was applied to a single reaction of PCR, which ran 38 cycles. As a positive control, PCR with 2.5 ng of mouse genomic DNA was also performed. For the qPCR (Supplementary Figure S5), 25 pg of cDNA was applied to one reaction using a QuantiFast SYBR Green PCR Kit (Qiagen) in a real time PCR machine ABI7900HT (Applied Biosystems). The primers used are shown in Supplementary Table S1.
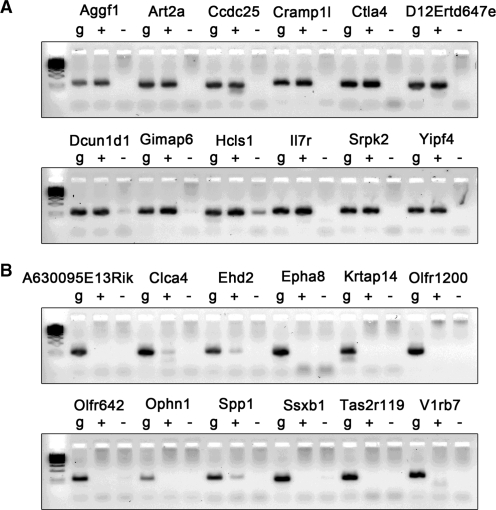
Figure 3.
The expression profiles of the genes selected from the SEVENS assay. (A) RT–PCR was performed for the genes enriched in fraction 2 (shown with bold letters in Table 1). (B) RT–PCR was performed for the genes enriched in fraction 18 (shown with bold letters in Table 2). Because the primers for PCR were designed to anneal to a coding region with a single exon, genomic DNA (g) was utilized as a positive control for the PCR reaction. The plus (+) or minus (–) character means a PCR reaction following cDNA synthesis with or without RT, respectively.
Sedimentation velocity method followed by normalization in the size of the DNA (the SEVENS assay)
A schema of this assay is shown in Figure 1. Mouse T cells (2 × 107) or livers that were perfused with PBS and ground on Parafilm (67–74 mg) were first crosslinked with 1 ml of 0.75% formaldehyde in PBS for 10 min at room temperature to preserve chromatin structure. Similar results were obtained with 1% formaldehyde, but the final DNA recovery was lower. The crosslinking reaction was stopped by the addition of 0.125 mM glycine. The cells were then washed with ice-chilled PBS twice, and solubilized with 250 µl of SDS lysis buffer [SLB: 1% SDS, 50 mM Tris–HCl (pH 8.0) and 10 mM EDTA, Complete Protease Inhibitor Cocktail (#4693124, Roche)]. The crosslinked soluble chromatin was sonicated for 5 s using a Fisher Dismembrator Model 100 (at level ‘2’) to yield a wide MW size range of the DNA (0.1 to >20 kb). The lysates were then spun down briefly (700g, for 5 min) to remove debris, and layered onto a 9.5 ml sucrose gradient (6–40%) in 1.1% Triton X-100, 0.01% SDS, 16.7 mM Tris–HCl (pH 8.0), 1.2 mM EDTA, 167 mM NaCl, and Complete Protease Inhibitor Cocktail in a polyallomer centrifuge tube (#331374, Beckman). Ultracentrifugation was then executed at 38 krpm for 2.5 h at 4°C using a Beckman swing rotor SW40Ti. Samples before centrifugation must be kept at room temperature in order to avoid crystallizing the SDS. Nineteen 0.5-ml fractions were collected from the gradient by picking them up from top to bottom using a micropipette. A 25-µl aliquot of each fraction was pooled as a mixed sum of all fractions (Sum). The rest of each fraction, as well as the Sum, were heated at 65°C overnight to reverse the crosslinking, and then treated with RNase A and Proteinase K, followed by extraction with phenol/chloroform. After precipitation by ethanol supplemented with 10 µg of glycogen, the DNA purified from each fraction or the Sum was loaded onto a 2% low melting point agarose gel in a Tris–acetate–EDTA (TAE) buffer and the gel cut to collect DNA ranging in size from 0.4–0.5 kb. A few experiments were also done with DNA of approximately 1 kb in size with comparable results. A MinElute spin column was utilized to purify the DNA from the gel block, and the recovered DNA was checked for size distribution using a SYBR green I-stained agarose gel (Figure 1D). For analyses using qPCR, 250 pg of DNA from each fraction or the Sum, which was quantified with a PicoGreen Kit (Invitrogen), was added to a qPCR reaction. The fold enrichment of the specific DNA sequence in each fraction relative to the amount in the Sum was then calculated, and the results are plotted as the log2 value versus fraction number. Primers for the specific DNA sequences used in these assays are shown in Supplementary Table S1. For microarray analyses, the recovered DNA was amplified once using a GenomePlex WGA2 Kit (Sigma), and applied to a mouse 385K MM8 RefSeq promoter array (full service by Roche NimbleGen).
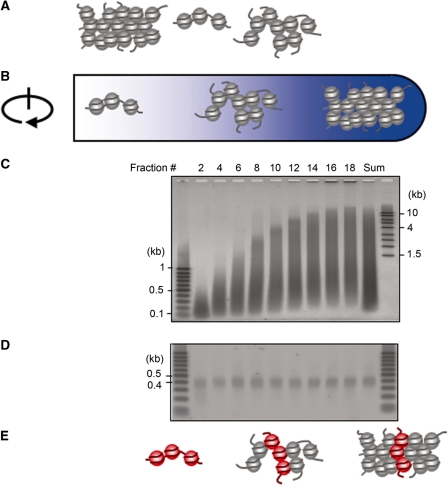
Figure 1.
The schema of the SEVENS assay. (A) Chromatin is crosslinked with formaldehyde and sonicated mildly, (B) Chromatin is fractionated by size in a sucrose density gradient, (C) The DNA is purified from each fraction as well as an equal mixture of all the fractions (Sum), and then separated by length in an agarose gel, (D) DNA ranging in length from 0.4 to 0.5 kb (covering 2–3 nucleosomes) is recovered from the gel and (E) A qPCR is performed. The fold enrichment of the region of interest (the amount in each fraction/the amount in the Sum) is calculated. When longer DNA (e.g. 0.8–0.9 kb) was tested, similar results were obtained (data not shown).
DNase accessibility assay
Four million T cells were lysed with 1 ml of 0.2% Nonidet P40 in ice-cold nuclear isolation buffer [NIB; 7.5 mM Tris–HCl (pH 7.5), 7.5 mM NaCl, 2.5 mM MgCl2, 30 mM KCl, 0.05 mM EGTA, 0.5 mM DTT, Complete Protease Inhibitor Cocktail] plus 10% sucrose. The lysate was layered on 2 ml of NIB supplemented with 0.05% Nonidet P40 and 40% sucrose, and subjected to centrifugation at 40 000g at 4°C for 20 min. The nuclei in the pellet were washed twice with ice-cold NIB containing 0.05% Nonidet P40, 10% sucrose and 10% glycerol. The pellet was then re-suspended with 500 µl of DNase digestion buffer [10 mM Tris–HCl (pH 7.5), 15 mM NaCl, 2.5 mM MgCl2, 0.5 mM CaCl2, 0.5 mM DTT, 0.05% Nonidet P40, 10% sucrose]. The suspension was divided into aliquots of 160 µl. After pre-warming at 37°C for 5 min, the nuclei in each aliquot were treated with 10 units of DNase I (New England Biolabs) at 37°C for 30 min. Non-digested nuclei were also prepared. Next, SDS was added to the suspension at a final concentration of 1%. DNA was extracted from the nuclei with phenol/chloroform at least four times following treatment with proteinase K, and precipitated with ethanol plus 10 µg of glycogen. The DNA pellet was suspended with 1× NE Buffer 2 (New England Biolabs) and treated with RNase A and the Rsa I restriction enzyme to optimize recovery for PCR (this four-base cutter does not digest the regions amplified in the subsequent PCR). DNA was purified in a MinElute spin column, and quantified using a PicoGreen Kit. For qPCR, 1.25 ng of DNase-treated or non-treated DNA was subjected to a single reaction. The DNase accessibility of the specific promoter sequence was expressed as a percentage of resistance to digestion, which is calculated from the amount of the DNase-treated DNA relative to that of the non-treated DNA. The primers used are shown in Supplementary Table S1.
RESULTS
Profiles of the DNA and proteins recovered from sedimentation velocity centrifugation fractions
For preparation of chromatin for this new method, mouse T cells were first treated with 0.75% formaldehyde to preserve chromatin structure (Figure 1). This crosslinking step allowed chromatin to be solubilized with a strong ionic detergent, SDS. The suspension was then mildly sonicated to generate DNA fragments distributed from 0.1 to >20 kb. These were observed by agarose gel electrophoresis following loading of the DNA after reverse crosslinking of each fraction (Figure 1C). The ‘Sum’ is a pool of equal aliquots of each fraction showing the size distribution and amounts of the sheared fragments in the total recovered sample, excluding the pellet. Ultracentrifugation on the sucrose density gradient yielded 19 chromatin fractions, which were collected from the top to the bottom of a centrifuge tube using a micropipette. For this report, only the even numbered fractions were analyzed. Figure 1C shows that larger DNA (>4 kb) was observed running mostly in the lower fractions of the gradient (fractions 12–18), while shorter DNA (<1 kb) was found throughout all fractions of the gradient. This is very different from what was observed in previous reports using non-crosslinked chromatin, where shorter DNA was not observed at all in the lower fractions (2,3,5). A microscopic examination using DNA-staining with Hoechst dye showed that particles found in the bottom fraction (fraction 18) were indeed larger than those in fraction 10 (Supplementary Figure S1A and B). Co-staining with anti-histone H3 identified many of these particles as chromatin (Supplementary Figure S1C). We conclude from these experiments that the gradient fractionation took place mainly on the basis of chromatin size.
When proteins in each fraction of the gradient were examined by PAGE, histone molecules were detected as the major proteins found throughout all the fractions (Supplementary Figure S2). In contrast, other proteins routinely found in heterochromatin structures, such as polycomb or methyl CpG-binding proteins, were mostly located in the upper fractions of the gradient when analyzed by immunoblotting (Supplementary Figure S3). These latter proteins appear to be crosslinked in either small chromatin fragments, which are sedimenting slowly or not crosslinked at all. Therefore, nucleosomes are most likely to be the major contributors to the large chromatin structures sedimenting quickly in the sucrose gradient, although we cannot rule out the contribution of small amounts of transcription factors binding at individual DNA promoters. Furthermore, some of the crosslinking could arise as a result of higher order structures such as scaffolds that form during chromatin looping.
Because these crosslinkable structures (such as neighboring nucleosomes) must be close to each other for the small crosslinker formaldehyde to work, the frequency of effective crosslinking should reflect the local density of the chromatin. Our shearing conditions, however, produce fragments of varying length from 0.1 to >20 kb (Figure 1C). In order to compare the frequency of heavily versus lightly crosslinked chromatin around a given promoter, we needed to isolate the trapped small promoter fragments from the large partially sheared pieces of DNA that also harbor the promoter, because the large fragments could contain the promoter in either crosslinked state. In fact, when the chromatin recovered from fraction 18 was sheared again and applied to a second sucrose density gradient, additional small pieces of DNA moved into the slowly sedimenting fractions (data not shown). To circumvent this problem, we recovered trapped DNA only ranging in size from 0.4–0.5 kb by de-crosslinking the DNA from each gradient fraction and loading it onto an agarose gel. A pooled ‘Sum’ containing an equal aliquot from each recovered fraction was also prepared and size-fractionated in the same manner. Note that our sonication conditions sheared 0.9–1.4% of the soluble DNA found in the Sum to 0.4–0.5 kb fragments, from which the promoter regions of an active gene, Actb (β-actin), and a silent gene, Bdnf, found in unactivated T cells were recovered at similar levels (6.3–6.7%) (see next section for further discussion of these genes). Finally, the fold enrichment of a specific DNA sequence in each gradient fraction relative to the amount in the Sum was calculated using a microarray analysis or a quantitative PCR (qPCR) analysis as described below. Thus, the assay we have developed seems to be a good strategy for examining chromatin structure at high spatial resolution. We have designated this method the SEVENS assay (sedimentation velocity centrifugation followed by normalization in the size of the DNA).
Correlation of the gradient distribution with the gene expression profile
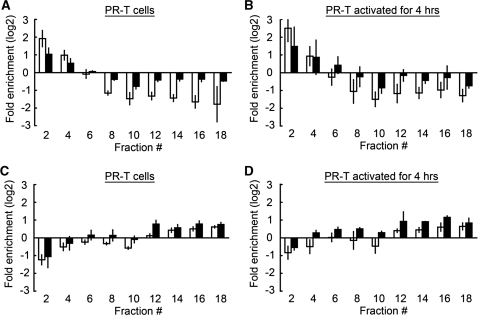
In order to evaluate the validity of this new assay, we assessed the fractional distribution of promoter regions spanning the transcription start site (TSS) of the active genes, Actb and Cd3d (CD3δ) in mouse T cells. We performed qPCR to detect these promoters in each fraction and calculated the fold-enrichment (using a log2 scale) compared to the pooled sum of all fractions. As shown in Figure 2A, the promoters of these two genes in pre-activated resting T cells were preferentially enriched in the slowly sedimenting fractions of the gradient and excluded from the faster sedimenting fractions. These patterns were also observed in T cells after activation for 4 h using anti-CD3 and anti-CD28 antibodies (Figure 2B). This suggests that there were fewer crosslinkable molecules in the vicinity of these active promoters resulting in smaller sized particles resistant to sedimentation. Despite the fact that β-actin mRNA is expressed 100-fold more than that of CD3δ (data not shown), both promoters showed a similar fractional distribution in the gradient, although the quantitative enrichment in the slowly sedimenting fractions did appear to be somewhat greater for Actb in both resting and 4-h-activated PR-T cells. Next, we examined promoters of two repressed genes, Bdnf and Adad1. These two promoters in mouse T cells were distributed in a reverse pattern to that of the active genes (Figure 2C and D). Their promoter regions accumulated predominantly in the faster sedimenting fractions of the gradient, suggesting that in T cells these regions were composed mostly of heavily crosslinked chromatin, which sedimented faster because of its larger size. The difference between active and repressed genes did not appear to be due to the number of nucleosomes at each promoter as there was no correlation with the results in a chromatin immunoprecipitation (ChIP) assay using an anti-pan-histone H3 mAb (Supplementary Figure S4). For example, comparable levels of histone H3 were found at the Cd3d and Bdnf promoters. This suggests that the number of wrapped nucleosomes is unlikely to be the sole determinant for the structural effects seen in the SEVENS assay.
Figure 2.
The SEVENS assays for active and repressed control genes. The resting (A and C) and 4 h-activated (B and D) mouse T cells were applied to the SEVENS assay. (A and B) The promoter region of the active genes, Actb (open columns) and Cd3d (closed columns), were enriched in the slowly sedimenting fractions of the gradient and excluded from the faster fractions. (C and D) The promoter region of the repressed genes, Bdnf (open columns) and Adad1 (closed columns), was distributed in a reverse pattern from that of the active genes.
In order to determine how extensive the inverse correlation was between transcribable chromatin and accessibility to formaldehyde crosslinking, a whole genome analysis using a promoter microarray (NimbleGen MM8 RefSeq array) was performed. The DNA recovered from fraction 2 or the Sum as a reference was labeled with Cy5 or Cy3, respectively, and then hybridized to the array. The relative intensity of the Cy5 signal to the Cy3 reference in one or more of three independent experiments showed enrichment in the slowly sedimenting fraction of DNA from around the proximal promoter and TSS (from −300 to +300 bp) of 993 loci (Supplementary Table S2). Because the Actb promoter could not be detected in this assay, the analysis using the microarray would seem to be less sensitive than that using qPCR. Thirty nine genes that were enriched in this fraction in all three experiments are listed in Table 1, among which were the ubiquitously expressed histone genes themselves (Hist1h2be, Hist1h3d and Hist1h3f) and linage-specific genes such as the T-cell surface molecule Ctla4. This suggests that the proximal promoter of these active genes consists of fewer crosslinkable structures. In contrast, when the DNA recovered from the fastest sedimenting fraction (fraction 18) was tested in the array, an enrichment of the proximal promoter regions of 1297 genes was observed (Supplementary Table S3). The list in Table 2 shows 57 of those genes that were strongly enriched in this fraction in all three of the experiments. In this group there were 20 Olfr (olfactory receptor) genes, which are not expressed at all in T cells. This suggests that repressed genes are composed of structures in which their promoter regions are more readily crosslinked by formaldehyde.
Table 1.
Genes enriched in Fraction 2
| 2700007P21Rik 5930403N24Rik Aggf1 AI451617 Arhgap15 Art2a Art2b Ccdc25 Ccdc28b Ccdc77 Chm Cramp1l Ctla4 D12Ertd647e Dcun1d1 Dnajb4 Gimap6 Hcls1 Hist1h2be Hist1h3d Hist1h3f Hsd11b1 Il7r Lilrb4 Pip5kl1 Ptgds2 Ptp4a2 Rabggtb Srpk2 Tmem115 Tmem69 Tnrc6b Tomm40l Tubgcp5 Vps24 Vps33b Wdr59 Yipf4 Zfp143 |
Expression of the genes with bold letters was tested in Figure 3A.
Table 2.
Genes enriched in Fraction 18
| 1700010D01Rik 4930433I11Rik 4930583K01Rik A630095E13Rik BC023179 Cfl1 Clca4 Crtam Dcir2 Dnaja4 Ehd2 Epha8 Fbxl19 Gzmg Krtap14 Krtap16-3 Krtap16-8 LOC384813 LOC432867 Ly86 MGC118210 Myo18a Olfr1084 Olfr1097 Olfr1197 Olfr1200 Olfr1240 Olfr1311 Olfr1337 Olfr1408 Olfr295 Olfr358 Olfr484 Olfr547 Olfr589 Olfr617 Olfr642 Olfr733 Olfr749 Olfr782 Olfr784 Olfr860 Oog1 Ophn1 Pira3 Pira6 Pla2g2d Spp1 Srbd1 Ssxb1 Ssxb2 Tas2r119 Tmem98 Ube2q Ugt1a1 V1rb7 V1rg10 |
Expression of the genes with bold letters was tested in Figure 3B.
To verify the relationship between transcription and chromatin structure seen in the SEVENS assay, RT–PCR analyses were also performed. As shown in Figure 3A, mRNAs for all 12 genes selected from the list in Table 1 were detected at a similar level to that of the control genomic DNA (see the figure legend). In contrast, the PCR for 12 genes from Table 2 did not show many mRNA signals, although there was a low level of expression for the Clca4, Ehd2 and Spp1 genes (Figure 3B). These observations confirm that the extent of formaldehyde crosslinking at the promoter is correlated with transcriptional activity.
Conversion of chromatin structure at the Il2 promoter to a less crosslinkable form following T-cell activation
When T cells are activated with anti-CD3 and anti-CD28 antibodies, the Il2 (interleukin 2) gene, which encodes a well-known growth and differentiation factor for the T cell itself (6), begins to be transcribed (Supplementary Figure S5). To acquire direct evidence for the correlation between chromatin structure seen in the SEVENS assay and active transcriptional status, we investigated the Il2 promoter region during its induction using qPCR after the SEVENS fractionation. We first confirmed that comparable levels of extraction of the Il2 promoter occurred into the chromatin fraction of the Sum consisting of 0.4–0.5 kb DNA. Before T-cell activation this value was 3.6% of the total soluble chromatin and after 3 h of activation 3.9%. When the SEVENS assay was performed with unactivated T cells, the Il2 promoter region (PCR position: −77 to +39) was evenly distributed throughout all fractions of the gradient (Figure 4A). This is in contrast to the constitutively active genes (Actb and Cd3d) and the non-transcribed genes (Bdnf and Adad1) (Figure 2). Because the distribution of the Il2 promoter in the liver, where IL-2 is not expressed, was similar to that of the repressed control genes examined in T cells (Figure 4B), the possibility that the Il2 locus was somehow intrinsically different from that of all other loci in its behavior in the SEVENS assay was ruled out. Thus, the Il2 locus can assume a readily crosslinkable conformation in tissues where it is not expressed, and interestingly seems to assume an intermediate (poised) state following T-cell differentiation. This may facilitate more rapid IL-2 production once T-cell activation ensues.
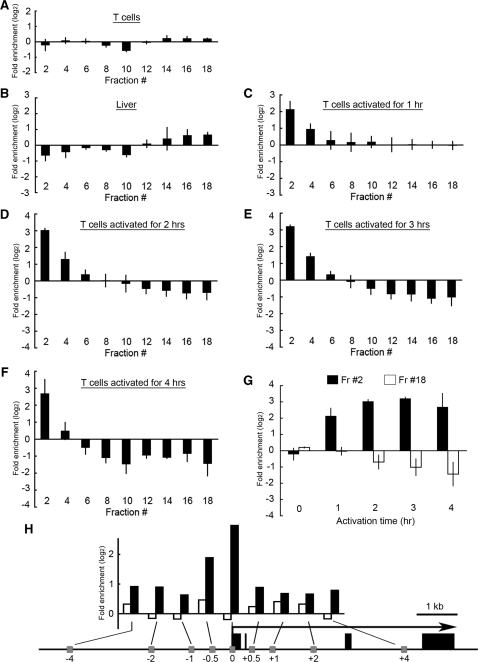
Figure 4.
SEVENS assays for the Il2 promoter region in mouse T cells and liver. (A–F) Chromatin from unactivated (A), 1-h (C), 2-h (D), 3-h (E), or 4-h (F) activated T cells, or liver (B), were run in the SEVENS assay. The fractional distribution of the Il2 promoter (PCR position: −77 to +39) in the sucrose gradient is shown. (G) The fold enrichments of the Il2 promoter in fraction 2 (closed columns) or fraction 18 (open columns) are summarized from the results in unactivated and hourly-activated T cells. (H) The SEVENS assay for unactivated (open columns) or 3-h activated PR-T cells (closed columns) was performed to assess the fold enrichment in fraction 2 of the regions upstream or downstream of the Il2 TSS, which are shown in the lower panel. The numbers denote the relative position to the TSS and the black boxes denote exons.
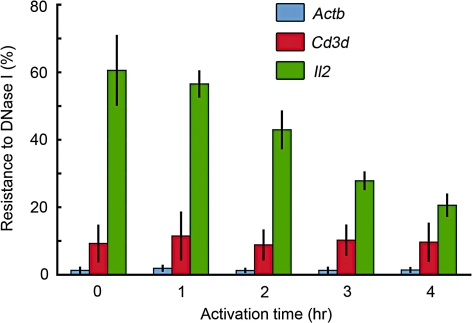
In order to look for chromatin structural changes around the Il2 promoter following T-cell stimulation, we examined activated T cells with the SEVENS assay at 1-h intervals after activation (Figure 4C–F). The cells activated for only 1 h showed a 4-fold (‘2’ for the log2 value in the figure) enrichment of the Il2 promoter in fraction 2 as compared to the Sum of all fractions. This level increased to 8-fold at 2 h, and a maximum was achieved at 3 h. There was a corresponding depletion of the Il2 promoter from fractions 16–18 in the lower half of the gradient. Figure 4G summarizes the kinetics of this shift in fraction distribution, demonstrating that a conversion of the chromatin around the Il2 promoter into a less crosslinkable form was first observed at 1 h, and was almost complete by 2 h. This is consistent with the positive correlation between transcription and promoter enrichment in the slowly sedimenting fractions of the gradient as described above. Because the level of acetylation of histone H3 and H4 around the Il2 promoter does not change significantly during this induction process, as shown in Supplementary Figure S6 and consistent with previous reports (7–9), such modifications are unlikely to be the ones affecting the distribution of the Il2 promoter in the gradient. Interestingly, when the same chromatin was analyzed using a DNase l accessibility assay, significant changes were not observed until 2 h after T-cell activation, and increase in the sensitivity was still occurring at 4 h (Figure 5). The different kinetics of the SEVENS assay from that of the more conventional DNase l method suggests that earlier chromatin structural changes are being detected with the SEVENS assay.
Figure 5.
DNase I accessibility assay.The amount of each promoter region resistant to DNase I digestion was calculated. A decrease in the resistance at the Il2 promoter (PCR position: −77 to +39) was observed following T-cell activation, suggesting an increase in the accessibility to DNase I. In contrast, both the Actb and Cd3d promoter regions did not show any significant changes during this activation period. The Bdnf promoter was not digested under these experimental conditions (data not shown).
Finally, to investigate how broadly the change to a less crosslinkable state occurred around the Il2 locus, we performed the SEVENS assay using primers upstream and downstream of the Il2 TSS (Figure 4H). When T cells were examined before activation, the amounts from all the regions tested in the top fraction (fraction 2) were close to that in the Sum (open columns). After activation for 3 h, however, the region from the Il2 TSS up to −0.5 kb was enriched about 4-fold in the top fraction (closed columns). In contrast, enrichment outside this region in either direction was consistently less than 2-fold. This suggests that the chromatin changes were most pronounced at the Il2 proximal promoter, where many transcription factors are known to bind to regulate II2 expression. Thus, the SEVENS assay appears to have achieved our goal of developing a method for the fine structure analysis of chromatin changes around the promoter region of genes undergoing transcriptional activation.
DISCUSSION
In order to analyze chromatin structure biochemically, chromatin is often crosslinked with formaldehyde, and then sheared by sonication or digested by enzymatic reaction. A ChIP assay requires such chromatin, which is usually crosslinked and sheared well for fine structure mapping (10,11). Dedon et al. have applied such chromatin to sedimentation velocity centrifugation on a sucrose gradient, and observed a simple single peak distribution of the chromatin in the gradient (12). We obtained a similar result showing that such chromatin was retained mostly in the uppermost fraction under our gradient conditions (data not shown). This indicates that chromatin prepared in this manner cannot be fractionated on a sucrose gradient (regardless of its structure), although its buoyant density can be measured as reported previously (13–15). Non-crosslinked and mildly-sheared chromatin can also be applied to such gradients if an optimum concentration of a cation is included. This strategy has revealed that the β-globin locus in chicken erythrocytes sediments slower than the bulk DNA, suggesting an open state of the chromatin at this active locus (2,3). Gilbert et al. have also used chromatin fractionated by such a method to investigate the relationship between transcriptional activity and chromatin structure in a whole genome (5). However, the spatial resolution in their strategy is inadequate for the assessment of local chromatin structure.
In this report, chromatin was crosslinked with 0.75% formaldehyde and gently sheared to yield a wide range of DNA (0.1 to >20 kb). After sedimentation on a sucrose gradient, the size-distribution of the DNA purified from each fraction following reverse crosslinking was broadly smeared (Figure 1C), which is different from that in previous reports using non-crosslinked chromatin (2,3,5). In particular, in our experiments the short fragments (<1 kb) were seen throughout the entire gradient. This suggests that the crosslinking reaction caused some of the small sheared chromatin to be trapped in large particles and to sediment more rapidly toward the bottom of the tube. A microarray analysis of the distribution of a few thousand promoters in the SEVENS assay suggested that those promoters migrating slowly in the gradient tended to be actively transcribing while those trapped in the faster migrating fractions were generally transcriptionally repressed. There are a number of reasons why this pattern of promoter distribution might come about. One technical reason could be the particular concentration of formaldehyde we used for crosslinking (0.75%). When we ran the assay with 1% formaldehyde instead, our four standard genes (Actb, Cd3d, Bdnf and Adad1) showed comparable distribution patterns to those seen when 0.75% was used, although only 50–60% of the DNA was recovered. In contrast, with 0.5% formaldehyde the crosslinking was much less and the repressed promoters migrated only to the middle of the gradient despite the continued enrichment of the active promoters in the upper fractions of the gradient (unpublished observations). These observations suggest that the concentration of formaldehyde is critical in determining the fractional distribution of each promoter, but that this is not related to an issue of solubility.
A second possible reason for the promoter distribution pattern is the nucleotide sequences of the promoters themselves, which might have different susceptibilities to crosslinking. This cannot be the sole basis for our results, however, because the same promoter (Il2) can be found in different fractions of the gradient depending on the activation status of the cell. Nonetheless, the sequences could determine what proteins are bound to the DNA and in particular we considered whether molecules such as polycomb and methyl CpG-binding proteins, which preferentially bind to heterochromatin (16,17), might be responsible for good crosslinking and thus direct repressed promoters into the faster sedimenting fractions. This was not the case, however, and in fact those molecules were found mostly in the slower sedimenting fractions of the gradient (Supplementary Figure S3). In addition, because the structural changes at the Il2 promoter precede recruitment of the RNA polymerase complex and certain transcription factors (unpublished observations), these DNA-binding proteins are also not directly correlated with the structural difference.
A third possible reason is higher-order structures that may be responsible for the crosslinking of small chromatin fragments to larger structures in the nucleus. Some inducible genes can be tethered to other distant loci through a chromatin loop (18). These contacts could be covalently stabilized by our crosslinking conditions; these might be amenable to further analysis in a chromosome conformation capture (3C) assay. The only proteins we found in large abundance throughout the entire gradient were histones (Supplementary Figure S2). This suggests that nucleosome structure may be a critical parameter for determining the fractional distribution of the promoters. The number of nucleosomes is known to vary with the transcriptional status of a gene (19), but we could not correlate the amounts of histone H3 in a ChIP assay with the position of the promoter in the gradient (Supplementary Figure S4). In addition, our preliminary experiments on the Il2 promoter, which is known to shed a nucleosome on transcriptional activation (7), showed that eviction of both histone H1 and H3 is an event that occurs after the structural changes seen with our SEVENS assay. Because neighboring nucleosomes can be folded by their direct interaction (20,21), we suspect that internucleosomal crosslinking is primarily responsible for the patterns we observe in the gradients. In this scenario, the nucleosomes would be in close proximity on repressed promoters, and thus could be heavily crosslinked, while active promoters would open up and become less amenable to formaldehyde crosslinking. Finally, an enhanced crosslinking mechanism could be working uniquely with promoters in repressed chromatin, which tends to be packaged at very high density in selective regions of the nucleus.
The SEVENS assay we have developed provides a new strategy for examining changes in the chromatin microenvironment at high spatial resolution. Note that the structural changes observed with this assay appear to be distinct from those measured in the nuclease accessibility assay, which occurred more slowly when the Il2 promoter region was observed during T-cell activation (Figure 4G versus Figure 5). This suggests that chromatin remodeling takes place in a multi-step process to facilitate transcriptional activation. The idea that chromatin can assume intermediate higher order structures during gene activation has been previously suggested based on agarose multigel electrophoresis experiments with the mouse mammary tumor virus (MMTV) promoter (22). Interestingly, the chromatin at the unactivated Il2 promoter also showed an intermediate state in the SEVENS assay (Figure 4A), with an equal distribution among chromatin fragments of various sizes. It is possible that for the resting state of inducible genes the chromatin is dynamically and spontaneously changing between various open and compact states. In such a model we might be taking a snapshot with the SEVENS assay of the equilibrated chromatin in various cells as a mixture of many configurations. This idea is supported by previous reports showing a dynamic equilibrium state for reconstituted oligonucleosomes (23,24), and the ability of the steroid-transcriptionally-activated MMTV promoter to reassume a more compact structure with changes in in vitro magnesium concentration (22). Taken together with our recent results for the Ifng (Interferon-γ) promoter in resting T cells, which showed a similar broad distribution in the sucrose density gradient (data not shown), these intermediate profiles of chromatin structure could reflect a poised state for inducible genes, whose promoter could locally and flexibly unwind in a reversible manner, for example, in a two-start helix model for a 30-nm fiber (25). Examination of more inducible genes will be required to determine just how general such a poised state could turn out to be.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Division of Intramural Research; National Institute of Allergy and Infectious Diseases; National Institutes of Health. Funding for open access charge: Laboratory of Cellular and Molecular Immunology, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Gary Felsenfeld, Carl Wu, Wendy Bickmore, Keiko Ozato and Nevil Singh for constructive criticism of the original version of this manuscript. They are also grateful to Dr Ellen Rothenberg for her helpful suggestions for the revised version.
REFERENCES
- 1.Noll H, Noll M. Sucrose gradient techniques and applications to nucleosome structure. Methods Enzymol. 1989;170:55–116. doi: 10.1016/0076-6879(89)70043-4. [DOI] [PubMed] [Google Scholar]
- 2.Kimura T, Mills FC, Allan J, Gould H. Selective unfolding of erythroid chromatin in the region of the active beta-globin gene. Nature. 1983;306:709–712. doi: 10.1038/306709a0. [DOI] [PubMed] [Google Scholar]
- 3.Caplan A, Kimura T, Gould H, Allan J. Perturbation of chromatin structure in the region of the adult beta-globin gene in chicken erythrocyte chromatin. J. Mol. Biol. 1987;193:57–70. doi: 10.1016/0022-2836(87)90626-7. [DOI] [PubMed] [Google Scholar]
- 4.Sojka DK, Bruniquel D, Schwartz RH, Singh NJ. IL-2 secretion by CD4+ T cells in vivo is rapid, transient, and influenced by TCR-specific competition. J. Immunol. 2004;172:6136–6143. doi: 10.4049/jimmunol.172.10.6136. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Leonard WJ. Type I cytokines and interferons and their receptors. In: Paul WE, editor. Fundamental Immunology. 6th edn. Philadelphia, PA: Lippincott Williams & Wilkins; 2008. pp. 706–749. [Google Scholar]
- 7.Chen X, Wang J, Woltring D, Gerondakis S, Shannon MF. Histone dynamics on the interleukin-2 gene in response to T-cell activation. Mol. Cell. Biol. 2005;25:3209–3219. doi: 10.1128/MCB.25.8.3209-3219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adachi S, Rothenberg EV. Cell-type-specific epigenetic marking of the IL2 gene at a distal cis-regulatory region in competent, nontranscribing T-cells. Nucleic Acids Res. 2005;33:3200–3210. doi: 10.1093/nar/gki637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Kametani Y, Katano I, Habu S. T-cell specific enhancement of histone H3 acetylation in 5′ flanking region of the IL-2 gene. Biochem. Biophys. Res. Commun. 2005;331:589–594. doi: 10.1016/j.bbrc.2005.03.216. [DOI] [PubMed] [Google Scholar]
- 10.Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 11.Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 2000;25:99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- 12.Dedon PC, Soults JA, Allis CD, Gorovsky MA. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein-DNA interactions. Anal. Biochem. 1991;197:83–90. doi: 10.1016/0003-2697(91)90359-2. [DOI] [PubMed] [Google Scholar]
- 13.Ip YT, Jackson V, Meier J, Chalkley R. The separation of transcriptionally engaged genes. J. Biol. Chem. 1988;263:14044–14052. [PubMed] [Google Scholar]
- 14.Reneker JS, Brotherton TW. Discrete regions of the avian beta-globin gene cluster have tissue-specific hypersensitivity to cleavage by sonication in nuclei. Nucleic Acids Res. 1991;19:4739–4745. doi: 10.1093/nar/19.17.4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz YB, Kahn TG, Pirrotta V. Characteristic low density and shear sensitivity of cross-linked chromatin containing polycomb complexes. Mol. Cell. Biol. 2005;25:432–439. doi: 10.1128/MCB.25.1.432-439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 17.Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell. 2005;19:667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 18.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 19.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 21.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 22.Georgel PT, Fletcher TM, Hager GL, Hansen JC. Formation of higher-order secondary and tertiary chromatin structures by genomic mouse mammary tumor virus promoters. Genes Dev. 2003;17:1617–1629. doi: 10.1101/gad.1097603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen JC, Ausio J, Stanik VH, van Holde KE. Homogeneous reconstituted oligonucleosomes, evidence for salt-dependent folding in the absence of histone H1. Biochemistry. 1989;28:9129–9136. doi: 10.1021/bi00449a026. [DOI] [PubMed] [Google Scholar]
- 24.Schwarz PM, Hansen JC. Formation and stability of higher order chromatin structures. Contributions of the histone octamer. J. Biol. Chem. 1994;269:16284–16289. [PubMed] [Google Scholar]
- 25.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.