Abstract
Dramatic progress in the development of next-generation sequencing technologies has enabled accurate genome-wide characterization of the binding sites of DNA-associated proteins. This technique, baptized as ChIP-Seq, uses a combination of chromatin immunoprecipitation and massively parallel DNA sequencing. Other published tools that predict binding sites from ChIP-Seq data use only positional information of mapped reads. In contrast, our algorithm MICSA (Motif Identification for ChIP-Seq Analysis) combines this source of positional information with information on motif occurrences to better predict binding sites of transcription factors (TFs). We proved the greater accuracy of MICSA with respect to several other tools by running them on datasets for the TFs NRSF, GABP, STAT1 and CTCF. We also applied MICSA on a dataset for the oncogenic TF EWS-FLI1. We discovered >2000 binding sites and two functionally different binding motifs. We observed that EWS-FLI1 can activate gene transcription when (i) its binding site is located in close proximity to the gene transcription start site (up to ∼150 kb), and (ii) it contains a microsatellite sequence. Furthermore, we observed that sites without microsatellites can also induce regulation of gene expression—positively as often as negatively—and at much larger distances (up to ∼1 Mb).
INTRODUCTION
The appearance of next-generation sequencing technologies has propelled forward the development of new techniques among which ChIP-Seq has become an important method for genome-wide discovery of binding sites for DNA-associated proteins and in particular for TFBSs. ChIP–Seq consists of the immunoprecipitation of protein–DNA complexes followed by massively parallel sequencing of short ends of immunoprecipitated DNA (1–3). This technique succeeded the ChIP-on-chip technique (4) and has nearly replaced the latter because of the increased accuracy in identification of TFBSs (2).
At the completion of a ChIP-Seq experiment, millions of short (∼35–50 bp) directional DNA tags are obtained, which can be positioned or aligned to the reference genome for the sample organism (Supplementary Figure S1). Each short tag represents an extremity of a longer DNA fragment (∼200–400 bp depending on the experiment) isolated from the immunoprecipitation. Thus, in the analysis of the short representative tags, it is important to take this experimental fact into consideration to identify the full length of the original fragment that gave rise to the tag. By extending each tag, it is then possible to identify areas of overlap, which represent the location of the protein binding event. The density profile of DNA fragment coverage can then be calculated and ‘peaks’ corresponding to putative binding sites can be extracted. This idea was elegantly implemented in the FindPeaks software (5). However, the accuracy of peak calling can be considerably improved by incorporating information about genomic sequences of peaks in addition to coverage depth information.
In this article we present an algorithm implemented in the MICSA software (Motif Identification for ChIP-Seq Analysis) that is based on the idea that functional binding sites of transcription factors (TFs) should contain a consensus motif (or a set of motifs). Consensus motifs are the composite sequences of DNA for which a DNA-binding protein, such as a TF or restriction enzyme, has a high affinity. Such motifs can be identified from the small subset of peaks with a high DNA fragment coverage.
The MICSA algorithm is innovative in the context of ChIP–Seq data analysis for simultaneous: (i) de novo TFBS motif identification and (ii) functional binding site prediction using information about motif occurrences in peaks along with coverage depth information. Here, motif identification is not a post-processing step as in other ChIP-Seq analysis pipelines (6) but a key element which allows keeping even low peaks if they have a strong motif occurrence.
Since MICSA checks for motif occurrences in all peaks including those with very low coverage depth, there is no need in the explicit selection of threshold on DNA tag/fragment coverage. The only parameter that remains to be specified is the maximal number of expected false positive hits among selected peaks or the maximal false discovery rate (FDR).
Using the procedure developed by Kharchenko et al. (7), we compared the peak identification performance of MICSA and 10 other published tools (5–14). The dataset selected for the comparison was generated by Johnson et al. (2) for the neuron-restrictive silencer factor (NRSF). MICSA showed a considerable increase in the performance over 10 other approaches. To increase the statistical basis we performed the same comparison procedure for selected algorithms on other ChIP-Seq datasets, including those for GA-binding protein (GABP) (10), signal transducer and activator of transcription 1 (STAT1) (9) and CCCTC-binding factor (CTCF) [ENCODE project, the Broad Institute and the Bradley E. Bernstein lab at the Massachusetts General Hospital/Harvard Medical School (15)]. The results of the comparison indicated that use of MICSA for ChIP-Seq data analysis allows us to significantly reduce the number of false positive predictions for TFBSs.
The MICSA package was also used on our ChIP-Seq data (16). Immunoprecipitation was performed with a specific antibody directed against the oncogenic TF EWS-FLI1 (17) to obtain biological insight into the functioning of this TF, which is known to be the major oncogene in Ewing sarcoma. Using our technique, based on motif identification, we confirmed the existence of two consensus motifs, one representing a (GGAA)n microsatellite, and the second containing the RCAGGAARY consensus sequence (16) (R = A/G, Y = T/C). Further analysis of the EWS-FLI1 data, together with expression arrays, suggested that EWS-FLI1 bound to (GGAA)n microsatellites can activate transcription of neighboring genes; while EWS-FLI1 bound to RCAGGAARY sites may, depending on genes, activate or repress transcription. Our analysis confirmed five known direct target genes of EWS-FLI1 and has also predicted many new genes that are putatively regulated directly by EWS-FLI1.
The algorithm we developed is pioneering in its use of motif information when predicting sites of specific binding for TFs from ChIP-Seq data. It allows identification of several motifs which, as shown at the EWS-FLI1 example, can possibly carry different biological function.
MATERIALS AND METHODS
Candidate peak identification
Candidate peaks are identified by FindPeaks (5) which is included in the MICSA package. The java class DeleteRegions is then used to eliminate peaks in satellite regions which are often a source of noise in ChIP-Seq data. We propose to use blacklist regions masked as centromeric repeats by RepeatMasker (http://www.repeatmasker.org) in UCSC genome browser (18). Files are provided in the MICSA package.
Files with candidate peaks are then processed by the java class FilterPeaks in order to filter out false peaks occurring both in ChIP and control datasets because of biases due to PCR errors. MICSA needs to be supplied with a reference genome from which it would extract DNA sequences of peaks.
Classes
The whole set of candidate peaks is decomposed into classes. Class Ci contains regions from which i overlapping DNA fragments were immunoprecipitated during ChIP experiment. For peaks from class Ci the probability pi to be a false binding site can be evaluated using control data: pi = min{1,(#peaks in control class C`i)/(#peaks in ChIP class Ci)}. Peaks highly enriched in mapped DNA tags are likely to be true positives, while peaks with low DNA tag coverage can probably be false since lots of peaks with the same values of coverage could be identified in the control data. Thus, pi will be smaller for higher values of i.
Here our hypothesis is that the normalized number of peaks in the control dataset is an estimate of the number of false binding sites in our ChIP dataset. For the same purpose, other methods use, for example, Monte–Carlo simulations (5) or Poisson approximation (8). We believe that using the control data is more appropriate in this case since it takes into account the sequencing bias and guarantees not to underestimate the number of false binding sites.
Motif identification
We use a subset of the highest peaks from classes with zero pi value to identify over-represented motifs. Moreover, we do not consider whole peak sequences but only sequences at the location of the maximal enrichment in each peak. The MEME motif finding tool (19) is then run automatically on this set of sequences in order to identify the most over-represented motif. Sequences from this set which do not contain the identified motif are subject to the second MEME run. Finally, one uses the top significance motifs, each in the form of a position-specific scoring matrix (PSSM) with the minimal threshold value, which occur in the areas of maximal enrichment of the high peaks. It is only one motif in the case when the protein does not undergo allosteric modifications which could change its binding motif, e.g. distance between its half-sites; one obtains more than one motif in the case when such change is possible. A large part of genome is scanned to yield the real frequencies of extracted motifs.
Optimization
In what follows below, we consider separately peaks on different chromosomes. For motif M and class Ci we will call peaks which have an occurrence of M with a PSSM score above TM,i within the top peak area defined by (h–δM,i), where h is a peak height, i.e. maximal number of overlapping DNA fragments coming from this region. The question is how to choose TM,i and δM,i for each particular class. It can be done through the calculation of expected number of false binding sites selected by the peak calling procedure given the null hypothesis of the Markov(0) model for nucleotide distribution in peaks. For each pair (TM,i, δM,i) and the class Ci we can evaluate the expectation Ei(TM,i, δM,i) of number of peaks called by chance. This value is equal to a sum of the motif P-values of each peak in Ci . The motif P-value means a probability to observe a motif by chance in a sequence of given length. We use the Poisson approximation for the P-values: P-value ≈1 – (1 – MotifProbability)TextLength – MotifLength + 1. Here, the motif probability, which is the probability of observing a motif occurrence above a given PSSM threshold on a given position, is considered to be equal to the genomic frequency of the motif with the given threshold. Finally, since pi is the estimate of the probability of any given peak in class Ci to be a false binding site, the product Ei(TM,i, δM,i)·pi estimates the total number of false binding sites which would be called by this selection procedure. The number of selected peaks Si in class Ci also depends on (TM,i, δM,i). Our optimization procedure maximizes ΣSi so that ΣEipi stays below the predefined threshold on the total number of false positives.
Score calculation
A score is reported for each peak selected during the optimization procedure. It is equal to the product of pi of corresponding class and the smallest motif P-value among motif P-values of all motif occurrences in the peak region. The second term is small if the motif is situated near the location of the maximal enrichment of the peak and its sequence is close to the consensus.
Additional methods
Information about ChIP-Seq library construction and sequencing for EWS-FLI1 data was published in (16). More detail on the optimization procedure is available in Supplementary Data.
Software availability
The MICSA algorithm is implemented as a Java package with a graphical user interface in Java. It is freely available for nonprofit use at http://bioinfo-out.curie.fr/projects/micsa/. All data presented in this study (mock control and ChIP-Seq data, and peak call coordinates) are available at the same website.
RESULTS
Theoretical framework
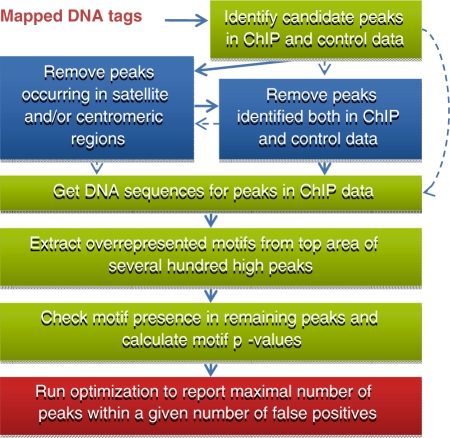
The key idea of MICSA is to use the information about TFBS motif occurrences to predict true binding sites from ChIP-Seq data. The MISCA workflow consists of four main steps (Figure 1): (i) identify all candidate peaks, (ii) identify TFBS motifs from a small subset of peaks, (iii) find motif occurrences in candidate peaks, and (iv) optimize the peak calling output by calculating statistics that take into account both motif occurrence and depth of coverage information. The last step roughly corresponds to filtering out of insignificant peaks without motif occurrences.
Figure 1.
Main steps of the MICSA pipeline.
The first step of the MICSA algorithm consists in identifying all regions that are enriched in mapped DNA tags (Figure 1). Such areas of enrichment, also called peaks, represent potential binding sites for the protein of interest and can often exceed tens of thousands of locations. To detect these regions we use the previously developed tool FindPeaks (5). One of the reasons that we use FindPeaks is that it generates UCSC compatible custom ‘WIG’ track files from aligned-read files. We calculate the false discovery rate (FDR) predictor to estimate the proportion of false peaks to called peaks for each dataset. FDR is estimated using both the ChIP and mock control data. Generally, the FDR estimate for high peaks (peaks with greater depth of DNA tag or fragment coverage) is smaller than for low ones since the former are less likely to occur randomly or in control data.
The second step is de novo motif identification. DNA sequences of peaks are extracted from the reference genome using position information. In order to limit the amount of time required, we only process three hundred peaks with high DNA fragment coverage. Moreover, the motif extraction is done only for central area of these peaks to avoid getting false motifs that are over-represented, such as those of cooperating TFs. The MICSA package utilizes the expectation maximization MEME algorithm (19) to detect the strongest motifs in peak sequences.
In the third step, discovered motifs are identified for all regions of enrichment located by FindPeaks (peaks file). The combined information on motif location and position-specific scoring matrix (PSSM) scores is then used to calculate a motif P-value for each peak (see ‘Materials and Methods’ section). In general, the P-value will be small if the motif occurrence closely resembles the consensus (and thus has a high PSSM score) or if the motif occurs close to the region of maximum enrichment of the peak. In contrast, the motif P-value will be high if the motif occurs in the periphery of the peak and the motif observed has a low PSSM score. The final peak score appearing in the MICSA output is a product of the motif P-value described above and the estimated FDR. Supposing that, for a given threshold on the peak height, the number of false binding sites in the ChIP sample is equal to the normalized number of peaks in the control sample, and nucleotides along such peak sequences are distributed according to the Markov(0) model, then MICSA’s peak score estimates the probability of the given peak to be a false binding site and have a motif occurrence just by chance.
In the fourth step we select binding sites out of candidate peak dataset based on motif occurrence and depth of coverage information. The raw peak candidate prediction set contains a large number of false positive predictions. To filter them out, other methods commonly determine a threshold on DNA tag/fragment coverage (6,10,13). However, our experience and that of other researchers (C. Wadelius, personal communication) shows that even regions with relatively low DNA tag coverage often contain functional binding sites. Here we propose an approach based on the presence of motif information in peak sequences. We use this information to retain additional peaks containing strong motifs, especially those with low DNA tag coverage. For each subset of peaks with a given depth of DNA fragment coverage, we choose a criterion for peak retention based on motif strength and position within the peak. We then evaluate a number of false positive peaks that we expect to satisfy the criterion (see ‘Materials and Methods’ section). We use an optimization procedure that applies different criteria in an attempt to maximize the total number of selected peaks for all subsets without having the estimated total number of false positive peaks passing the selection exceed the user defined threshold. MICSA outputs the list of peaks which are selected by the optimization procedure and ranked according to their scores and associated motifs. As a result of the optimization, high peaks are usually kept by MICSA even if there is no strong motif hit. In other words, though the motif based filtering is applied by MICSA to all peaks, it is only effective on lower ones (Supplementary Figure S2).
Performance of MICSA
In order to assess relative performances of MICSA and other existing tools, we used the algorithm comparison method developed by Kharchenko et al. (7). The tested programs were MACS (8), PeakSeq (9), QuEST (10), wdt (7), Useq (11), F-Seq (12), CisGenome (6), ERANGE/ChIPSeqMini (13), SISSRs (14) and FindPeaks (5). Each application was used with its default parameters, according to the instructions given in the manual (see Supplementary Table S1 for command lines). In cases where only a small number of peaks were extracted, parameters were modified in order to increase the number of identified peaks. ChIP-Seq data for the NRSF (2,20) were selected for testing because they had already been widely used by other groups to validate ChIP-Seq analysis software (2,6–8,11,14). Additionally, we run MICSA on datasets for GABP (10), STAT1 (9) and CTCF [ENCODE project, the Broad Institute and the Bradley E. Bernstein lab at the Massachusetts General Hospital/Harvard Medical School (15)].
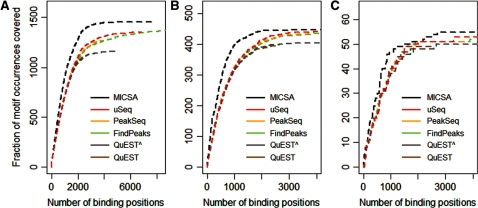
For NRSF we used two positive TFBS sets to assess the sensitivity of each of the different methods. The first one is a list of 3000 high-scoring motif instances designed using canonical sequence binding motifs for NRSF by Kharchenko et al. (7). The second is composed of 83 binding sites verified by qPCR (2). To compare the methods’ sensitivities, we selected increasing numbers of top peaks returned by each method and analyzed the fraction of peaks containing the motif of interest (Figure 2 and Supplementary Figure S3). MICSA clearly outperforms other methods on both tested datasets, with almost any threshold on a number of called peaks. We compared algorithm performances considering the best 3000 peaks called by each program. For the first positive set of 3000 high-scoring motif instances we found that 1422 of them were identified by MICSA (Figure 2A). According to this test we could rank the other tools as uSeq (1254), wdt (1229), PeakSeq (1227), F-Seq (1217), FindPeaks 3.3 (1216), CisGenome (1203), MACS (1195), SISSRs (1194), QuEST 2.0 (1132), ERANGE 3.1 (1118) (Figure 2A, Supplementary Figure S3a and Supplementary Table S2). However, it is unlikely that all 3000 best NRSF matrix matches are true functional binding sites. Thus, we reduced the positive set to the best 500 motif instances with the highest score. Using this smaller positive set, within 3000 best peaks, MICSA was able to successfully identify 447 out of all 500 instances (Figure 2B). Other tools can be ranked accordingly: uSeq (437), MACS (435), F-Seq (432), CisGenome (431), PeakSeq (431), wdt (430), FindPeaks 3.3 (428), SISSRs (424), ERANGE 3.1 (412) and QuEST 2.0 (402) (Figure 2B, Supplementary Figure S3b and Supplementary Table S3).
Figure 2.
Performance comparison of MICSA with FindPeaks, PeakSeq, QuEST and uSeq. As a positive set of binding sites of NRSF we used (A) 3000 best matches of the canonical NRSF matrix in the human genome, (B) 500 best matches of the canonical NRSF matrix in the human genome, (C) 83 q-PCR verified NRSF-binding sites in the human genome. Peaks extracted by each algorithm were ranked according to in-built scores or P-values. For each number of top peaks the frequency of identified positive sites among them was plotted. ‘ToolName^ ’ means that the default parameters of the tool were modified to make it report more peaks.
When evaluating the tool sensitivity on the set of 83 qPCR verified NRSF-binding sites, comparable results were obtained: MICSA (55 sites), MACS (52), CisGenome (52), FindPeaks 3.3 (51), SISSRs (51), F-Seq (51), uSeq (51), wdt (51), PeakSeq (51), QuEST 2.0 (50), ERANGE 3.1 (50) (Figure 2C, Supplementary Figure S3c and Supplementary Table S4). However, this can be probably explained by the fact that only regions that were highly enriched in mapped reads were tested by qPCR. It is noteworthy that MICSA was able to identify the maximum number of experimentally tested regions within the dataset of 3000 reported peaks.
Motifs identified by MICSA during the analysis procedure nicely corresponded to the known NRSF-binding motifs (Supplementary Figure S4).
In the comparison above MICSA was the only tool in which DNA motif information was utilized. Since the CisGenome package also contains a module for identification of enriched motifs we added to the comparison the subset of peaks identified by CisGenome which also contain strong hits for enriched motifs. Though motifs identified by CisGenome were close to those identified by MICSA, the latter called more peaks from the positive datasets (Supplementary Figure S5).
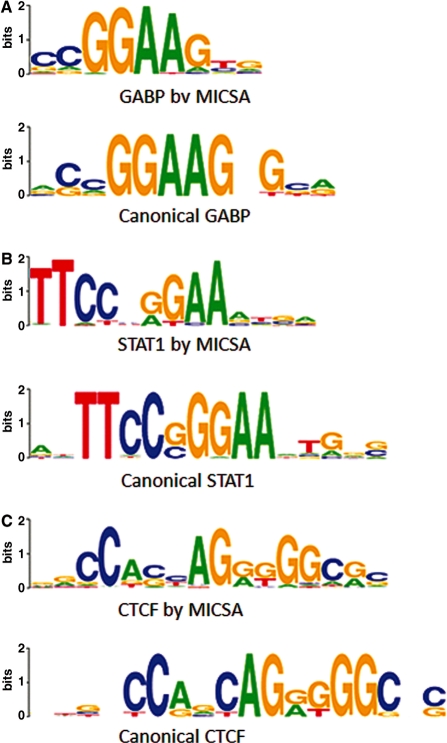
We run MICSA on datasets for GABP (10), STAT1 (9) and CTCF [ENCODE project, the Broad Institute and the Bradley E. Bernstein lab at the Massachusetts General Hospital/Harvard Medical School (15)] and compared the reported sets of peaks with those reported by other peak calling tools: FindPeaks, uSeq, QuEST and PeakSeq. To compare algorithms between each other we applied the same procedure (7) as previously for NRSF. We used canonical sequence motifs for binding by GABP (Genomatix, http://www.genomatix.de), STAT1 (21) and CTCF (22) to create positive sets of 3000 peaks. For any considered TF, within a given number of peaks selected by each algorithm there were more peaks from the positive set in the output of MICSA than in the output of any other program (Supplementary Figure S6). For all three TFs, binding motifs identified by the MICSA pipeline were highly similar to the canonical motifs of the same TF (Figure 3).
Figure 3.
Binding motifs identified by MICSA in ChIP-Seq data for GABP, STAT1 and CTCF resemble canonical motifs. (A) GABP motif logos [Weblogos (32)], canonical motif from (Genomatix, http://www.genomatix.de), (B) STAT1 motif logos (21), (C) motif logos for CTCF (22).
The results of the test above might appear to be anticipated in advance. Indeed, MICSA is the only algorithm to use motif information in prediction and the motifs identified by MICSA perfectly matched to the known binding motifs for the considered TFs. So we performed an additional test to assess MICSA’s performance. We took the latest ChIP-Seq data for NRSF [ENCODE project, Myers Lab at the HudsonAlpha Institute for Biotechnology (15)]. There the sequencing depth was increased to obtain about 13 million of uniquely mapped reads instead of 2 million in the Johnson dataset (2). Since the depth of sequencing had increased, we expected peaks selected by MICSA from the old dataset to appear in the new dataset and to have a read coverage greater than peaks selected by FindPeaks from the old dataset but rejected by MICSA. Indeed, there are 4572 peaks among 7780 selected by MICSA which depth increased more than or exactly twice, while there are 11 748 peaks among 22 116 peaks selected by FindPeaks. This constitutes the overall ratio of 59% against 53% (Supplementary Table S5). The difference is higher for low peaks and disappears with the increase of peak heights (Supplementary Figure S7). This test though not being based on the information about a priori known motifs, demonstrated the advantage of using MICSA for filtering low peaks.
Biological application of MICSA
To demonstrate the use of the MICSA package to obtain biological insight, we then applied the package to ChIP-Seq data for the oncogenic TF EWS-FLI1. EWS-FLI1 is a chimeric protein produced by a fusion of the EWS and FLI1 genes. This abnormal TF is a key oncogene in Ewing sarcoma (17,23). To investigate EWS-FLI1 DNA binding, we reanalyzed ChIP-Seq data previously collected by our team (16). The DNA fragments retained on complexes immunoprecipitated by an FLI1-specific antibody were processed by the Illumina/Solexa cluster station and 1G analyzer for the A673 Ewing cancer cell line and aligned with the Maq (24) software with a maximum two mismatches to the unmasked human reference genome (NCBIv36, hg18) (18,25). A control was obtained using the same anti-FLI1 antibody in a rhabdoid tumor cell line (MON) that does not express EWS-FLI1 or ETS family TFs.
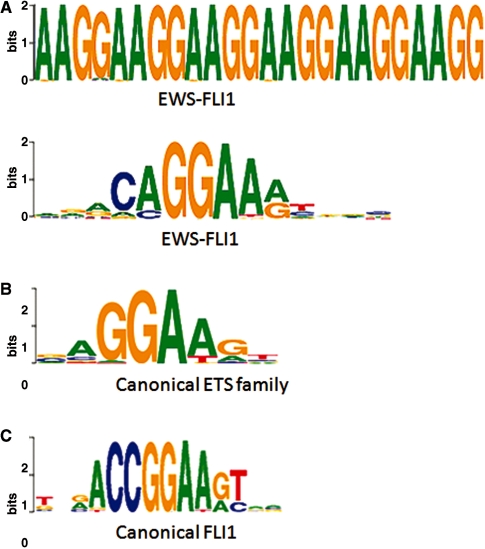
A key characteristic of these data is that the total amount of sequenced DNA is insufficient for straight-forward identification of the majority of binding sites. Indeed, our previous analysis of these data showed a very limited number of regions of EWS-FLI1 specific binding (246). Upon re-analysis with the MICSA tools, we were able to discover 2264 sites with an expectation FDR of 5%. MICSA was also able to identify two known consensus motifs occurring in the most highly enriched regions of called peaks (16). The first one represents a (GGAA)≥6 microsatellite and it is found in 496 peak sequences; the second motif, found in 1768 peak sequences, corresponds to the consensus RCAGGAARY (R = A/G, Y = T/C) (Figure 4A).
Figure 4.
Motifs identified by MICSA in EWS-FLI1 ChIP-Seq data resemble but are not identical to the canonical binding motif of FLI1. (A) Consensus motifs identified by MICSA [Weblogos (32)], (B) canonical motif for ETS family of TFs including the TF FLI1 (26), (C) canonical motif for the TF FLI1 (27).
Since the single RCAGGAARY consensus motif resembles the known binding motifs for the ETS TF family (26) (Figure 4B), we will refer to it as the ETS motif. Interestingly, the extracted ETS motif, although resembling the consensus motif of FLI1 (27) (CCGGAARY) (Figure 4C), does not completely coincide with the latter, even though EWS-FLI1 shares the same DNA-binding domain as FLI1.
We compared the locations of discovered peaks with gene organization and expression data (28) for the Ewing cancer cell line A673 in both the presence and absence of EWS-FLI1 using a random set of peaks as a control (Figure 5). To create the random set we randomly selected 2264 locations in the annotated part of the human genome (NCBIv36, hg18) (18,25). From the expression data, we extracted a list of putative target genes of EWS-FLI1: 557 genes downregulated by EWS-FLI1 and 577 upregulated genes (fold change >|2| with a Welsh P-value <0.01). These are genes modulated by EWS-FLI1 in A673 and SK-N-MC Ewing cell lines.
Figure 5.
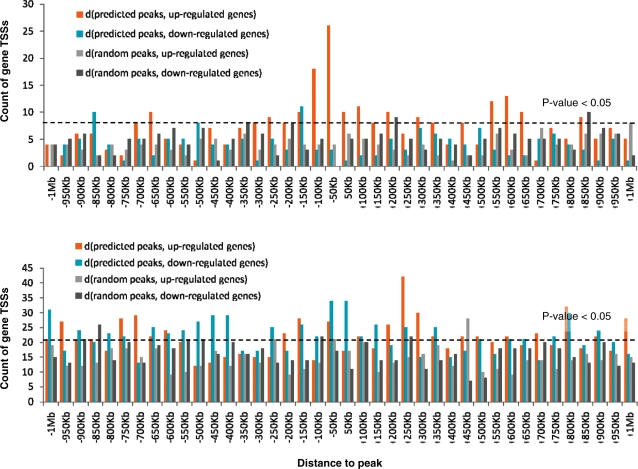
Histogram of distances between predicted/random peaks and genes up/downregulated by EWS-FLI1. (A) Predicted sites containing (GGAA)n microsatellites; (B) ETS sites (site without microsatellites). EWS-FLI1 binding to GGAA microsatellites results in significant expression activation of neighboring genes. EWS-FLI1 binding to single ETS sites can produce both negative and positive effects on transcription of neighboring genes. The P-values were directly evaluated by Monte-Carlo simulations of random peaks. Distances from the TSSs of modulated genes to random peaks (iterative trials) and to predicted sites were calculated. The P-values correspond to the probability to get at least the observed number of distances falling within a given 50-kb window, under the hypothesis that peaks are randomly distributed and their coordinates are independent of coordinates of TSSs of EWS-FLI1 modulated genes. Bars above the dashed line correspond to a P-value <0.05.
Our analysis revealed the tendency of sites bearing microsatellites to upregulate neighboring genes [sites found from 150-kb upstream to 50-kb downstream of gene transcription start sites (TSSs)] (Figure 5A and Supplementary Figure S8), while sites with the ETS motif do not seem to have a definite activator function (Figure 5B). ETS sites show some transcriptional inhibitory influences on gene expression when located in the first 50-kb downstream of the TSSs. However, when ETS-sites are found further away from genes (within 1 Mb upstream or downstream but not in the first 50-kb downstream TSS), both activatory and inhibitory influences are observed for EWS-FLI1 transcriptional activity. Among other hypotheses, this could be explained by competitive binding of EWS-FLI1 and native repressor or activator TFs.
Our analysis confirmed binding sites for five known direct target genes of EWS-FLI1: C-Myc, CCND1, TGFbRII (29), CAV1 (30) and IGF1 (31). However, in two cases out of five, a binding site was identified inside the gene and not in the promoter region. MICSA predicted many new possibly direct targets of EWS-FLI1 (Supplementary Table S6). Among them we find PPP1R1A, LBH, FAS, CAV2 and NBL1. This information will aid in the construction of a detailed and accurate regulation network for this particular type of cancer.
Our motivating idea was that low peaks without motifs are likely to be false positives while low peaks with motifs may indicate real binding events. To test it we performed ChIP–qPCR for 16 interesting genes having a low peak of EWS-FLI1 within 50-kb upstream of gene TSS (Supplementary Table S7). To create a control set we took seven peaks which had been discarded by MICSA as low peaks without strong motif occurrence. The heights of selected peaks vary from 3.9 to 8 for the set of peaks selected by MICSA and from 4 to 8 for the control set. For tested genes from MICSA’s set we got a clear positive response for CCND1, GYG2 and PAPPA, and a positive trend for AKAP7 and SLCO5A1 (Supplementary Figure S9). Interestingly, none of the 7 peaks from the control set was found to be positive in our experiment. Though we performed the experiments on a limited set of peaks, we believe that the results clearly reinforce our idea that MICSA is a useful tool to distinguish between possible binding events in the case of low peaks.
DISCUSSION
To our knowledge, MICSA is the first tool developed for peak identification in ChIP-Seq data that uses an approach combining knowledge about DNA fragment coverage in ChIP and control experiments along with motif discovery. MICSA is able to automatically identify overrepresented motifs in a single run, as well as to use motif occurrence probabilities to enhance the result set returned. MICSA achieves a higher accuracy in identifying regions of TF binding in comparison to other methods. For example, no other tested tool was able to identify more than 45% of predicted motif occurrences (Figure 2) within the top 3000 selected peaks. With default parameters, only Useq (11) (with the best 6073 peaks), MACS (8) (with 6450 peaks), FindPeaks (5) (with 7097 peaks) and F-Seq (12) (with 8576 peaks) managed to obtain the same coverage. However, that required a significantly larger set of selected peaks. This example shows that using MICSA helps to avoid inclusion of thousands of low peaks which do not carry TF-binding motifs and thus are likely to be false binding sites.
Two other major advantages of MICSA, in addition to its greater accuracy, are the score calculation for each reported peak and evaluation of the number of false positives in the total output.
MICSA uses two previously published tools: FindPeaks (5) for candidate peak calling and MEME (19) for de novo motif finding. They were chosen as the best performing from the point of view of speed and result quality. Additionally, FindPeaks allows elimination of duplicate tags due to PCR errors and supports many input formats including: both Maq’s .map and mapview formats (24), ELAND, ELAND Extended and BED format.
Because of its high sensitivity, MICSA can be used on medium quality datasets with low average DNA fragment coverage. EWS-FLI1 data (16) represents one such example. In spite of non-perfect initial data quality, we were able to get rich biological results and some insights in the function of EWS-FLI1 in Ewing cancer cells.
For the EWS-FLI1 TF we identified two different types of motifs carrying different biological functions, i.e. microsatellites (GGAA)n and single RCAGGAARY motif (R = A/G, Y = T/C). Interestingly, none of them completely coincide with the known binding motif of FLI1, which has the same DNA-binding domain as EWS-FLI1. This may be a result of the presence in EWS-FLI1 of an additional EWS domain. The observed motif difference provides evidence that de novo motif finding is an important issue in ChIP-Seq data analysis. Our results suggest that EWS-FLI1 binding to a site bearing (GGAA)n microsatellites can activate gene expression if the site occurs within 150-kb upstream and 50-kb downstream region from gene TSS. Occasionally, even more distant sites bearing (GGAA)n microsatellites appear to moderate the activator function of EWS-FLI1. EWS-FLI1 binding to a site without microsatellites can, depending on the gene, activate or repress transcription (sites within 1-Mb upstream 1-Mb downstream of gene TSS). This change of regulatory function depending on binding motif provides an insight into the molecular mechanisms of EWS-FLI1 function. One of the hypotheses is a conformation change induced by dimerization of EWS-FLI1 on microsatellites. Verification of this hypothesis was out of the scope of the article, however, if true, this would indicate a potentially new way to target Ewing cancer by disrupting dimerization.
In conclusion, the MICSA package proposes solutions for a great number of problems including peak calling with predefined false positives number, peak score calculation and de novo motif identification and should be a useful tool in ChIP-Seq data analysis of TFBSs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Curie Institute, the « Ligue Nationale contre le Cancer » (V.B., E.B., D.S., N.G., F.T. and O.D. are members of labeled team, the project was also supported by the CIT program ‘Carte d’Identité des Tumeurs’); Institut National de la Santé et de la Recherche Médicale and the Agence Nationale de la Recherche (SITCON project). Funding for open access charge: Institut Curie, Paris, France.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank A. Zinovyev, P. Kharchenko, I.V. Kulakovskiy and N. Rajewsky for valuable discussion and help, M. Hue for the idea of the optimization procedure and K. Bleakley for the proofreading of the manuscript.
REFERENCES
- 1.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 3.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 4.Buck MJ, Lieb JD. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83:349–360. doi: 10.1016/j.ygeno.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Fejes AP, Robertson G, Bilenky M, Varhol R, Bainbridge M, Jones SJ. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24:1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kharchenko V, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat. Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nix DA, Courdy SJ, Boucher KM. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics. 2008;9:523. doi: 10.1186/1471-2105-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyle AP, Guinney J, Crawford GE, Furey TS. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008;24:2537–2538. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 14.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guillon N, Tirode F, Boeva V, Zynovyev A, Barillot E, Delattre O. The oncogenic EWS-FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PLoS ONE. 2009;4:e4932. doi: 10.1371/journal.pone.0004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.May WA, Gishizky ML, Lessnick SL, Lunsford LB, Lewis BC, Delattre O, Zucman J, Thomas G, Denny CT. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc. Natl Acad. Sci. USA. 1993;90:5752–5756. doi: 10.1073/pnas.90.12.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 20.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 21.Horvath CM, Wen Z, Darnell J.E., Jr A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 22.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riggi N, Suva ML, Suva D, Cironi L, Provero P, Tercier S, Joseph JM, Stehle JC, Baumer K, Kindler V, et al. EWS-FLI-1 expression triggers a Ewing's sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res. 2008;68:2176–2185. doi: 10.1158/0008-5472.CAN-07-1761. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 26.Wingender E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief Bioinform. 2008;9:326–332. doi: 10.1093/bib/bbn016. [DOI] [PubMed] [Google Scholar]
- 27.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11:421–429. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 29.Fukuma M, Okita H, Hata J, Umezawa A. Upregulation of Id2, an oncogenic helix-loop-helix protein, is mediated by the chimeric EWS/ets protein in Ewing sarcoma. Oncogene. 2003;22:1–9. doi: 10.1038/sj.onc.1206055. [DOI] [PubMed] [Google Scholar]
- 30.Tirado OM, Mateo-Lozano S, Villar J, Dettin LE, Llort A, Gallego S, Ban J, Kovar H, Notario V. Caveolin-1 (CAV1) is a target of EWS/FLI-1 and a key determinant of the oncogenic phenotype and tumorigenicity of Ewing's sarcoma cells. Cancer Res. 2006;66:9937–9947. doi: 10.1158/0008-5472.CAN-06-0927. [DOI] [PubMed] [Google Scholar]
- 31.Cironi L, Riggi N, Provero P, Wolf N, Suva ML, Suva D, Kindler V, Stamenkovic I. IGF1 is a common target gene of Ewing's sarcoma fusion proteins in mesenchymal progenitor cells. PLoS ONE. 2008;3:e2634. doi: 10.1371/journal.pone.0002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.