Abstract
Postreplication repair (PRR) is a pathway that allows cells to bypass or overcome lesions during DNA replication1. In eukaryotes, damage bypass is activated by ubiquitylation of the replication clamp PCNA through components of the RAD6 pathway2. Whereas monoubiquitylation of PCNA allows mutagenic translesion synthesis by damage-tolerant DNA polymerases3-5, polyubiquitylation is required for an error-free pathway that likely involves a template switch to the undamaged sister chromatid6. Both the timing of PRR events during the cell cycle and their location relative to replication forks, as well as the factors required downstream of PCNA ubiquitylation, have remained poorly characterised. Here we demonstrate that the RAD6 pathway normally operates during S phase. However, using an inducible system of DNA damage bypass in budding yeast, we show that the process is separable in time and space from genome replication, thus allowing direct visualisation and quantification of productive PRR tracts. We found that both during and after S phase ultraviolet radiation-induced lesions are bypassed predominantly via translesion synthesis, whereas the error-free pathway functions as a backup system. Our approach has for the first time revealed the distribution of PRR tracts in a synchronised cell population. It will allow an in-depth mechanistic analysis of how cells manage the processing of lesions to their genomes during and after replication.
Traditional analyses of PRR, involving measurements of the amount and size of DNA synthesised after exposure of cells to genotoxic agents, have indicated that replication of damaged templates does not lead to prolonged replication fork arrest, but causes an accumulation of daughter strand gaps in the genome7-9. Electron microscopy revealed gaps on both daughter strands in the vicinity of replication forks, suggesting re-priming downstream of lesions even on the leading strand10. In vertebrate cells, a partitioning of damage bypass into fork- and gap-associated events was reported11. These notions raise the question as to what extent DNA damage bypass is coupled to replication progression. In order to address theis issue, we designed a system that would allow us to activate the RAD6 pathway at will by placing RAD18, encoding the ubiquitin ligase responsible for PCNA monoubiquitylation and a limiting factor for PRR12, under control of inducible promoters (Fig. S1). The GAL1-10 promoter afforded overexpression of RAD18 and enforced constitutive PCNA ubiquitylation (Fig. S2a, b), whereas a truncated version13 (GALS-RAD18) and a doxycycline-repressible construct14 (Tet-RAD18) resulted in near wild-type (WT) protein levels, and PCNA modification required treatment with a DNA-damaging agent such as ultraviolet (UV) radiation (Fig. S2a, c, d). All constructs conferred UV sensitivities comparable to WT RAD18 when induced (Fig. S3).
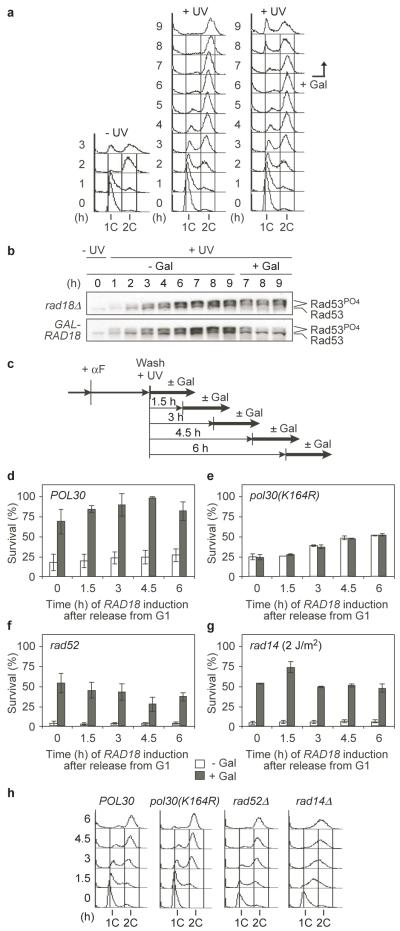
When synchronised uninduced GAL-RAD18 cultures were treated with low UV doses at the G1/S boundary, cells passed through S phase and accumulated in G2/M with an activated checkpoint (Fig. 1a, b, S4). Induction of RAD18 at this stage resulted in progression to the next G1 phase and deactivation of the checkpoint (Fig. 1a, b). In order to determine whether this was due to productive damage bypass, we induced RAD18 at various times during the cell cycle and compared survival (Fig. 1c). Surprisingly, we found that the extent to which viability was restored was independent of the timing of RAD18 expression (Fig. 1d, h). Use of GALS-RAD18 confirmed that the rescue was not due to forced overexpression of RAD18 (Fig. S5). In order to verify that the effect reflected the PRR pathway mediated by PCNA ubiquitylation, we repeated the experiment in a ubiquitylation-deficient PCNA mutant, pol30(K164R)2. As expected, RAD18 induction did not promote survival above uninduced controls (Fig. 1e, h). In contrast, neither homologous recombination (HR, represented by RAD52) nor nucleotide excision repair (NER, represented by RAD14) was required for rescue of survival by RAD18 (Fig. 1f, g, h). Instead, viability in the absence of Rad18 was significantly reduced in both mutants, reflecting the synergistic relationship between PRR and HR or NER, respectively. Hence, ubiquitin-mediated DNA damage bypass can effectively be delayed without adverse effects on viability until after the bulk of genome replication is completed.
Figure 1. Ubiquitin-dependent DNA damage bypass can be delayed until after genome replication.
a Cell cycle profiles of synchronised GAL-RAD18 cultures either unirradiated (left) or treated with 10 J/m2 UV (middle, right). RAD18 expression was induced by galactose (Gal, right). b Time course of Rad53 phosphorylation in rad18Δ and GAL-RAD18, treated as above. c Experimental scheme for GAL-RAD18 induction during and after S phase. αF: alpha factor. d-g Survival of the indicated GAL-RAD18 strains (UV dose: 10 J/m2 – rad14Δ: 2 J/m2). Error bars represent standard deviations from 3 experiments. d POL30; e pol30(K164R); f rad52Δ; g rad14Δ. h Cell cycle profiles at the time of RAD18 induction.
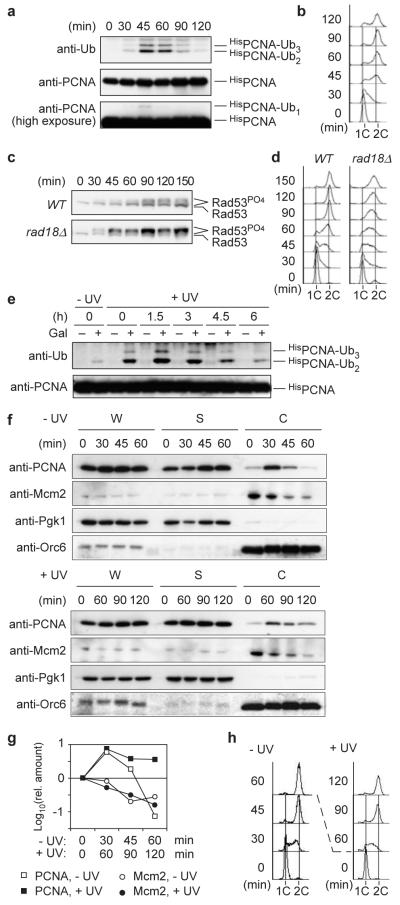
These observations, together with the notion that expression of REV1, encoding one of the damage-tolerant polymerases, peaks in G2/M15, raised the possibility that PRR is naturally delayed until after S phase. However, when G1-synchronised WT cells were irradiated and released, PCNA ubiquitylation was maximal in S phase, suggesting that damage bypass is at least initiated during replication (Fig. 2a, b). Moreover, whilst WT cells only showed a transient checkpoint signal around G2/M, PRR defects resulted in strong checkpoint activation early during S phase (Fig. 2c, d), indicating that the RAD6 pathway normally contributes to damage processing, thereby effectively preventing checkpoint signalling during replication.
Figure 2. PRR normally operates during S phase, but chromatin-bound PCNA can be ubiquitylated in G2/M.
a Ubiquitylation of HisPCNA in WT cells after release from G1 arrest (UV dose: 20 J/m2). b Cell cycle profile of the above culture. c Time course of Rad53 phosphorylation in WT and rad18Δ cells treated as above. d Cell cycle profiles of the above cultures. e HisPCNA ubiquitylation in GALS-RAD18 cells treated as described in Fig. 1c. f Distribution of PCNA and Mcm2 in whole-cell extracts (W), soluble (S) and chromatin-associated (C) fractions prepared from G1-irradiated cultures (± 20 J/m2) at the indicated times after release. Pgk1 and Orc6 served as controls for soluble and chromatin-bound proteins, respectively. g Quantification of PCNA and Mcm2 in the chromatin fractions. h Cell cycle profiles of the above cultures. Since irradiation slows down cell cycle progression, different time scales were used for damaged versus undamaged cells in panels f-h in order to relate comparable cell cycle stages to each other.
Ubiquitylation of PCNA requires the clamp to reside on DNA12,16,17. Hence, damage to G2-arrested cells does not normally result in modification12. However, when GALS-RAD18 cells were irradiated in G1 and allowed to complete the cell cycle in the absence of Rad18, induction of PRR in G2/M resulted in PCNA ubiquitylation (Fig. 2e). Consistent with the modification, fractionation of cell extracts revealed a prolonged association of PCNA in G2/M with chromatin in response to DNA damage inflicted at the beginning of S phase (Figs. 2f-h). The association pattern was unchanged in rad18Δ, indicating that ubiquitylation is not responsible for the localisation pattern of PCNA (Fig. S6). Dissociation of the replication factor Mcm2 over the course of the cell cycle suggested that the chromatin-bound PCNA was not engaged at active replication forks at the late time points (Figs. 2f-h, S6). We interpret this finding to indicate that DNA damage left unrepaired during S phase causes some PCNA to remain associated with daughter-strand gaps even after bulk DNA synthesis is completed, and that this residual pool of PCNA is the target for ubiquitylation in G2/M. Alternatively, PCNA may have been reloaded and subsequently ubiquitylated at PRR sites.
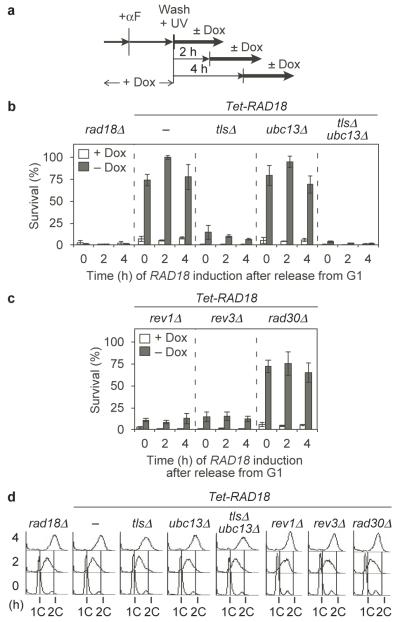
Having established that ubiquitin-dependent PRR can rescue cell viability irrespective of its timing, we wished to determine which of its two branches, TLS or error-free damage avoidance, was mainly responsible for survival during and after S phase, respectively. We therefore analysed viability of Tet-RAD18 cells deficient in all three damage-tolerant polymerases (rev1Δ rev3Δ rad30Δ, designated as tlsΔ) and/or UBC13 (Fig. 3a). Whereas the former strain should be devoid of TLS activity, deletion of UBC13 should selectively prevent PCNA polyubiquitylation and thereby error-free damage avoidance2, and combination of the mutations should completely abolish PRR. Lack of UBC13 had no measurable effect on survival during S phase or in G2/M, whereas viability was drastically reduced in tlsΔ, suggesting that throughout the cell cycle TLS is much more important for tolerance to UV lesions than error-free bypass (Fig. 3b, d). Examination of individual polymerase mutants revealed major contributions of Rev1 and polymerase ζ, encoded by REV3 (Fig. 3c, d). Further sensitisation in tlsΔ ubc13Δ beyond the level of tlsΔ indicated that polyubiquitin-dependent PRR might still play a role in a backup function (Fig. 3b, S7). Hence, both TLS and error-free PRR appear capable of acting during S phase and in a postreplicative manner.
Figure 3. UV-induced lesions are bypassed predominantly by TLS.
a Experimental scheme for Tet-RAD18 induction during and after S phase (UV dose: 10 J/m2). b, c Survival of the indicated strains, relative to unirradiated controls. Standard deviations were derived from 4 experiments. d Cell cycle profiles of the indicated strains at the time of plating.
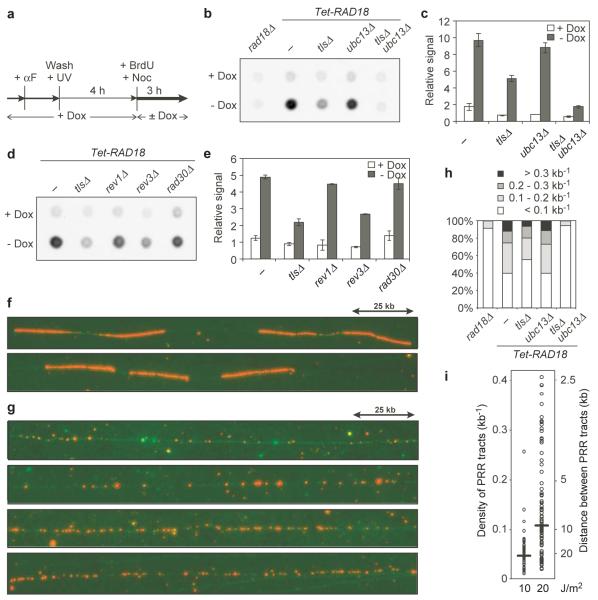
The notion that PRR can effectively be uncoupled from genome replication allowed us to visualise and quantify bypass-dependent DNA synthesis by means of 5-bromodeoxyuridine (BrdU) labelling18. In order to observe PRR-associated DNA synthesis, we irradiated uninduced Tet-RAD18 cells in G1, allowed passage through S phase and induced RAD18 in the presence of BrdU when cells had reached the G2/M arrest (Fig. 4a). Nocodazole was used to prevent DNA synthesis due to entry into the next cell cycle. Analysis of a control strain, rad18Δ, revealed virtually no BrdU incorporation (Fig. 4b), while induction of RAD18 resulted in de novo DNA synthesis as judged by the appearance of a BrdU signal. Inactivation of TLS caused a much stronger reduction in BrdU incorporation than deletion of UBC13, and no signal above the background was observed in tlsΔ ubc13Δ (Fig. 4b, c). These results are consistent with the effects of TLS and Ubc13 on survival, again indicating that TLS accounts for the majority of PRR. Quantification of the signals also showed that among the three single polymerase mutants, only rev3Δ resulted in an appreciable PRR defect, suggesting significant redundancy among the polymerases, but a dominant role of polymerase ζ in postreplicative processing of UV lesions (Fig. 4d, e). The discrepancy between the survival defect of rev1Δ and its ability to incorporate BrdU is surprising, but it might indicate ubiquitin-independent contributions of this polymerase to damage processing, which would affect overall damage sensitivity but would not be detectable by RAD18 induction. Consistent with this model, a rad18Δ tlsΔ mutant is more UV-sensitive than rad18Δ alone19, and similar findings have been reported for vertebrate cells11.
Figure 4. Quantification and visualisation of PRR tracts in G2/M-arrested cells.
a Experimental scheme for labelling of PRR tracts in Tet-RAD18 cells (UV dose: 20 J/m2). b Dot blot for detection of BrdU incorporation. c Quantification of BrdU signals with s.d. from a minimum of 3 independent experiments. d Dot blot for detection of BrdU incorporation in single TLS polymerase mutants. e Quantification of the above signals. f Fluorescence microscopy images of DNA fibres (green) labelled with BrdU (red) in HU-treated S phase cells. g Fluorescence microscopy images of DNA fibres labelled postreplicatively with BrdU in Tet-RAD18 cells. h Density distribution of BrdU patches. i Dose dependence of BrdU patch densities. Horizontal bars indicate median values.
The distribution of PRR tracts along chromosomal DNA was revealed by DNA fibre analysis. As a control, we visualised replication tracts in hydroxyurea (HU)-treated cells, detectable as strongly and continuously labelled regions mostly originating from bidirectional replication forks (Fig. 4f). Fibres observed after postreplicative labelling of irradiated Tet-RAD18 cells (Fig. 4a) looked strikingly different: DNA synthesis had occurred in numerous small patches dispersed along the chromosomes (Fig. 4g). Their almost complete absence in rad18Δ and in uninduced cells confirmed that they represented PRR and not replication tracts (Fig. S8a-c). Taking into account an average inter-origin distance of 40 to 100 kb20, the density of the patches also suggests that many of them do not originate from stalled forks. Whereas deletion of UBC13 had no notable effect on their distribution, inactivation of TLS caused some reduction in their density, and signals in the tlsΔ ubc13Δ mutant were again reduced to background (Fig. 4h, S8d). After irradiation with 10 and 20 J/m2, their density roughly correlated with the UV dose (Fig. 4i). Notably, at 40 J/m2 (Fig. S9a), the pattern became reminiscent of HU-stalled replication forks. At the same time, flow cytometry indicated that cells had not completed S phase by the time of RAD18 induction (Fig. S9b). Hence, it appeared that high UV doses might have impeded progression of replication forks to such an extent that replicative DNA synthesis and PRR could no longer be separated.
The finding that even low damage levels cause a G2/M checkpoint arrest in RAD18-deficient cells highlights the importance of PRR for completion of the cell cycle21. Based on previous measurements22,23 a UV dose of 20 J/m2 introduces approximately 0.3-0.5 cyclobutane pyrimidine dimers (CPDs)/kb into the budding yeast genome. The tract density determined by us therefore suggests that cells can process a significant fraction of all UV-induced lesions by PRR in G2/M. Although our data suggest that PRR is normally active during S phase, they show that damage tolerance is fully functional even when uncoupled from genome replication. A pronounced peak of REV1 expression during G2/M suggests that this mode of PRR may be biologically relevant15. The predominance of TLS over the error-free pathway is surprising and contrasts with previous reports based on the replication of plasmids containing a defined 6-4 photoproduct6 or exposure to chronic low-dose UV21. However, the balance between TLS and error-free bypass may depend as much on the nature of the lesion (mainly CPDs in our case) as on the overall amount of damage. Likewise, the dramatic UV sensitivity of TLS-deficient strains is unexpected, but may be explained by our use of synchronised cell populations exposed to damage immediately before the onset of replication. In this situation, alternative repair pathways might not be readily available before lesions are encountered by replication forks. Finally, although a prominent role of polymerase η might have been predicted based on its biochemical properties24, our analysis indicates some redundancy among the TLS polymerases. The unique importance of polymerase ζ could be connected to its role as an “extender polymerase” that cooperates with other TLS polymerases and can therefore not be replaced25-27. In contrast to traditional PRR assays that largely rely on unsynchronised cultures of extremely damage-sensitive, NER-deficient cells and do not address the relevance of the observed activity for survival7,9, our approach has permitted the visualisation of productive PRR in synchronised populations. Further characterisation of this experimental system should give insight into the structure of PRR tracts and the yet elusive mechanism of the error-free pathway of lesion bypass.
Methods Summary
Yeast strains
All experiments were performed in isogenic strains, listed in Table S1. Details about their construction are given in the additional Methods section.
Determination of cell survival and checkpoint activation
GAL-RAD18 cells were synchronised in G1, irradiated (254 nm) and released into S phase in lactate medium. At the indicated time points, RAD18 expression was induced for 2 h by addition of galactose. Survival relative to unirradiated cultures was determined by colony counting on glucose medium. Tet-RAD18 cells were synchronised, irradiated, and released in the presence of doxycycline. At the indicated times cells were plated directly onto medium with or without doxycycline for colony counting. Checkpoint activation was monitored by observing Rad53 phosphorylation in total cell extracts.
Detection of PCNA ubiquitylation
HisPCNA was isolated from HisPOL30 strains under denaturing conditions, and ubiquitylated forms were detected by Western blot as previously described2,4,28.
Chromatin binding assays
Fractionation of total cell extracts prepared by spheroplast lysis was achieved by centrifugation through a sucrose cushion essentially as described29.
Detection of BrdU-labelled DNA
Briefly, total DNA from cells grown in the presence of 0.4 mg/ml BrdU for 3 h, isolated by glass bead disruption and phenol/chloroform extraction, was denatured at 95°C, snap-cooled on ice and spotted onto nylon membrane. After UV-crosslinking the membrane was probed with an anti-BrdU antibody, followed by chemiluminescence detection. A detailed description is given in the additional Methods section.
DNA fibre spreading
Analysis of DNA fibres after spheroplast lysis of cells labelled for 3 h (or 90 min in case of HU-treated cells) with 0.4 mg/ml BrdU was performed essentially as described30. A full protocol is given in the additional Methods section.
Methods
Yeast strains and plasmids
The GAL-RAD18 strains were based on rad18Δ cells carrying RAD18 under control of the GAL1-10 promoter on an integrative plasmid. GALS-RAD18 was constructed by replacement of the RAD18 promoter in the endogenous locus with a truncated version of the GAL1 promoter (marked by KanMX)13. Tet-RAD18 was constructed by replacement of the RAD18 promoter with a TetO7 array (marked by KanMX) and integration of a construct encoding a TetR′-Ssn6 fusion14, rendering RAD18 repressible by doxycyline. For the purpose of BrdU incorporation, genes encoding exogenous thymidine kinase and a nucleoside transporter were introduced on an integrative plasmid, p306-BrdU-Inc18. Features were combined by subsequent transformations or by mating and tetrad dissection. All strains were grown at 30°C.
Determination of cell survival and checkpoint activation
GAL-RAD18 cells were pregrown in 2% lactate medium (pH 5.5) to exponential phase and synchronised in G1 with 10 μg/ml α-factor for 2.5 h. Cells were washed and resuspended in water, UV-irradiated at 254 nm and released into lactate medium. Incubation was continued in the dark to prevent photoreactivation. At the indicated time points, galactose (2%) or water was added. After further incubation for 2 h, cells were plated onto glucose (YPD) medium, and survival relative to unirradiated cultures was determined by colony counting after 3 days. Tet-RAD18 cells were pregrown in YPD containing 2 μg/ml doxycycline, synchronised and irradiated as above, and released into YPD with doxycycline to maintain RAD18 repression. At the indicated times cells were plated directly onto YPD with or without doxycycline for colony counting. Checkpoint activation was monitored by observing Rad53 phosphorylation with an anti-Rad53 antibody on Western blots of total cell extracts.
Detection of BrdU-labelled DNA
After BrdU labelling of the appropriate strains according to the relevant scheme (usually 3 h), total DNA isolated from ca. 5·107 cells was resuspended in Tris-EDTA (TE) buffer, and the concentration was adjusted to 500 ng/μl. After heat-denaturation and snap-cooling, aliquots of 1 μg were spotted onto Hybond N+ nylon membrane (GE Healthcare) and subjected to UV crosslinking in a Stratalinker™ (Stratagene) at 1.2·105 μJ. Subsequently, the membrane was equilibrated in phosphate-buffered saline (PBS) containing 0.1% Tween-20 (PBST), blocked with 5% non-fat milk in PBST for 30 min and probed with a monoclonal anti-BrdU antibody (B44, Becton-Dickinson, at 0.025 μg/ml) in PBST + 0.5% milk overnight. After washing and addition of secondary antibody (anti-mouse HRP conjugate in PBST + 0.5% milk), the blot was developed using enhanced chemiluminescence (GE Healthcare) and exposed to Hyperfilm™. In parallel, the signals were directly quantified using a Fuji LAS3000 imager. At least three independent experiments were performed to assure reproducibility. From a representative set, DNA samples were spotted a minimum of three times and developed together to achieve comparability and determine averages and standard deviations. Values were plotted relative to the average signal of all uninduced cultures, which was set to 1. Hence, the numbers are comparable only within each experiment.
DNA fibre spreading
For labelling of replication tracts, cells were treated with 200 mM HU for 90 min. For PRR tract labelling, cells were treated as described above for the quantification of BrdU signals. After the desired treatment, a culture of ca. 0.5·107 BrdU-labelled cells was treated with 0.1% sodium azide. Cells were harvested and incubated with 0.2 mg/ml Zymolyase-100T (Seikagaku) in 1 ml of 50 mM potassium phosphate, pH 7.5, 0.6 M sorbitol and 10 mM dithiothreitol for 1 h at 35°C. Cells were pelleted and resuspended in 200 μl of PBS. DNA was spread onto microscope slides by allowing a drop of the solution to run down a tilted slide as described 30. The dried slides were blocked with PBST + 2% bovine serum albumin (BSA) for 30 min. Rat anti-BrdU antibody (BU1/75, AbD Serotech, at 5 μg/ml) in PBST + 0.2% BSA was added for 3 h, and donkey anti-rat IgG coupled to Alexa 594 (Molecular Probes, at 5 μg/ml) was used for immunostaining (30 min). Total DNA was stained with YOYO-1 (Molecular Probes, at 1.3 μM) for 30 min, slides were washed with PBST + 0.2% BSA and covered with anti-fade reagent ProLong® Gold (Invitrogen). Fluorescence microscopy was performed with a 63x objective on an Axio Imager (ZEISS) equipped with a Hamamatsu CCD Camera. For quantification, a length of 1 μm was assumed to represent 2.59 kb of DNA30, and densities of PRR tracts were estimated with the Volocity software (Improvision) by determining average BrdU spot densities on isolated DNA fibres spanning lengths from 10 to 130 kbp. Total numbers of analysed fibres were as follows: Tet-RAD18 (10 J/m2) – 68; Tet-RAD18 (20 J/m2) – 117; Tet-RAD18 ΔTLS – 36; Tet-RAD18 ubc13Δ – 33; Tet-RAD18 ΔTLS ubc13Δ – 13; rad18Δ – 17. The values were used for determination of the density distribution (Fig. 4h) and the median tract densities (Fig. 4i).
Supplementary Material
Acknowledgements
We thank Oscar Aparicio for plasmid p306-BrdU-Inc and the lab of Vincenzo Costanzo for advice on DNA fibre analysis. This work was funded by Cancer Research UK.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
Author Information The authors declare no competing financial interests.
References
- 1.Lawrence C. The RAD6 DNA repair pathway in Saccharomyces cerevisiae: what does it do, and how does it do it? BioEssays. 1994;16:253–258. doi: 10.1002/bies.950160408. [DOI] [PubMed] [Google Scholar]
- 2.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 3.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 4.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe K, et al. Rad18 guides pol| to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Lawrence CW. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc. Natl. Acad. Sci. USA. 2005;102:15954–15959. doi: 10.1073/pnas.0504586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.di Caprio L, Cox BS. DNA synthesis in UV-irradiated yeast. Mutat. Res. 1981;82:69–85. doi: 10.1016/0027-5107(81)90139-1. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann AR. Postreplication repair of DNA in ultraviolet-irradiated mammalian cells. J. Mol. Biol. 1972;66:319–337. doi: 10.1016/0022-2836(72)90418-4. [DOI] [PubMed] [Google Scholar]
- 9.Prakash L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet. 1981;184:471–478. doi: 10.1007/BF00352525. [DOI] [PubMed] [Google Scholar]
- 10.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Edmunds CE, Simpson LJ, Sale JE. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol. Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 12.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by Replication Protein A. Mol. Cell. 2008;29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 14.Belli G, Gari E, Piedrafita L, Aldea M, Herrero E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998;26:942–947. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waters LS, Walker GC. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G(2)/M phase rather than S phase. Proc. Natl. Acad. Sci. USA. 2006;103:8971–8976. doi: 10.1073/pnas.0510167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases | and REV1. Proc. Natl. Acad. Sci. USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haracska L, Unk I, Prakash L, Prakash S. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc. Natl. Acad. Sci. USA. 2006;103:6477–6482. doi: 10.1073/pnas.0510924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viggiani CJ, Aparicio OM. New vectors for simplified construction of BrdU-Incorporating strains of Saccharomyces cerevisiae. Yeast. 2006;23:1045–1051. doi: 10.1002/yea.1406. [DOI] [PubMed] [Google Scholar]
- 19.Parker JL, Bielen AB, Dikic I, Ulrich HD. Contributions of ubiquitin- and PCNA-binding domains to the activity of Polymerase η in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:881–889. doi: 10.1093/nar/gkl1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacAlpine DM, Bell SP. A genomic view of eukaryotic DNA replication. Chromosome Res. 2005;13:309–326. doi: 10.1007/s10577-005-1508-1. [DOI] [PubMed] [Google Scholar]
- 21.Hishida T, Kubota Y, Carr AM, Iwasaki H. RAD6-RAD18-RAD5-pathway-dependent tolerance to chronic low-dose ultraviolet light. Nature. 2009;457:612–615. doi: 10.1038/nature07580. [DOI] [PubMed] [Google Scholar]
- 22.Resnick MA, Setlow JK. Repair of pyrimidine dimer damage induced in yeast by ultraviolet light. J. Bacteriol. 1972;109:979–986. doi: 10.1128/jb.109.3.979-986.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unrau P, Wheatcroft R, Cox B, Olive T. The formation of pyrimidine dimers in the DNA of fungi and bacteria. Biochim. Biophys. Acta. 1973;312:626–632. doi: 10.1016/0005-2787(73)90065-8. [DOI] [PubMed] [Google Scholar]
- 24.Prakash S, Johnson RE, Prakash L. Eukaryotic Translesion Synthesis DNA Polymerases: Specificity of Structure and Function. Annu. Rev. Biochem. 2004;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 25.Bresson A, Fuchs RPP. Lesion bypass in yeast cells: Pol| participates in a multi-DNA polymerase process. EMBO J. 2002;21:3881–3887. doi: 10.1093/emboj/cdf363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 27.Shachar S, et al. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papouli E, et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.