Abstract
DNA cytosine-5 methylation is a well-studied epigenetic pathway implicated in gene expression control and disease pathogenesis. Different technologies have been developed to examine the distribution of 5-methylcytosine (5mC) in specific sequences of the genome. Recently, substantial amounts of 5-hydroxymethylcytosine (5hmC), most likely derived from enzymatic oxidation of 5mC by TET1, have been detected in certain mammalian tissues. Here, we have examined the ability of several commonly used DNA methylation profiling methods to distinguish between 5mC and 5hmC. We show that techniques based on sodium bisulfite treatment of DNA are incapable of distinguishing between the two modified bases. In contrast, techniques based on immunoprecipitation with anti-5mC antibody (methylated DNA immunoprecipitation, MeDIP) or those based on proteins that bind to methylated CpG sequences (e.g. methylated-CpG island recovery assay, MIRA) do not detect 5hmC and are specific for 5mC unless both modified bases occur in the same DNA fragment. We also report that several methyl-CpG binding proteins including MBD1, MBD2 and MBD4 do not bind to sequences containing 5hmC. Selective mapping of 5hmC will require the development of unique tools for the detection of this modified base.
INTRODUCTION
In mammalian cells, DNA methylation is an enzymatic modification at the 5-position of cytosine present abundantly within the CpG dinucleotide sequence context. This DNA modification is inheritable and reversible without primary DNA base sequence changes resulting in possible epigenetic modulation of phenotype and gene expression (1,2). The de novo formation and maintenance of 5-methylcytosine (5mC) is catalyzed by DNA methyltransferase proteins (DNMTs) (3). The biological importance of 5mC as a major epigenetic modification has been recognized widely, and a variety of techniques for the study of DNA methylation have been developed and used over the past three decades. The most commonly used assays that distinguish 5mC from normal cytosine can be classified into several groups on the basis of their principles: (i) selective restriction enzyme digestion of unmethylated DNA, (ii) selective chemical conversion of unmethylated cytosine by sodium bisulfite treatment and (iii) selective affinity of antibodies or proteins towards 5mC (4–6).
In addition to 5mC, mammalian DNA contains very low levels of various modified DNA bases arising from DNA damage through normal metabolic activities and/or environmental factors, which are generally eliminated by DNA repair processes. However, recently Kriaucionis and Heintz reported that substantial amounts of a specific modified DNA base, 5-hyroxymethylcytosine (5hmC) are present in mouse Purkinje and granule neurons (7). Independently, another research group discovered the existence of an enzymatic activity involved in producing 5hmC from 5mC and carried out by the TET1 5-methylcytosine oxidase (8). In addition, 5hmC may be produced by the addition of formaldehyde to DNA cytosines by DNMT proteins (9).
5hmC might serve biologically important roles, or it might serve as an intermediate in direct DNA demethylation. For example, the oxidation of 5mC at methylated CpG sites is known to inhibit binding of the methyl-CpG-binding domain (MBD) of MeCP2, which is a transcriptional repressor, suggesting a potential regulatory role of 5hmC (10). Deamination of 5hmC will produce 5-hydroxymethyluracil (5hmU) and generate a mismatched base pair between 5hmU and guanine promoting DNA demethylation by potential DNA repair mechanisms (11,12). In other studies, a reversible enzymatic reaction catalyzed by DNMT proteins, leading to the release of formaldehyde from 5hmC and thus producing unmodified cytosine was proposed, suggesting that 5hmC might be an intermediate in direct DNA demethylation (9).
Since 5hmC is present in mammalian DNA at physiologically relevant levels and in a tissue-specific manner (7,8), there is an important need to determine how 5hmC can be distinguished from 5mC and normal cytosine. Here, we have addressed this question by comparing the ability of some of the most commonly used DNA methylation mapping techniques to detect 5mC and 5hmC, respectively.
MATERIALS AND METHODS
Synthesis of oligonucleotides containing modified cytosines
Production of modified base-containing synthetic DNA fragments using polymerase chain reaction (PCR) amplification was accomplished through the use of modified deoxycytidine triphosphates, 5-methyl-2′-deoxycytidine 5′-triphosphate (5mdCTP) (Fermentas; Glen Burnie, MD) and 5-hydroxymethyl-2′-deoxycytidine 5′-triphosphate (5hmdCTP) (Bioline; Taunton, MA). A starting amount of 0.5 ng of single-stranded 76-mer oligonucleotide (sequence 5′-CCTCACCATCTCAACCAATATTATATTACGCGTATATCGCGTATTTCGCGTTATAATATTGAGGGAGAAGTGGTGA-3′) containing three BstUI restriction sites (5′-CGCG) was used to generate 76 bp DNA amplicons by PCR reactions with reaction buffer containing 0.1 mM of each dNTP (or 5mdCTP or 5hmdCTP in place of dCTP), and Taq polymerase (Roche; Branchburg, NJ). PCR cycling conditions in 25 μl reaction volumes were as follows: 94°C for 2 min and then 22 cycles of PCR at 94°C for 20 s, 55°C for 25 s and 72°C for 30 s, followed by a final extension step at 72°C for 2 min, using the forward primer 5′-CCTCACCATCTCAACCAATA-3′ and the reverse primer 5′-TCACCACTTCTCCCTCAAT-3′. In order to effectively remove unmodified DNA templates from the final products, another 30 cycles of subsequent PCR amplifications were performed using 0.5 µl of first round PCR products in 50 μl of reaction volume under the same reaction conditions. PCR products were then purified using PCR purification kits (Qiagen; Valencia, CA). These three oligonucleotides containing C, 5mC or 5hmC at all three BstUI sites are referred to as C76, 5mC76 and 5hmC76, respectively. In addition, 76-mer oligonucleotides (sequence 5′-CCTCACCATCTCAACCAATATTATATTACGCGTATAACGCGTATTGCGC GCTATAATATTGAGGGAGAAGTGGTGA-3′) containing MluI (5′-ACGCGT), NruI (5′-ACGCGT) and HhaI (5′-GCGC) restriction sites were prepared as described above. The purified PCR products were digested with methylation-sensitive restriction enzymes. The digested PCR products were separated by electrophoresis on 3% Nusieve GTG agarose gels (Cambrex; Charles City, IA). Oligonucleotides referred to as 5mC5hmC76 contain both 5mC and 5hmC, with 5hmC bases at the central BstUI site and 5mC bases at the 5′ and 3′ BstUI sites. They were synthesized using 5-hydroxymethylcytosine phosphoramidite (Glen Research; Sterling, VA). A control oligonucleotide with 5mC at the 5′ and 3′ BstUI sites and normal C at the central BstUI site was also prepared and is referred to as 5mC76a. All restriction enzymes were obtained from New England Biolabs (Ipswich, MA).
Combined bisulfite restriction and bisulfite sequencing analysis
Bisulfite conversion and purification of 76-mers were accomplished using the EpiTect Bisulfite kit (Qiagen; Valencia, CA). Each purified 76-mer 0.5 µg were treated with sodium bisulfite and the obtained PCR products were subjected to combined bisulfite restriction analysis (COBRA) (13). Bisulfite modified DNAs were amplified using the following primers; the forward primer 5′-CCCTTTTATTATTTTAATTAATATTATATT-3′ and reverse primer 5′-TCACCACTTCTCCCTCAAT-3′. The reaction buffer contained all four regular dNTPs and Hotstart Taq polymerase (Qiagen) and the samples were incubated at 95°C for 15 min, and then 48 cycles of PCR at 94°C for 30 s, 45°C for 30 s and 72°C for 30 s were performed, followed by a final extension step at 72°C for 3 min. The PCR products were digested with the BstUI restriction enzyme, which cleaves only methylated DNA after bisulfite conversion. The digested PCR products were separated by electrophoresis on 3% Nusieve GTG agarose gels (Cambrex). For sequence analysis, the PCR products obtained after bisulfite conversion were purified using QIAquick PCR purification kits (Qiagen) and were then ligated into the pCR2.1 TA cloning vector (Invitrogen; Carlsbad, CA). Ten colonies for each cloned sample were sequenced and evaluated. Quantitative PCR with 1 ng of 76-mer templates was performed at 95°C for 3 min followed by 40 cycles at 95°C for 10 s and 50°C for 45 s. PCR was performed with primers as described above and the probe 5′-CGCGTATATCGCGTATTTCGCG-3′ with 5′-Cy5 and 3′-Iowa Black RQ-Sp modifications (IDT; Coralville, IA) using 0.6 units iTaq polymerase in an iQ5 real-time PCR cycler (Biorad; Hercules, CA). Data was analyzed with the iQ5 optical system software.
DNA immunoprecipitation with anti-5mC antibody
To test antibody affinity towards modified cytosines, immunoprecipitation with an antibody directed against 5-methylcytidine was carried out as described previously with some modifications (14). Each purified 76-mer was 32P-end-labeled with T4 polynucleotide kinase and [γ-32P]ATP and purified by G-50 spin columns (Roche). These end-labeled oligomers were denatured in TE buffer for 10 min at 98°C and immediately chilled on ice for 10 min. Approximately 1 × 105 cpm of each 76 bp oligomer and 1 μg of a mouse monoclonal anti-5mC antibody (Eurogentec; Seraing, Belgium) in a final volume of 200 μl IP buffer (10 mM sodium phosphate pH 7.0, 140 mM NaCl and 0.05% Triton X-100) were incubated for 2 h at 4°C on a rocking platform. To allow selective collection of immunocaptured 76-mers, the mixtures were then incubated with 7 μl of magnetic Dynabeads M-280 sheep antibody to mouse IgG (Dynal Biotech), pre-washed with PBS including 0.1% BSA, for 2 h at 4°C on a rocking platform and washed three times with 600 μl of IP buffer for 5 min at room temperature. The levels of immunocaptured oligomers were measured using a liquid scintillation counter (LS-6500, Beckman Coulter; Fullerton, CA) and the data were displayed as % by referring to the level of 5mC immunocaptured by anti-5mC antibody.
Binding of the MBD2b/MBD3L1 complex and other MBD proteins to modified cytosines
For gel mobility shift assays, His-tagged MBD2b, MBD3L1 and MBD4 proteins were prepared according to published procedures (15,16). Recombinant MBD4 was kindly provided by David Baker and Timothy O’Connor (City of Hope). Full-length MBD1 protein was obtained from Abnova (Taipei, Taiwan). The 32P-end-labeled probes were prepared as described above. Approximately 200 ng of recombinant proteins (MBD2b and MBD3L1 were pre-incubated on ice for 30 min) and 0.1 ng of probes (1 × 104 cpm) were incubated at room temperature for 40 min in binding buffer containing 20 mM HEPES, pH 7.9, 1 mM EDTA, 3 mM MgCl2, 2 mM dithiothreitol, 4% glycerol, 0.1% Triton X-100 and 125 ng of sonicated JM110 (dcm minus) Escherichia coli genomic DNA. The protein-DNA complexes were fractionated by electrophoresis on a 5% non-denaturing polyacrylamide gels in 1 × Tris-buffered EDTA at 4°C and visualized by autoradiography.
RESULTS
Preparation of modified oligonucleotides
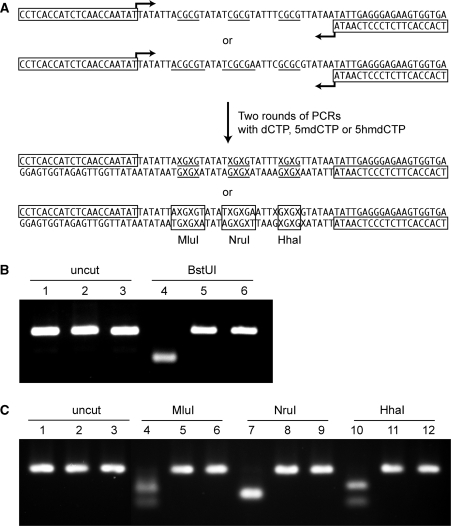
A 76-mer oligonucleotide sequence was designed for use in this study (Figure 1A). The synthesized DNA fragment contained three recognition sites for the methylation-sensitive restriction enzyme BstUI (5′-CGCG) allowing for restriction digest analysis and the incorporation of the modified bases 5mC and 5hmC only at CpG sites. In order to prepare the 76-mers including unmodified (C76), methylated (5mC76) or hydroxymethylated (5hmC76) cytosines at CpG sites, a PCR method was used allowing incorporation of only normal cytosines (by using dCTP) or modified cytosines (by using 5mdCTP or 5hmdCTP), similar as described by Kriaucionis and Heintz (7). We designed the 76-mers such that only the BstUI sites selectively contained the normal (C) or the specific modified cytosines (5mC or 5hmC) on newly synthesized DNA strands extended from primers. To assure that our final PCR products were depleted of starting template DNA, subsequent PCRs were carried out by using 0.5 μl of the first round PCR products as template. Oligonucleotides containing both 5mC and 5hmC were synthesized using phosphoramidite chemistry.
Figure 1.
Preparation and validation of modified oligonucleotides. (A) Sequence and preparation of the 76-mers used in the assays. The synthesized DNA fragments contain three BstUI sites (5′-CGCG, underlined). Two rounds of PCR were performed to obtain C76, 5mC76 or 5hmC76 containing C, 5mC or 5hmC at CpG sites. X indicates normal C, or modified bases 5mC or 5hmC. The boxed sequences indicate the PCR primers. Oligonucleotides containing both 5mC and 5hmC were synthesized chemically. (B) Analysis of PCR products C76, 5mC76 and 5hmC76 by BstUI cleavage. PCR products were prepared and digested with the methylation-sensitive restriction enzyme BstUI, then separated and visualized by electrophoresis on 3% Nusieve GTG agarose gels. Lane 1 (C76), lane 2 (5mC76) and lane 3 (5hmC76) show clean single bands for each of the PCR products. After BstUI digestion, C76 (lane 4) was fully digested, whereas samples in lane 5 (5mC76) and lane 6 (5hmC76) resisted digestion. (C) Analysis of PCR products C76, 5mC76 and 5hmC76 by cleavage with different methylation-sensitive restriction enzymes. C76 (lanes 1, 4, 7 and 10), 5mC76 (lanes 2, 5, 8 and 11) and 5hmC76 (lanes 3, 6, 9 and 12) were left untreated (lanes 1–3) or were incubated with MluI (lanes 4–6), NruI (lanes 7–9) or HhaI (lanes 9–12).
To analyze the final PCR products, C76, 5mC76 and 5hmC76 were separated and visualized by electrophoresis on 3% Nusieve GTG agarose gels (Figure 1B, lanes 1–3). Analysis of PCR products indicated one single PCR product band for each of the synthesized DNA fragments consisting of normal or specific modified cytosines. In order to further test if C, 5mC or 5hmC were present in the synthesized PCR fragments, we performed AflIII (5′TCGCGA) restriction enzyme digestion with these PCR products and thin layer chromatography (data not shown). The methylation-sensitive restriction enzyme BstUI cannot digest the fragment if the BstUI restriction site is modified by methylation on the 5-position of cytosine at CpGs. In Figure 1B, lanes 4–6, we observed that C76 was fully digested, while the 5mC76 and 5hmC76 oligomers resisted digestion by BstUI. These data show that the selectivity of the methylation-sensitive restriction endonuclease BstUI is affected by the presence of 5hmC within the restriction sites, which is consistent with previous reports for other methylation-sensitive restriction enzymes (8,17). Using a similar approach to synthesize templates, we also tested the methylation-sensitive restriction enzymes MluI, NruI and HhaI for reactivity towards target sequences containing 5hmC in the recognition sequences. These enzymes were also strongly inhibited by presence of 5hmC (Figure 1C).
Combined bisulfite restriction and bisulfite sequencing analysis
Bisulfite assays are widely used in DNA methylation studies due to the selective chemical reaction of sodium bisulfite with cytosine versus 5mc residues (18–20). Thus, it is important to test if the assay can distinguish 5hmC from C or 5mC.
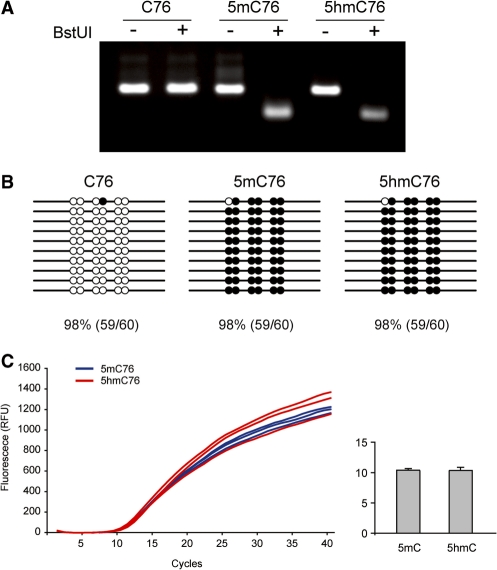
In total, 0.5 µg of purified 76-mers were treated with sodium bisulfite and the products were then amplified using normal dNTPs and Taq polymerase. Bisulfite-treated 5hmC76 as well as C76 and 5mC76 were successfully amplified, indicating that treatment of 5hmC-containing templates with sodium bisulfite does not affect PCR amplification. Using these DNAs amplified from bisulfite-treated 76-mers, we first performed a COBRA assay using BstUI (5′-CGCG), and observed that 5mC76 and 5hmC76 were clearly digested, but C76 fully resisted digestion with BstUI (Figure 2A). This indicates that 5mC and 5hmC were not converted to uracil during bisulfite treatment and hence retained the BstUI restriction sites, allowing for digestion of the PCR products.
Figure 2.
Comparison of the reactivity of sodium bisulfite towards 5mC and 5hmC. (A) COBRA assay. The PCR products obtained after sodium bisulfite treatment of the C-, 5mC- and 5hmC-containing templates were analyzed by the BstUI combined bisulfite restriction analysis (COBRA) method. Bisulfite-converted 76-mers were PCR amplified and digested with the restriction enzyme BstUI (5′-CGCG), which produces digestion products only when restriction sites are not converted by bisulfite. The electrophoresis on 3% Nusieve GTG agarose gels shows that 5mC76 and 5hmC76 were clearly digested, but C76 fully resisted digestion with BstUI. (B) Bisulfite sequencing. The PCR products obtained after bisulfite conversion were cloned into the pCR2.1 TA cloning vector (Invitrogen) and ten individual clones were sequenced. The 76 base pairs of C76, 5mC76 and 5hmC76 contain six cytosines or modified cytosines (5mC or 5hmC) on each strand. The modified (unconverted) cytosines are depicted as solid black circles while the unmodified cytosines are shown as open circles. Each CpG site was counted separately. (C) Real-time PCR with templates containing 5mC and 5hmC. Blue (5mC76) and red (5hmC76) curves are for three independent reactions. In the column graph, the ct value for the two templates is shown with standard deviation.
The result was further verified by bisulfite sequencing analysis. The amplified PCR fragments following bisulfite treatment were cloned into pCR2.1 TA cloning vectors and ten individual clones were sequenced (Figure 2B). The 76 base pairs of C76, 5mC76 and 5hmC76 contain six cytosines or modified cytosines (5mC or 5hmC) on each strand, and sequencing data showed that 98% (59/60) of the 5mC or 5hmC was read by polymerase as cytosine during PCR amplification subsequent to bisulfite treatment, while 98% (59/60) of unmodified cytosines were converted to uracils on C76 and were read as thymines in the sequencing reads. These results indicate that bisulfite treatment can distinguish 5mC and 5hmC from cytosine but cannot distinguish between 5mC and 5hmC. To address the issue whether templates containing 5hmC may be amplified less efficiently than templates containing 5mC, we performed quantitative PCR reactions with the different templates. Manual PCR over a wide range of cycle numbers initially indicated that templates with 5hmC and 5mC were amplified with similar kinetics but slightly less efficiently than templates containing C (data not shown). Real-time PCR indicated clearly that the 5hmC- and 5mC-containing templates had similar amplification efficiencies (Figure 2C).
DNA immunoprecipitation with anti-5mC antibody
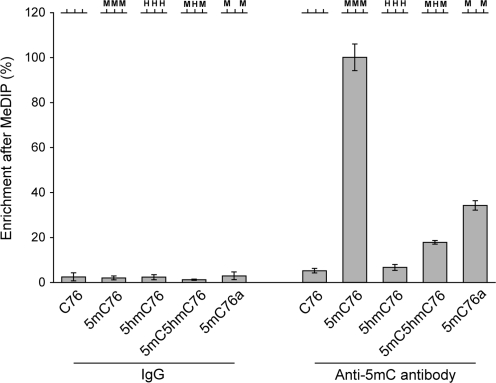
In addition to sequence-specific enzymatic cleavage and chemical conversion of 5hmC, affinity-based detection of 5hmC was investigated with a monoclonal antibody directed against 5-methylcytidine. This antibody is used commonly in the MeDIP procedure (14). To selectively collect a population of 76-mers recognized by the antibody, 32P-end-labeled 76-mers containing normal or modified cytosines were subjected to immunoprecipitation (IP) with anti-5mC antibody and then immunocaptured by using a secondary antibody conjugated to magnetic beads. The immunocaptured 76-mers were subjected to liquid scintillation counting. As a negative control we used normal mouse IgG for IP with C76, 5mC76 or 5hmC76. As seen in Figure 3, we observed a high affinity of the anti-5mC antibody towards 5mC on 5mC76 relative to C76 and 5hmC76. Comparatively, the affinity of the anti-5mC antibody to C76 or 5hmC76 is similar to that of control IgG. The results indicate that the anti-5mC antibody has a high selective affinity to 5mC but not to cytosine or 5hmC in DNA. We also tested if the antibody can still recognize a DNA fragment that contains both 5mC and 5hmC. The antibody can recognize such a fragment (5mC5hmC76 in Figure 3) albeit with lower efficiency compared to the same fragment that lacks 5hmC and contains C at the 5hmC positions (5mC76a in Figure 3).
Figure 3.
DNA immunoprecipitation with anti-5mC antibody. Immunoprecipitation with an antibody against 5-methylcytidine was carried out to test the antibody’s affinity towards modified cytosines. The levels of immunocaptured 76-mers were measured using liquid scintillation counting. As a control, normal mouse IgG was used for immunoprecipitation. The oligonucleotides C76, 5mC76 and 5hmC76 were synthesized by PCR and contain C, 5mC or 5hmC at three BstUI sites as schematically indicated at the top of the Figure (see Figure 1 for sequence). Oligonucleotides 5mC5hmC76 and 5mC76a were prepared by chemical synthesis and contain 5hmC or C at the central BstUI site. Experiments were done in triplicates and the standard deviation is shown.
Methyl-CpG-binding proteins and 5hmC binding
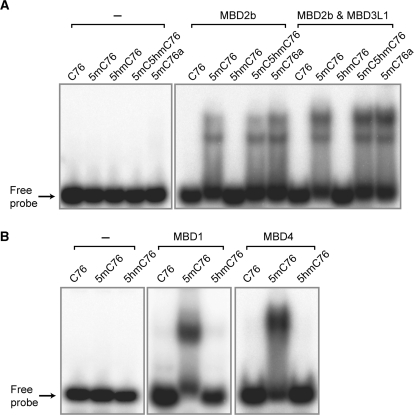
Among the methylated-CpG binding domain (MBD) family of proteins, MBD2b has the highest affinity to methylated CpG sites with an ability to effectively distinguish methylated from unmethylated CpGs (21). We proceeded to test the binding of MBD2b to 5hmC-containing oligomers. Using recombinant His-tagged MBD2b protein, we tested the affinity of this protein to 32P-end-labeled 76-mers containing normal or modified cytosines at CpG sites. Gel mobility shift assays using a 5% non-denaturing polyacrylamide gel were performed. As seen in Figure 4A, recombinant MBD2b protein can only bind to 5mC76 but not to C76 or 5hmC76 indicating that MBD2b can only identify 5mC and that its binding to 5mC can be inhibited by oxidation of 5mC to 5hmC at CpG sites. MBD2b is able to bind to an oligonucleotide that contains both 5mC and 5hmC (indicated as 5mC5hmC76 in Figure 4). In addition to MBD2b, we also tested the binding of full-length MBD1 and MBD4 proteins towards oligonucleotides containing both modified bases. Binding of these MBD proteins also is strongly inhibited by presence of 5hmC (Figure 4B).
Figure 4.
Affinity of MBD proteins towards 5mC- and 5hmC-containing oligomers. (A) Binding of MBD2b and the MBD2b/MBD3L1 complex (MIRA complex) to modified cytosines. 76-mer oligonucleotides containing symmetrically modified cytosines, 5mC or 5hmC at CpG sites were incubated with recombinant MBD2b alone (200 ng of protein) or with the MBD2b/MBD3L1 complex (100 ng of each protein). The mobility shift assay was carried out using a 5% non-denaturing polyacrylamide gel. The oligonucleotides C76, 5mC76 and 5hmC76 were synthesized by PCR and contain C, 5mC or 5hmC at three BstUI sites (see Figure 1). Oligonucleotides 5mC5hmC76 and 5mC76a were prepared by chemical synthesis and contain 5hmC or C at the central BstUI site (see Figure 3). (B) Binding of MBD1 and MBD4 to modified cytosines. These proteins bind effectively to methylated CpG sequences but do not bind to the same sequences containing 5hmC in place of 5mC.
The binding affinity of MBD2b to 5mC at CpG sites is enhanced by formation of a complex with MBD3L1, a protein with substantial homology to MBD2 and MBD3 but lacking the MBD (15,22). The complex of MBD2b and MBD3L1 is used in the methylated-CpG island recovery assay (MIRA) technique, a method used for genome-scale analysis of mammalian DNA methylation patterns (16,22,23). Therefore, the binding affinity of the MBD2b/MBD3L1 complex to 5mC and 5hmC was tested. In Figure 4A, we show that the complex has little or no affinity for C76 or 5hmC76, suggesting that MBD2b and MBD3L1 form a protein complex, which can only recognize 5mC at CpG sequences, but not 5hmC at the same sites. The complex can still bind to an oligonucleotide that contains both 5mC and 5hmC (Figure 4A).
DISCUSSION
The recent discovery of substantial amounts of the modified DNA base 5-hydroxymethylcytosine in certain mammalian cell types (7,8) has raised awareness of previously undiscovered DNA modifications with potential physiological significance. The similarity of 5mC and its oxidation product 5hmC suggests that investigations into the methodology used for detecting 5mC in mammalian DNA need to be conducted. We show here that two established mapping techniques for 5mC in mammalian genomes, MeDIP and MIRA (14,16), are in fact specific for 5mC. Both the anti-5mC antibody and the methylated CpG binding complex consisting of MBD2b and MBD3L1 cannot recognize the oxidized base. Also the MBD2b protein alone, as well as full-length MBD1 and MBD4 (Figure 4) and the MBD domain of MeCP2 (10) specifically bind to 5mC and binding does not occur when 5mC is oxidized. Lack of binding of MBD family proteins to 5hmC will likely have biological significance in vivo. The 5mC oxidation pathway carried out by the TET1 protein (8,24) may be relevant for reactivation of gene expression from methylation-silenced promoters by displacing bound transcriptional repressors of the MBD family type.
An important finding of our study is that the most commonly used technique for DNA methylation mapping, sodium bisulfite sequencing and its derivative approaches such as the COBRA assay, cannot distinguish between 5mC and 5hmC. Earlier studies have shown that 5hmC can react with bisulfite and, instead of promoting the usual cytosine deamination process, the reaction gives rise to cytosine 5-methylenesulfonate as the product. Cytosine 5-methylenesulfonate was only very slowly deaminated by treatment with bisulfite (25). It was suggested that this adduct may interfere with PCR and sequencing reactions (26). However, we show here that bisulfite-treated DNA templates containing 5hmC can be amplified efficiently (Figure 2) and that, analogous to 5mC, 5hmC does not undergo conversion to a deaminated cytosine ring that would be read as a T base after bisulfite treatment and PCR. Thus, although the biological significance of 5mC and 5hmC at CpG sequences may be completely different, as exemplified by the inability of MBD family proteins to bind to 5hmC, the readout of bisulfite sequencing for these two modified bases is exactly identical (Figure 2).
A specific methodology for detection of 5hmC will need to be developed. For mapping purposes, not requiring single base resolution, an antibody specific for hydroxymethylated cytosine can be prepared. If a particular stretch of DNA contains both 5mC and 5hmC, antibodies against both modified bases will need to be used. In addition, for single base resolution mapping of 5hmC, the requirements are more challenging. Specific chemical or enzymatic cleavage of 5hmC coupled to ligation-mediated PCR (27) may be one possibility. More indirect approaches, by which 5hmC bases or 5hmC-containing molecules would first be removed, and then the bisulfite sequencing data before and after removal of 5hmC would be compared, are also conceivable. This could be done, for e.g. by selecting 5mC-containing DNA molecules by MeDIP or MIRA. Alternatively, the identification of enzymatic activities that either remove 5hmC from DNA by base excision repair, or remove the hydroxymethyl group from the modified base, may be required, in particular if 5mC and 5hmC are present in the same DNA strand. In summary, our data indicate that significant limitations exist for interpreting data obtained from commonly used techniques to map mammalian CpG methylation.
FUNDING
Funding for open access charge: National Institutes of Health (grant AG036041).
Conflict of interest statement. Under a licensing agreement between City of Hope and Active Motif (Carlsbad, CA) the methylated-CpG island recovery assay (MIRA) technique was licensed to Active Motif and G.P.P. is entitled to a share of the royalties received by City of Hope from sales of the licensed technology.
ACKNOWLEDGEMENTS
The authors thank David Baker and Timothy O’Connor for a gift of recombinant MBD4 protein and Piotr Swiderski for oligonucleotide synthesis.
REFERENCES
- 1.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 2.Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet. Cell. Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 3.Bestor TH. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 4.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 5.Hahn MA, Pfeifer GP. Methods for genome-wide analysis of DNA methylation in intestinal tumors. Mutat. Res. 2009 doi: 10.1016/j.mrfmmm.2009.10.005. doi:10.1016/j.mrfmm.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tost J. DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Methods Mol. Biol. 2009;507:3–20. doi: 10.1007/978-1-59745-522-0_1. [DOI] [PubMed] [Google Scholar]
- 7.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liutkeviciute Z, Lukinavicius G, Masevicius V, Daujotyte D, Klimasauskas S. Cytosine-5-methyltransferases add aldehydes to DNA. Nat. Chem. Biol. 2009;5:400–402. doi: 10.1038/nchembio.172. [DOI] [PubMed] [Google Scholar]
- 10.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boorstein RJ, Chiu LN, Teebor GW. Phylogenetic evidence of a role for 5-hydroxymethyluracil-DNA glycosylase in the maintenance of 5-methylcytosine in DNA. Nucleic Acids Res. 1989;17:7653–7661. doi: 10.1093/nar/17.19.7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rusmintratip V, Sowers LC. An unexpectedly high excision capacity for mispaired 5-hydroxymethyluracil in human cell extracts. Proc. Natl Acad. Sci. USA. 2000;97:14183–14187. doi: 10.1073/pnas.97.26.14183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 15.Jiang CL, Jin SG, Pfeifer GP. MBD3L1 is a transcriptional repressor that interacts with methyl-CpG-binding protein 2 (MBD2) and components of the NuRD complex. J. Biol. Chem. 2004;279:52456–52464. doi: 10.1074/jbc.M409149200. [DOI] [PubMed] [Google Scholar]
- 16.Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- 17.Huang LH, Farnet CM, Ehrlich KC, Ehrlich M. Digestion of highly modified bacteriophage DNA by restriction endonucleases. Nucleic Acids Res. 1982;10:1579–1591. doi: 10.1093/nar/10.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark SJ, Statham A, Stirzaker C, Molloy PL, Frommer M. DNA methylation: bisulphite modification and analysis. Nat. Protoc. 2006;1:2353–2364. doi: 10.1038/nprot.2006.324. [DOI] [PubMed] [Google Scholar]
- 20.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraga MF, Ballestar E, Montoya G, Taysavang P, Wade PA, Esteller M. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003;31:1765–1774. doi: 10.1093/nar/gkg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rauch T, Pfeifer GP. Methylated-CpG island recovery assay: a new technique for the rapid detection of methylated-CpG islands in cancer. Lab. Invest. 2005;85:1172–1180. doi: 10.1038/labinvest.3700311. [DOI] [PubMed] [Google Scholar]
- 23.Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc. Natl Acad. Sci. USA. 2009;106:671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayatsu H, Shiragami M. Reaction of bisulfite with the 5-hydroxymethyl group in pyrimidines and in phage DNAs. Biochemistry. 1979;18:632–637. doi: 10.1021/bi00571a013. [DOI] [PubMed] [Google Scholar]
- 26.Loenarz C, Schofield CJ. Oxygenase catalyzed 5-methylcytosine hydroxylation. Chem. Biol. 2009;16:580–583. doi: 10.1016/j.chembiol.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Pfeifer GP, Drouin R, Riggs AD, Holmquist GP. In vivo mapping of a DNA adduct at nucleotide resolution: detection of pyrimidine (6-4) pyrimidone photoproducts by ligation-mediated polymerase chain reaction. Proc. Natl Acad. Sci. USA. 1991;88:1374–1378. doi: 10.1073/pnas.88.4.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]