Abstract
Background
Multiple mechanisms regulate cancer-associated telomerase activity at the level of human telomerase reverse transcriptase (hTERT) transcription which may serve as novel targets for anticancer approaches.
Materials and Methods
The effects of prolonged all-trans retinoic acid (ATRA) exposure on hTERT regulation in estrogen receptor-negative SK-BR-3 breast cancer cells were examined.
Results
ATRA had a profound effect on the morphology and proliferation rate of the SK-BR-3 cells. ATRA also hindered the ability of these cancer cells to grow independently, rendering them more like normal somatic cells. The effect of ATRA on the decrease of telomerase activity was found to be associated with a rapid decrease in histone H3-lysine 9 acetylation (H3-K9-Ac) of the hTERT promoter. Extended-exposure to ATRA in these cells also caused the initiation of a putative compensatory mechanism, counteracting the induced surge in apoptosis.
Conclusion
A rapid decrease of H3-K9 acetylation at the hTERT promoter could be an important mechanism by which ATRA shuts down telomerase activity and mediates its antitumor effects in estrogen receptor-negative breast cancer cells.
Keywords: Breast cancer, retinoic acid, epigenetic, histone deacetylation, telomerase, hTERT.
Retinoids, naturally occurring derivatives of vitamin A, regulate processes such as differentiation, development, cell growth and apoptosis and have a profound role in cancer initiation and progression. Perhaps the most pharmacologically potent retinoid is all-trans retinoic acid (ATRA) which asserts its effect on target cells by regulating gene transcription (1). Once inside the cell, ATRA binds to its nuclear receptor, retinoic acid receptor (RAR), and is transported into the nucleus as a homodimer-ligand complex. The ATRA-RAR complex binds to retinoic acid response elements (RAREs) in the promoter region of a wide range of genes and provides the means to regulate the transcription of downstream genes. There are three members of the RAR subfamily, RARα, RARβ and RARγ, with the expression of each depending on cell type and stage of development. RARα is ubiquitously expressed, but the expression patterns of RARβ and RARγ are more limited.
Multiple breast cancer cell lines exhibit limited proliferation in the presence of retinoids that appears to be mediated by an induction of cellular differentiation and/or apoptosis (2–6). It has long been thought that more highly differentiated, estrogen receptor (ER)-positive breast cancer cell lines respond to retinoid treatment because they express high levels of RARα, but that under-differentiated, ER-negative (ER(−)) cells are refractory to retinoic acids due to their lack of RARα. SK-BR-3 breast cancer cells are ER(−) and express high levels of RARα and RARγ, but undetectable levels of RARβ (7). SK-BR-3 cells therefore serve as an excellent model system to study the effects of ATRA treatments on differentiation and the mechanisms of human telomerase reverse transcriptase (hTERT) inhibition in breast cancer cells because of their sensitivity to retinoids and representative gene expression profile shared with many breast carcinomas.
Materials and Methods
Cell culture and ATRA treatment
SK-BR-3 cells (American Type Culture Collection, Manassas, VA, USA) were cultured in McCoy’s 5A 1X media supplemented with 10% fetal bovine serum and 1X APS (amphotericin B, penicillin and streptomycin). Cells treated with 2 μM ATRA dissolved in ethanol (as determined in optimal dose response studies) were grown concurrently with controls treated with an equal volume of vehicle (ethanol). All the cells were incubated at 37 °C and 5% CO2 and harvested using 0.25% (v/v) trypsin-EDTA.
Apoptosis assessment
ATRA-treated cells were harvested and processed on days 0, 3, 6, 9 or 12 of treatment. The harvested cell pellets were resuspended in 1X annexin-binding buffer as prepared in a Vibrant Apoptosis Detection kit (Invitrogen, Carlsbad, CA, USA). After incubation with labeled dyes, the cells were analyzed on a fluorescence activated cell sorter (FACS) calibur flow cytometer (Becton Dickinson, San Jose, CA, USA) with CellQuest software (BD Biosciences, San Jose, CA, USA).
Soft agar assay
For the various treatment conditions, 5000 cells/well of 6-well plates were seeded in a semi-solid top layer of 0.3% agar noble/culture media and plated on a bottom layer of 0.6% agar noble/culture media. The top three wells of the 6-well plate were treated with 2 μM ATRA and the bottom three wells were treated with an equal volume of ethanol as controls. After 14 days, the cultures were fixed with 3.7% formaldehyde, washed with PBS and stained with 0.005% crystal violet. Images were captured with a Nikon 990 Coolpix digital camera (Nikon Corporation, Tokyo, Japan) and colonies of greater than 30 cells were counted.
TRAP (telomeric repeat amplification protocol) assay
TRAP assays were performed using a TRAPeze XL kit (Millipore, Temecula, CA, USA) as previously described (8). Briefly, the cells were harvested using trypsin-EDTA, pelleted, and washed with PBS. Protein extracts were flash frozen in liquid N2 and stored at −80°C until assayed. A standard TSR8 56-bp internal control (IC) provided by the manufacturer was included in all assays (8). Samples for PCR amplification were visualized on a 10% non-denaturing polyacrylamide gel stained with SYBR Green and analyzed with Kodak Digital Science software (Rochester, NY, USA).
Bisulfite modification
Genomic DNA was extracted with a DNeasy Tissue kit (Qiagen Sciences, Germantown, MD, USA) and treated with a MethylEasy ™ DNA Bisulfite Modification kit (Human Genetic Signatures, Sydney, NSW, Australia). A nested PCR amplification was performed with primers specific to the hTERT promoter region [8]. The PCR products were gel extracted from a 0.8% agarose gel and sequenced.
Chromatin immunoprecipitation (ChIP) assay
Cells (1 × 106) were crosslinked with 3.7% formaldehyde as reported previously (8). The cell pellets were lysed, sonicated, and samples were pre-cleared with a protein-G/salmon sperm slurry and incubated overnight with anti-acetylated histone 3 lysine 9 (H3-K9-Ac) (9) (Millipore), at 4°C. The complexes were pulled down with agarose beads, washed with a series of buffers from an EZ ChIP kit (Millipore), and eluted. After the crosslinks were reversed, samples were purified for PCR consisting of 15 cycles (94°C for 5 min, followed by 30 cycles at 94°C for 30 sec, 61°C for 45 sec, and 72°C for 45 sec and a final incubation at 72°C for 5 min). Amplicons were resolved on a 2% agarose gel and visualized after staining with ethidium bromide.
Results
Effect of ATRA on cellular morphology in ER(−) cells
SK-BR-3 cellular morphology was monitored during the course of extended ATRA exposure. As the cells were exposed to 2 μM ATRA, they displayed gradual morphological changes from a relatively spherical shape in non-treated cells (Figure 1A) to a more branched formation (Figures 1B and 1C) as the treatment progressed. Apoptotic bodies, nuclear inclusion granules, and cytoplasmic shrinkage were also observed by day 6. By day 12 (Figure 1C), high numbers of cytoplasmic vacuoles were evident and the cells were visibly fewer in number. Non-treated cells maintained consistent morphology throughout the 12 days of concurrent culture and appeared as shown in Figure 1A.
Figure 1.
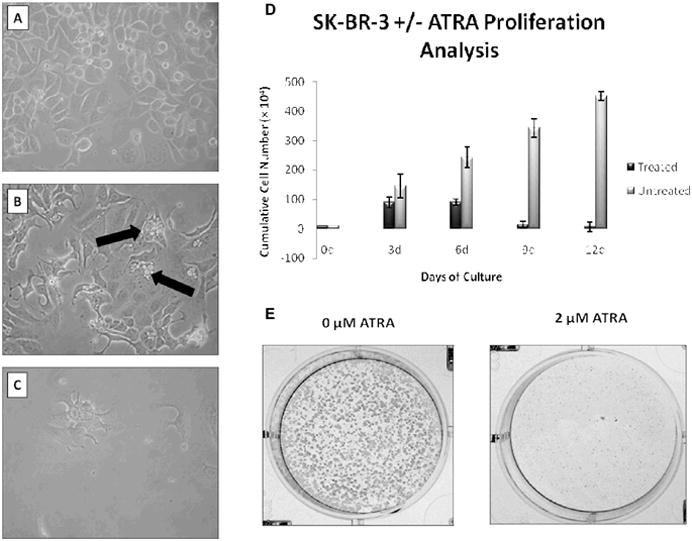
Effect of ATRA exposure on morphology and proliferation in cultured breast cancer cells. Representative photographs, at 200X magnification of SK-BR-3 cells not exposed to ATRA (A) or exposed to ATRA for 6 (B) or 12 days (C). Black arrows in (B) indicate cells undergoing apoptosis. (D) Graphical representation of the means of cell counts taken during 12-day ATRA treatment. Means are +/− s.d. of triplicate experiments. (E) Representative SK-BR-3 cells colony formation.
Effects of ATRA on proliferation of ER(−) breast cancer cells
As early as day 3 of ATRA treatment, the SK-BR-3 cells only grew at about 60% of the rate of the untreated cells. This downward trend continued throughout the remainder of the treatment (Figure 1D), with treated cells proliferating at about 38%, 3.4% and 1.5% of untreated cells by days 6, 9 and 12, respectively. Extended exposure to only one treatment of ATRA (Figure 1E) appeared similar in efficacy to continued treatments with fresh ATRA being added every 3 days (data not shown).
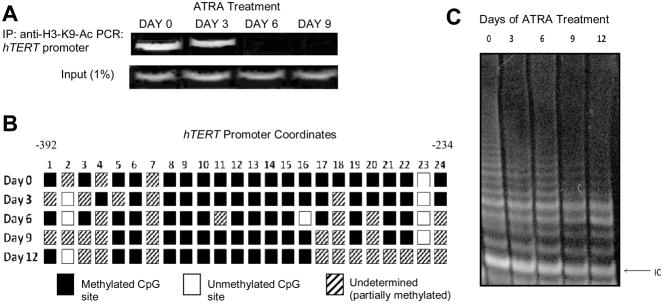
Effect of ATRA on histone acetylation and DNA methylation in the hTERT promoter of SK-BR-3 cells
By day 3 of treatment, H3-K9-Ac (9) levels were diminishing at the hTERT promoter (Figure 2A). By day 6, there was no noticeable acetylation at H3-K9 of the hTERT promoter. Carcinogenesis increases DNA methyltransferase (DNMT) activity [10] although no significant change in hTERT promoter methylation was observed (Figure 2B). There was a noticeable increase, however, in undetermined sequences (i.e., partially methylated CpGs) from day 0 to day 12 of ATRA treatment.
Figure 2.
Effect of ATRA on histone acetylation and DNA methylation at the hTERT promoter and telomerase activity. (A) Chromatin immunoprecipitation. Chromatin complexes were precipitated with anti-H3-K9-Ac and resultant purified DNA was amplified by PCR with primers specific for the hTERT promoter. “Input” represents 1% of samples removed prior to the immunoprecipitation and amplified by hTERT primers. (B) Schematic of the 24 CpG sites in the hTERT promoter region represented by boxes. Underdetermined: methylation status could not be determined (which generally indicates partially methylated CpGs). (C) TRAP assay. The height and intensity of the ladder correlates with telomerase activity. IC: internal control.
Telomerase activity with ATRA exposure in ER(−) breast cancer cells
Telomerase activity steadily decreased from day 0 to day 12 in the ATRA-treated cells (Figure 2C). Day 0 was treated with ethanol as a control and days 3, 6, 9 and 12 that were also treated with ethanol were assayed, but showed continuous telomerase activity (data not shown). The ATRA-induced suppression of telomerase activity correlated temporally with decreased H3-K9 acetylation, but residual telomerase activity was observed for a time after there was no more H3-K9 acetylation of the hTERT promoter possibly due in part to stability of the telomerase enzyme.
Effect of ATRA on the anchorage-independence of SK-BR-3 cells
The efficiency of the ATRA-treated cells to form colonies in soft agar was significantly less than in the untreated cells. Additionally, the colonies observed in the ATRA-treated cultures were smaller and more densely packed than the untreated controls (Figure 3A). After 14 days in culture the ATRA-treated cells displayed more anchorage dependence than the untreated SK-BR-3 cells (Figure 3B).
Figure 3.
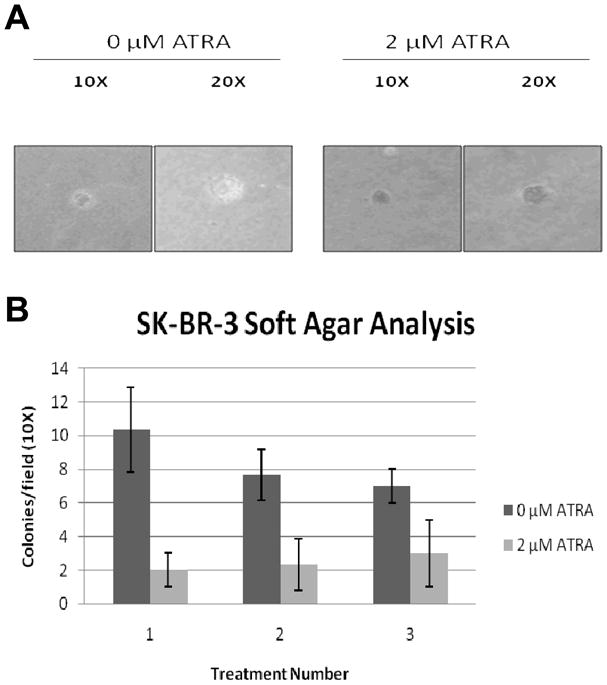
Effect of ATRA on anchorage-independent growth. (A) Representative colonies in soft agar assay. (B) Quantification of colony counts. Treatment number: separate ATRA treatments at the indicated concentrations. Means +/− s.d. of triplicate experiments.
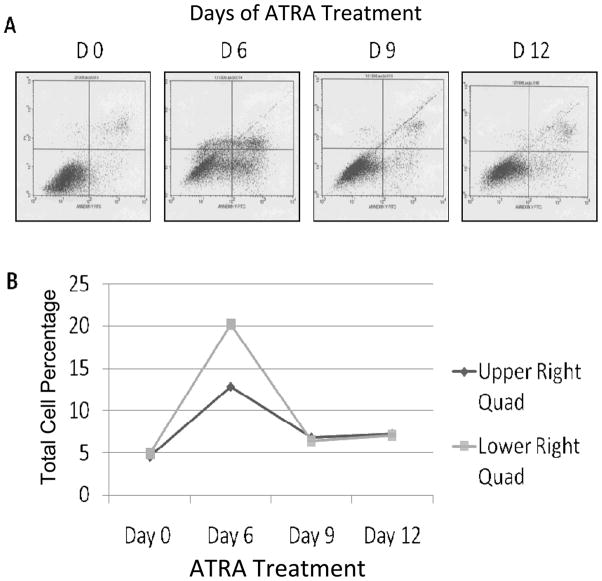
Effect of ATRA on apoptosis of SK-BR-3 breast cancer cells
Early and late stages of apoptosis were also monitored with FACS (Figure 4A) and the total apoptosis increased drastically from day 0 to day 6 and then dropped by day 12 (Figure 4B).
Figure 4.
Effect of ATRA on cellular apoptosis measured by FACS. (A) The lower left quadrant represents live cells, the upper and lower right quadrants represent late and early apoptotic cells respectively and the upper left quadrant indicates dead (necrotic) cells. (B) Graphical representation of cell numbers in the early and late apoptotic quadrants.
Discussion
ATRA-induced differentiation tends to be a gradual process (6); therefore, studies need to be in the order of days instead of hours to fully understand the effects of ATRA on cellular proliferation, apoptosis and the inhibition of telomerase activity. The present histone analysis of the hTERT promoter during prolonged exposure to ATRA illustrated for the first time how quickly chromatin structure can be remodeled to aid in locking hTERT in a repressed state and how telomerase activity persisted in decreasing amounts longer than the epigenetic mechanisms acted to remove hTERT from the cellular expression profile. A study on RARα expression in SK-BR-3 cells found increased levels of apoptosis after 96 h of retinoid exposure (11) whereas no similar effects were observed until day 6 of ATRA treatment in the present study. Interestingly, the levels of apoptosis tapered off after day 6 and decreased to levels closer to that originally observed in the untreated cells by days 9 and 12 of ATRA exposure. There may be compensatory mechanisms that counteract the harmful effects of ATRA treatment that have a delayed initiation. A decline in apoptosis due to fewer cells actively undergoing apoptosis is unlikely since levels were normalized to the number of cells harvested and pooled with the spontaneously detached cells.
While the general paradigm for DNA methylation correlates gene promoter methylation and gene silencing, the hTERT promoter is hypermethylated in many telomerase-positive tumors and hypomethylated in most normal somatic cells (10). Others have reported that the hTERT promoter remains hypomethylated in tumors and cell lines (13–14) and that heterogeneity of hTERT alleles might contribute to some of the conflicting methylation results (14). However, the present findings did not show a strong increase or decrease in methylation at the hTERT promoter during ATRA treatment in the ER(−) breast cancer cells but instead indicated that ATRA may have more of an impact on histone acetylation of the hTERT promoter than DNA methylation in these cells.
The elucidation of control mechanisms that influence telomerase activity in cancer could lead to the development of more specific therapeutics which obliterate telomerase expression and serve to eliminate neoplasias that depend on telomerase activity.
ATRA treatment of ER(−) SK-BR-3 cells involves down-regulation of telomerase activity, contributing to a decrease in the proliferation of these cells and a rapid decrease of H3-K9 acetylation at the hTERT promoter could be an important mechanism that shuts down telomerase activity in the presence of retinoids.
Acknowledgments
This work was supported in part by grants from the NCI (R01 CA129415) and Susan G. Komen for the Cure. We thank J. Tyson DeAngelis for assistance with the study.
References
- 1.Yang L, Tin-U C, Wu L, Brown P. Role of retinoid receptors in the prevention and treatment of breast cancer. J Mammary Gland Biol Neoplasia. 1999;4:377–388. doi: 10.1023/a:1018718401126. [DOI] [PubMed] [Google Scholar]
- 2.Roman S, Clarke C, Hall R, Alexander I, Sutherland R. Expression and regulation of retinoic acid receptors in human breast cancer cells. Cancer Res. 1992;52:2236–2242. [PubMed] [Google Scholar]
- 3.van der Burg B, van der Leede BM, Kwakkenbos-Isbrucker L, Salverda S, de Laat SW, van der Saag PT. Retinoic acid resistance of estradiol-independent breast cancer cells coincides with diminished retinoic acid receptor function. Mol Cell Endocrinol. 1993;91:149–157. doi: 10.1016/0303-7207(93)90267-n. [DOI] [PubMed] [Google Scholar]
- 4.Hansen NJ, Wylie RC, Phipps SM, Love WK, Andrews LG, Tollefsbol TO. The low-toxicity 9-cis UAB30 novel retinoid down-regulates the DNA methyltransferases and has anti-telomerase activity in human breast cancer cells. Int J Oncol. 2007;30:641–650. [PMC free article] [PubMed] [Google Scholar]
- 5.Krupitza G, Hulla W, Harant H, Dittrich E, Kallay E, Huber H, Grunt T, Dittrich C. Retinoic acid induced death of ovarian carcinoma cells correlates with c-myc stimulation. Int J Cancer. 1995;61:649–657. doi: 10.1002/ijc.2910610511. [DOI] [PubMed] [Google Scholar]
- 6.Love WK, Berletch JB, Andrews LG, Tollefsbol TO. Epigenetic regulation of telomerase in retinoid-induced differentiation of human leukemia cells. Int J Oncol. 2008;32:625–631. [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzgerald P, Teng M, Chandraratna RA, Heyman RA, Allegretto EA. Retinoic acid receptor alpha expression correlates with retinoid-induced growth inhibition of human breast cancer cells regardless of estrogen receptor status. Cancer Res. 1997;57:2642–2650. [PubMed] [Google Scholar]
- 8.Liu L, Saldanha SN, Pate MS, Andrews LG, Tollefsbol TO. Epigenetic regulation of human telomerase reverse transcriptase promoter activity during cellular differentiation. Genes Chromosomes Cancer. 2004;41:26–37. doi: 10.1002/gcc.20058. [DOI] [PubMed] [Google Scholar]
- 9.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 10.Casillas MA, Lopatina N, Andrews LG, Tollefsbol TO. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol Cell Biochem. 2003;252:33–43. doi: 10.1023/a:1025548623524. [DOI] [PubMed] [Google Scholar]
- 11.Schneider SM, Offterdinger M, Huber H, Grunt TW. Activation of retinoic acid receptor alpha is sufficient for full induction of retinoid responses in SK-BR-3 and T47D human breast cancer cells. Cancer Res. 2000;60:5479–5487. [PubMed] [Google Scholar]
- 12.Guilleret I, Yan P, Grange R, Braunschweig R, Bosman FT, Benhattar J. Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int J Cancer. 2002;101:335–341. doi: 10.1002/ijc.10593. [DOI] [PubMed] [Google Scholar]
- 13.Renaud S, Loukinov D, Abdullaev Z, Guilleret I, Bosman FT, Lobanenkov V, Benhattar J. Dual role of DNA methylation inside and outside of CTCF-binding regions in the transcriptional regulation of the telomerase hTERT gene. Nucleic Acids Res. 2007;35:1245–1256. doi: 10.1093/nar/gkl1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zinn RL, Pruitt K, Eguchi S, Baylin SB, Herman JG. hTERT is expressed in cancer cell lines despite promoter DNA methylation by preservation of unmethylated DNA and active chromatin around the transcription start site. Cancer Res. 2007;67:194–201. doi: 10.1158/0008-5472.CAN-06-3396. [DOI] [PubMed] [Google Scholar]