Abstract
Carcinus maenas, commonly known as the European green crab, is one of the best-known and most successful marine invasive species. While a variety of natural and anthropogenic mechanisms are responsible for the geographic spread of this crab, its ability to adapt physiologically to a broad range of salinities, temperatures and other environmental factors has enabled successful establishment in these new habitats. To extend our understanding of hormonal control in C. maenas, including factors that allow for its extreme adaptability, we have undertaken a mass spectral/functional genomics investigation of the neuropeptides used by this organism. Via a strategy combining MALDI-based high resolution mass profiling, biochemical derivatization, and nanoscale separation coupled to tandem mass spectrometric sequencing, 122 peptide paracrines/hormones were identified from the C. maenas central nervous system and neuroendocrine organs. These peptides include 31 previously described Carcinus neuropeptides (e.g. NSELINSILGLPKVMNDAamide [β-pigment dispersing hormone] and PFCNAFTGCamide [crustacean cardioactive peptide]), 49 peptides only described in species other than the green crab (e.g. pQTFQYSRGWTNamide [Arg7-corazonin]), and 42 new peptides de novo sequenced here for the first time (e.g. the pyrokinins TSFAFSPRLamide and DTGFAFSPRLamide). Of particular note are a collection of 25 FMRFamide-like peptides (including 9 new isoforms sequenced de novo) and a collection of 25 A-type allatostatin peptides (including 10 new sequences reported for the first time) in this study. Both peptide families are among the most diverse families, each containing a large number of isoforms in arthropod species. Also of interest was the identification of two SIFamide isoforms, GYRKPPFNGSIFamide and VYRKPPFNGSIFamide, the latter peptide known previously only from members of the astacidean genus Homarus. Using transcriptome analyses, 15 additional peptides were characterized, including an isoform of bursicon β and a neuroparsin-like peptide. Collectively, the data presented in this study not only greatly expand the number of identified C. maenas neuropeptides, but also provide a framework for future investigations of the physiological roles played by these molecules in this highly adaptable species.
Keywords: Carcinus maenas, European green crab, matrix-assisted laser desorption/ionization-Fourier transform mass spectrometry (MALDI-FTMS), electrospray ionization quadrupole time-of-flight tandem mass spectrometry (ESI-Q-TOF MS/MS), expressed sequence tag (EST), tblastn, blastp, transcriptomics, neuropeptide, neurohormone, peptide sequencing, supraoesophageal ganglion, brain, thoracic ganglia, sinus gland (SG), pericardial organ (PO)
1. Introduction
The European green crab, Carcinus maenas, is native to the Atlantic coasts of Europe and North Africa. Due to its ability to tolerate a wide variety of environmental conditions, particularly variations in salinity and temperature, this species is capable of utilizing a multitude of habitats in its native Eastern North Atlantic (Cohen and Carlton, 1995). This extreme adaptability has also enabled C. maenas to rapidly expand its range over the last two-hundred years, via both natural and anthropogenic routes, to the East Coast of North America from Nova Scotia to Maryland, the West Coast of North America from California to British Columbia, as well as to the waters of Hawaii, Australia, Japan and South Africa (Carlton and Cohen, 2003; Grosholz and Ruiz, 1996; Hidalgo et al., 2005; Yamada and Hauck, 2001). This crab’s voracious predation of many infaunal species in these invaded areas has resulted in dramatic changes in the structure of many of the resident invertebrate communities, thereby making C. maenas one of the world’s most problematic marine invasive species (Grosholz, 2005; Lafferty and Kuris, 1996).
In crustacean species, as in most multicellular organisms, locally-released paracrines and circulating hormones play critical roles in affecting the physiological processes that allow for adaptation to changing environmental conditions (Madsen, 1990; Morris and Airriess, 1998). Three classes of hormones are generally recognized based on chemical composition: amines, steroids and peptides, the latter class the largest and most diverse of these groups (Strand, 1999). While a variety of tissues may contribute to the complement of circulating hormones (Fu et al., 2005a; Huybrechts et al., 2003), in arthropods the nervous system is one of the main hormone producers.
To understand hormonal control in any organism, it is first necessary to characterize the molecules it uses as signaling agents. Biological mass spectrometry and functional genomics have become prominent methods for the elucidation of peptide hormones, especially for large-scale peptidomic analyses (Dowell et al., 2006; Fu et al., 2005a; Huybrechts et al., 2003). In this study, our strategy to characterize the neuropeptidome of C. maenas combined transcriptomics with two distinct mass spectral techniques: matrix-assisted laser desorption/ionization Fourier transform mass spectrometry (MALDI-FTMS)-based high resolution mass profiling and nanoflow liquid chromatography coupled to electrospray ionization quadrupole time-of-flight tandem mass spectrometry (nanoLC-ESI-Q-TOF MS/MS). Specifically, MALDI-base profiling was used to screen tissue fragments or tissue extracts for known peptides via accurate mass measurements, while nanoscale biochemical separation/derivatization coupled to ESI-Q-TOF MS/MS was used to de novo sequence novel peptides from tissue extracts. Using this combined approach, 122 peptides were identified from the central nervous system (CNS) and neuroendocrine organs of C. maenas, including 91 new to this species. Moreover, 15 additional peptides were identified and characterized by functional genomic analyses, most described here for the first time. Collectively, our data not only greatly expand the catalog of peptide hormones known to be present in C. maenas, but also provide a foundation for future studies of peptide function in this highly adaptable species. Some of these data have appeared previously in abstract form (Li et al., 2007).
2. Materials and methods
2.1. Animals and tissue collection
2.1.1. Animals
European green crabs, Carcinus maenas, were collected by hand from multiple locations on Mount Desert Island, Maine. In total, approximately 300 individuals were used in this study. To ensure a broad coverage of the neuropeptidome, the animals used for pooled tissue mass spectral analysis included both males and females, pre-molt and recent post-molt individuals, green and red color morph animals, individuals collected from/exposed to either high (32 ppt) or low (15 ppt) salinity environments, and animals sacrificed at different time points in the day/night and tidal cycles (including air exposed individuals).
2.1.2. Tissue collection
The CNS (i.e. the supraoesophageal ganglia [brain] and the fused thoracic neuromeres [thoracic ganglia]) and the primary neuroendocrine organs of C. maenas (i.e. the sinus gland [SG] and the pericardial organ [PO]) were dissected free from ice-anesthetized animals in chilled (approximately 10 °C) physiological saline (composition in mM: 440 NaCl; 11 KCl; 13 CaCl2; 26 MgCl2; 10 HEPES acid; pH 7.4 [adjusted with NaOH]). Following dissection, tissue samples were either immediately assayed via direct tissue mass spectral analysis or placed in acidified methanol (methanol/glacial acetic acid/deionized water, 90:9:1) and stored at −80 °C until utilized for peptide extraction or direct tissue mass spectral analysis.
2.2. Mass spectral analyses
2.2.1. Tissue extraction and HPLC fractionation
For some experiments, tissues were pooled, homogenized with a handheld ground glass tissue homogenizer, and extracted with acidified methanol (see 2.1.2). Extracts were dried in a Savant SC 110 SpeedVac concentrator (Thermo Electron Corporation, West Palm Beach, FL) and resuspended in approximately 100 µl of 0.1% formic acid. The resuspended extracts were then vortexed and briefly centrifuged, and the resulting supernatants subsequently fractionated via high performance liquid chromatography (HPLC).
HPLC separations were performed using a Rainin Dynamax HPLC system equipped with a Dynamax UV-D II absorbance detector (Rainin Instrument Inc., Woburn, MA). The mobile phases included deionized water containing 0.1% formic acid (Solution A) and acetonitrile (HPLC grade, Fisher Scientific) containing 0.1% formic acid (Solution B). For each separation run, 20 µl of extract was injected onto a Macrosphere C18 column (2.1 mm i.d. × 250 mm length, 5 µm particle size; Alltech Assoc. Inc., Deerfield, IL). The separation consisted of a 120 minute gradient of 5%-95% Solution B with fractions automatically collected every two minutes using a Rainin Dynamax FC-4 fraction collector.
2.2.2. Formaldehyde derivatization
For some experiments, peptides in an HPLC fraction were derivatized with formaldehyde prior to mass spectral analysis. Specifically, 0.3 µl of a fraction was spotted on the MALDI plate, followed by the addition and mixing of 0.3 µl of 26 mM sodium cyanoborohydride (Sigma-Aldrich, St. Louis, MO), and subsequent addition of 0.3 µl of formaldehyde (20% in H2O vol/vol, Sigma-Aldrich). The droplet was left at room temperature for 5 minutes and then 0.3 µl of 50 mM ammonium bicarbonate solution was added to the reaction mixture. Finally, 0.3 µl of a saturated 2,5-dihydroxybenzoic acid (DHB; ICN Biomedical Corp., Costa Mesa, CA) matrix (150 mg/mL in a 50:50 v/v mixture of deionized water and purge and trap grade methanol (Sigma-Aldrich)) was added to the droplet and allowed to crystallize at room temperature.
2.2.3. MALDI-FTMS and sustained off resonance irradiation-collision induced dissociation
MALDI-FTMS experiments were performed on an IonSpec ProMALDI Fourier transform mass spectrometer (Lake Forest, CA) equipped with a 7.0 Tesla actively-shielded superconducting magnet. This FTMS instrument contains a high pressure MALDI source where the ions from multiple laser shots can be accumulated in the external hexapole storage trap before the ions are transferred to the ICR cell via a quadrupole ion guide. A 337 nm nitrogen laser (Laser Science, Inc., Franklin, MA) was used for ionization/desorption. The ions were excited prior to detection with a radio frequency sweep beginning at 7050 ms with a width of 4 ms and amplitude of 150 V base to peak. The filament and quadrupole trapping plates were initialized to 15 V, and both were ramped to 1V from 6500 to 7000 ms to reduce baseline distortion of peaks. Detection was performed in broadband mode from m/z 108.00 to 4500.00.
Peptide fragmentation was accomplished by sustained off resonance irradiation-collision induced dissociation (SORI-CID). An arbitrary waveform from 2000 ms to 2131 ms with a ±10 Da isolation window was introduced to isolate the ion of interest. Ions were excited with SORI burst excitation (2.648V, 2500–3000 ms). A pulse of nitrogen gas was introduced through a pulse valve from 2500 to 2750 ms to introduce collision activation.
Tissues from two animals were used for the direct tissue analysis. Tissue fragments were desalted by a brief rinse in a solution of DHB prepared in deionized water (10 mg/mL). The tissue was then placed onto the MALDI sample plate along with 0.3 µl of saturated DHB matrix (prepared as described in 2.2.2) before allowing the DHB spot to crystallize at room temperature.
Off-line analysis of HPLC fractions (prepared as described in 2.2.1) was performed by spotting 0.3 µL of saturated DHB on the MALDI sample plate and adding 0.3 µL of the HPLC fraction of interest. The resulting mixture was allowed to crystallize at room temperature, with subsequent MALDI-FTMS analysis performed as described above.
2.2.4. Capillary LC-ESI-Q-TOF MS/MS
Nanoscale LC-ESI-Q-TOF MS/MS was performed using a Waters capillary LC system coupled to a Q-TOF Micro mass spectrometer (Waters Corp., Milford, MA). Chromatographic separations were performed on a C18 reverse phase capillary column (75 µm internal diameter × 150 mm length, 3 µm particle size; Micro-Tech Scientific Inc., Vista, CA). Three mobile phases were used: deionized water with 5% acetonitrile and 0.1% formic acid (A), acetonitrile with 5% deionized water and 0.1% formic acid (B) and deionized water with 0.1% formic acid (C). A 6.0 µL aliquot of an HPLC fraction (see 2.2.1) was injected and loaded onto the trap column (PepMap™ C18; 300 µm column internal diameter × 1 mm, 5 µm particle size; LC Packings, Sunnyvale, CA) using mobile phase C at a flow rate of 30 µL/min for 3 minutes. Following injection, the stream select module was switched to a position in which the trap column became in line with the analytical capillary column, and a linear gradient of mobile phases A and B was initiated. A splitter was added between the mobile phase mixer and the stream select module to reduce the flow rate from 15 µL/min to 200 nL/min.
The nanoflow ESI source conditions were set as follows: capillary voltage 3200 V, sample cone voltage 35 V, extraction cone voltage 1 V, source temperature 120°C, cone gas (N2) 10 L/hr. A data-dependent acquisition was employed for the MS survey scan and the selection of precursor ions and subsequent MS/MS of the selected parent ions. The MS scan range was from m/z 300 to 2000 and the MS/MS scan was from m/z 50 to 1800. The MS/MS de novo sequencing was performed with a combination of manual sequencing and automatic sequencing by PepSeq software (Waters Corp.).
2.2.5. Figure production
MALDI-FTMS figures were produced by converting the initial spectra obtained using IonSpec version 7.0 software (IonSpec Corp.) to a bitmap image using Boston University Data Analysis (BUDA) software (version 1.4; Boston University, Boston, MA). The BUDA files were then pasted into Fireworks MX 2004 (Macromedia, Inc., San Francisco, CA) and resampled to improve the resolution. All MS/MS figures were produced using a combination of Fireworks MX 2004 and Microsoft Windows Paint tool (Microsoft Corporation, Redmond, WA).
2.3. Protein homology searches
In silico proteomic searches were conducted to identify de novo sequenced peptides that did not fit into any known neuropeptide family. Specifically, the online program blastp (National Center for Biotechnology Information [NCBI], Bethesda, MD; http://www.ncbi.nlm.nih.gov/BLAST/) was used to search the extant NCBI crustacean protein database, using the unknown peptide sequences as queries. For all searches, the blastp database was set to non-redundant protein sequences (i.e. nr) and restricted to crustacean sequences (i.e. taxid: 6657). For each of the proteins putatively identified via blastp, the BLAST score and BLAST-generated E-value for significant alignment are provided in the appropriate subsection of the Results.
2.4. Functional genomics
2.4.1. Database searches
Transcriptome searches were conducted using methods modified from several recent publications (Christie, 2008a; Christie, 2008b; Christie et al., 2008). Specifically, the online program tblastn (NCBI; http://www.ncbi.nlm.nih.gov/BLAST/) was used to mine for expressed sequence tags (ESTs) encoding putative C. maenas neuropeptide precursors using queries of known arthropod prepro-hormone sequences. For all searches, the program database was set to non-human, non-mouse ESTs (EST_others) and restricted to C. maenas transcripts (taxid: 6759). All hits were fully translated and checked manually for homology to the target query, as well as for typical peptide precursor features, including start and stop codons (i.e. a full-length prepro-hormone), the presence of a signal sequence, and pro-hormone convertase processing sites (see 2.4.2). For each of the putative neuropeptide-encoding transcripts identified, the BLAST score and BLAST-generated E-value for significant alignment are provided in the appropriate subsection of the Results.
2.4.2. Peptide prediction
Translation of EST nucleotide sequences was performed using the Translate tool of ExPASy (Swiss Institute of Bioinformatics, Basel, Switzerland; http://www.expasy.ch/tools/dna.html). Signal peptide prediction was done via the online program SignalP 3.0, using both Neural Networks and Hidden Markov Models algorithms (Center for Biological Sequence Analysis, Technical University of Denmark, Lyngby, Denmark; http://www.cbs.dtu.dk/services/SignalP/)(Dyrlov Bendtsen et al., 2004). Pro-hormone convertase cleavage sites were predicted based on the information presented in Veenstra (2000). Prediction of the sulfation state of Tyr residues was done using the online program Sulfinator (Swiss Institute of Bioinformatics; http://www.expasy.org/tools/sulfinator/) (Monigatti et al., 2002). When applicable, other post-translational modifications (e.g. cyclization of amino (N)-terminal Gln/Glu residues and carboxyl (C)-terminal amidation at Gly residues) were predicted by homology to known peptide isoforms.
3. Results
3.1. Mass spectral analysis of peptides in the nervous system of C. maenas
For mass spectral elucidation of the neuropeptides present in C. maenas central nervous system (CNS) and neuroendocrine organs, we have used a strategy combining MALDI-FTMS based high resolution mass profiling (direct tissue and off-line HPLC fraction analysis) and nanoscale biochemical separation coupled to ESI-Q-TOF MS/MS de novo sequencing. To facilitate de novo sequencing, some HPLC fractions were derivatized with formaldehyde prior to the ESI-Q-TOF mass spectrometric analysis. We have grouped the identified peptides into families of related isoforms, whenever possible, and these are presented below in alphabetical order based on family name.
3.1.1. A-type allatostatin
Twenty five peptides possessing -YXFGLamide C-termini (where X is a variable amino acid) were sequenced via ESI-Q-TOF MS/MS from the neural tissue of C. maenas (Table 1). This C-terminus classifies these peptides as members of the A-type allatostatin (A-type AST) family (Stay and Tobe, 2007). Of the peptides identified, EAYAFGLamide, GGPYAFGLamide, NPYAFGLamide, DPYAFGLamide, AGPYAFGLamide, EPYAFGLamide, AGPYSFGLamide, ASPYAFGLamide, SDMYSFGLamide, TGQYAFGLamide, APGPYAFGLamide, EYDDMYTEKRPKVYAFGLamide, YDDMYTEKRPKVYAFGLamide and GYEDEDEDRPFYALGLGKRPRTYSFGLamide are known C. maenas isoforms (Duve et al., 1997a). Two other peptides, ARPYSFGLamide and PADLYEFGLamide, are identical in structure to A-type ASTs previously identified in other arthropods, i.e. the moth Cydia pomonella and the crab Cancer borealis (Duve et al., 1997b; Fu and Li, 2005b), but are new discoveries in C. maenas. The remaining nine isoforms, AAPYAFGLamide, GKPYAFGLamide, EPYEFGLamide, RGPYAFGLamide, ARPYAFGLamide, FSGASPYGLamide, AASPYSFGLamide, LKAYDFGLamide, KLPYSFGLamide and TRPYSFGLamide, were de novo sequenced for the first time (Table 1). All of the A-type ASTs described in this study were identified in the brain and/or PO except PADLYEFGLamide, which was detected in thoracic ganglia and pericardial organ (PO).
Table 1.
Neuropeptides detected in the central nervous system and neuroendocrine organs of the European green crab Carcinus maenas.
| Neuropeptide | MALDI-FT | QTOF | |||
|---|---|---|---|---|---|
| Family | [M+H]+ | Sequence | MS | MS/MS | Tissue location |
| AST-A | |||||
| 769.39 | EAYAGFLamide | − | + | Br/PO | |
| 780.40 | NPYAFGLamide | − | + | Br/PO | |
| 780.40 | GGPYAFGLamide | − | + | Br | |
| 781.39 | DPYAFGLamide | − | + | Br | |
| 794.42 | AGPYAFGLamide | − | + | Br/PO | |
| 795.40 | EPYAFGLamide | − | + | Br/PO | |
| 808.44 | AAPYAFGLamide | − | + | Br/PO | |
| 810.41 | AGPYSFGLamide | − | + | Br/PO | |
| 824.43 | ASPYAFGLamide | − | + | Br/PO | |
| 851.48 | GKPYAFGLamide | − | + | Br/PO | |
| 853.41 | EPYEFGLamide | − | + | Br | |
| 879.48 | RGPYAFGLamide | + | + | Br/PO | |
| 893.50 | ARPYAFGLamide | − | + | PO | |
| 897.45 | FSGASPYGLamide | − | + | Br/PO | |
| 909.49 | ARPYSFGLamide | + | + | Br/PO | |
| 911.46 | AASPYSFGLamide | − | + | PO | |
| 918.40 | SDMYSFGLamide | − | + | PO | |
| 925.51 | LKAYDFGLamide | − | + | Br | |
| 926.47 | ATGQYAFGLamide | − | + | Br | |
| 939.50 | TRPYSFGLamide | − | + | Br/PO | |
| 923.53 | KLPYSFGLamide | − | + | Br | |
| 962.51 | APGPYAFGLamide | + | − | PO | |
| 1023.51 | PADLYEFGLamide | − | + | TG/PO | |
| 2224.07 | EYDDMYTEKRPKVYAFGLamide | + | − | PO | |
| 3150.53 | GYEDEDEDRPFYALGLGKRPRTYSFGLamide | + | − | PO | |
| AST-B | |||||
| 1031.26 | AWSNLGQAWamide | − | + | PO | |
| 1179.72 | AGWNKFQGSWamide | + | + | PO | |
| 1182.57 | TSWGKFQGSWamide | + | + | PO | |
| 1194.00 | VTWGKFQGSWamide | − | + | PO | |
| 1222.58 | GNWNKFQGSWamide | + | + | PO | |
| 1246.47 | GSNWSNLRGAWamide | + | + | PO | |
| 1252.58 | NNWSKFQGSWamide | + | + | PO | |
| 1258.37 | GVNWSNLRGAWamide | − | + | TG/PO | |
| 1107.58 | QWSSMRGAWamide | + | + | Br/PO | |
| 1260.71 | SGKWSNLRGAWamide | + | + | Br/PO | |
| 1293.63 | STNWSSLRSAWamide | + | + | TG/PO | |
| 1366.63 | NNNWSKFQGSWamide | + | + | Br/PO | |
| 1470.70 | VPNDWAHFRGSWamide | + | + | Br/TG/PO | |
| CCAP | |||||
| 956.38 | PFCNAFTGCamide | + | + | Br/TG/PO | |
| Corazonin | |||||
| 1369.63 | pQTFQYSRGWTNamide | + | − | Br | |
| CHH | |||||
| 8631.87 | PO CHH | + | − | PO | |
| 8525.70 | SG CHH-I | + | − | SG/PO | |
| CPRP | |||||
| RSTPGYGRMDRILAALKTSPMEPS | |||||
| 4061.07 | AALAVEHGTTHPLE | + | + | SG/PO | |
| RSTQGYGRMDRILAALKTSPMEP | |||||
| 4092.08 | SAALAVEHGTTHPLE | + | + | SG/PO | |
| 1552.83 | RSTQGYGRMDPIL | − | + | SG | |
| 1521.79 | RSTPGYGRMDRIL | − | + | SG | |
| 1694.96 | RSTQGYGRMDRILAA | − | + | SG | |
| 1663.89 | RSTPGYGRMDRILAA | − | + | SG | |
| 1629.83 | PSAALAVEHGTTHPLE | − | + | SG | |
| 2073.94 | SPMEPSAALAVEHGTTHPLE | − | + | SG | |
| 2175.06 | TSPMEPSAALAVEHGTTHPLE | − | + | SG | |
| FLPs | |||||
| 851.50 | RNFLRFamide | + | + | Br/TG/PO | |
| 863.45 | pQGNFLRFamide | + | + | PO | |
| 887.56 | PSLRLRFamide | − | + | Br/TG | |
| 905.51 | PSMRLRFamide | − | + | Br/TG | |
| 921.51 | PSM(O)RLRFamide | − | + | Br | |
| 938.53 | NRSFLRFamide | − | + | Br/TG/PO | |
| 954.53 | SRNYLRFamide | + | + | Br/TG/PO | |
| 965.54 | NRNFLRFamide | + | + | Br/TG/SG/PO | |
| 966.53 | DRNFLRFamide | + | + | Br/SG/PO | |
| 1019.59 | APRNFLRFamide | + | + | TG/PO | |
| 1022.56 | GNRNFLRFamide | + | + | Br/PO | |
| 1048.57 | APQGNFLRFamide | + | + | Br/PO | |
| 1104.61 | GAHKNFLRFamide | + | − | PO | |
| 1105.63 | SMPSLRLRFamide | + | + | Br/TG/SG | |
| 1121.63 | SM(O)PSLRLRFamide | − | + | TG | |
| 1124.63 | GLSRNYLRFamide | + | + | Br/TG/PO | |
| 1137.59 | DGNRNFLRFamide | + | + | Br/TG/PO | |
| 1147.64 | EMPSLRLRFamide | − | + | TG | |
| 1163.64 | EM(O)PSLRLRFamide | − | + | TG | |
| 1147.65 | APQRNFLRFamide | + | + | TG | |
| 1158.62 | YGNRSFLRFamide | + | + | Br/TG/PO | |
| 1271.65 | pQDLDHVFLRFamide | + | + | Br/TG/PO | |
| 1288.68 | QDLDHVFLRFamide | + | + | Br/TG/PO | |
| 1181.62 | SENRNFLRFamide | + | + | TG | |
| 1314.78 | DARTAPLRLRFamide | + | + | Br/TG | |
| Orcokinin | |||||
| 1098.52 | EIDRSGFGFA | + | − | PO | |
| 1228.56 | NFDEIDRSSFamide | − | + | Br | |
| 1198.55 | NFDEIDRSGFamide | + | + | Br | |
| 1256.55 | NFDEIDRSGFG | + | + | Br/TG | |
| 1270.57 | NFDEIDRSGFA | + | + | TG/PO | |
| 1286.56 | NFDEIDRSSFG | − | + | TG | |
| 1300.58 | NFDEIDRSSFA | − | + | Br | |
| 1360.62 | FDEIDRSGFGFA | − | + | PO | |
| 1403.62 | NFDEIDRSGFGF | + | + | TG/PO | |
| 1474.66 | NFDEIDRSGFGFA | + | + | Br/TG/SG/PO | |
| 1502.69 | NFDEIDRSGFGFV | + | + | TG/SG/PO | |
| 1503.68 | DFEDIERSGFGFV | + | + | TG/PO | |
| 1517.67 | NFDEIDRSGFGFN | + | − | PO | |
| 1532.70 | NFDEIDRSSFGFV | + | + | TG/SG/PO | |
| 1547.68 | NFDEIDRSSFGFN | + | + | Br/TG/SG/PO | |
| Orcomyotropin | |||||
| 1186.52 | FDAFTTGFGHS | + | + | Br/TG/SG/PO | |
| PDHs | |||||
| 1927.03 | NSELINSILGLPKVMNDAamide | + | + | SG | |
| Proctolin | |||||
| 649.37 | RYLPT | + | − | PO | |
| Pyrokinin | |||||
| 878.52 | LYFAPRLamide | − | + | Br | |
| 1024.56 | TSFAFSPRLamide | − | + | Br | |
| 1109.57 | TDGFAFSPRLamide | + | + | Br | |
| RPCH | |||||
| 930.44 | pQLNFSPGWamide | − | + | SG | |
| RYamide | |||||
| 784.41 | FVGGSRYamide | + | − | PO | |
| 832.41 | FYANRYamide | − | + | PO | |
| 959.47 | SGFYAPRYamide | + | + | PO | |
| 862.42 | FYSQRYamide | + | + | PO | |
| 976.46 | SGFYANRYamide | + | + | PO | |
| 977.45 | SGFYADRYamide | − | + | PO | |
| 1030.47 | pEGFYSQRYamide | + | + | PO | |
| 1114.47 | SSRFVGGSRYamide | + | + | PO | |
| 2091.76 | (X)YANRYamide | + | + | PO | |
| SIFamide | |||||
| 1161.65 | RKPPFNGSIFamide | + | + | Br | |
| 1381.74 | GYRKPPFNGSIFamide | + | + | Br/TG/SG | |
| 1423.78 | VYRKPPFNGSIFamide | − | + | TG | |
| Tachykinin | |||||
| 863.46 | PSGFLGMRamide | − | + | Br/TG/SG | |
| 934.49 | APSGFLGMRamide | + | + | Br/TG/SG | |
| 964.50 | TPSGFLGMRamide | + | + | Br/TG/SG | |
| 950.49 | APSGFLGM(O)Ramide | + | + | Br/TG | |
| 980.50 | TPSGFLGM(O)Ramide | + | + | Br/TG | |
| 992.50 | APSGFLGMRG | − | + | Br | |
| 766.40 | SGFLGMRamide | + | + | Br/TG | |
| Others | |||||
| 844.48 | HIGSLYRamide | + | + | Br/SG/PO | |
| 902.53 | KIFEPLR | − | + | TG | |
| 1259.71 | KIFEPLRDKN | − | + | TG | |
| 1372.79 | KIFEPLRDKNL | − | + | TG | |
| 916.55 | KIFEPLVA | − | + | TG | |
Br, brain; TG, thoracic ganglion; PO, pericardial organ; SG, sinus gland; AST, allatostatin; CCAP, crustacean cardioactive peptide; CHH, crustacean hyperglycemic hormone; CPRP, crustacean hyperglycemic hormone precursor related peptide; FLPs, FMRFamide-like peptides; PDH, pigment dispersing hormone; RPCH, red pigment concentrating hormone.
Previously known Carcinus maenas peptides are shown in normal font; peptides previously described from other decapods, but new to Carcinus maenas are shown in italics; novel peptides are shown in bold.
3.1.2. B-type allatostatin
Thirteen peptides possessing the C-terminal motif -W(X)6Wamide (X indicating variable amino acids), the hallmark of the B-type allatostatin (B-type AST) family (Stay and Tobe, 2007), were sequenced via ESI-Q-TOF MS/MS and/or MALDI-FTMS from the brain, thoracic ganglia and PO (Table 1). Five of the identified B-type ASTs, AWSNLGQAWamide, AGWNKFQGSWamide, VTWGKFQGSWamide, GSNWSNLRGAWamide, and GVNWSNLRGAWamide were de novo sequenced here for the first time (Table 1). The remaining eight peptides, TSWGKFQGSWamide, GNWNKFQGSWamide, NNWSKFQGSWamide, QWSSMRGAWamide, SGKWSNLRGAWamide, STNWSSLRSAWamide, NNNWSKFQGSWamide, and VPNDWAHFRGSWamide, were previously identified in the crabs Cancer productus and Cancer borealis (Fu et al., 2005a; Fu and Li, 2005b), but are described in C. maenas for the first time in our study (Table 1).
3.1.3. Corazonin
The peptide pQTFQYSRGWTNamide, commonly referred to as Arg7-corazonin (Veenstra, 1989), was identified via direct tissue MALDI-FTMS analysis of the brain and thoracic ganglia (Table 1). Internal calibration of the spectra containing this peptide showed mass measurement accuracy (MMA) of approximately 1.6 ppm, which strongly supported this attribution. While Arg7-corazonin has been identified previously from the PO of the crab Cancer borealis (Li et al., 2003), our report describes the first detection of this peptide in C. maenas.
3.1.4. Crustacean cardioactive peptide
The known C. maenas neuropeptide (Stangier et al., 1987) PFCNAFTGCamide, commonly referred to as crustacean cardioactive peptide (CCAP), was sequenced from the brain, thoracic ganglia and PO via both MALDI-FTMS and ESI-Q-TOF MS/MS (Table 1).
3.1.5. Crustacean hyperglycemic hormone (CHH) superfamily
Two peptides with masses similar to C. maenas SG CHH I, pEIYDTSCKGVYDRALFNDLEHVCDDCYNLYRTSYVASACRSNCYSNLVFRQCMDDLL MMDEFDQYARKVQMVamide, and C. maenas PO CHH, pEIYDTSCKGVYDRALFNDLEHVCDDCYNLYRTSYVASACRNNCFENEVFDVCVYQLY FPNHEEYLRSRDGLKG-OH, were detected in the SG and PO, respectively, via direct tissue MALDI-FTMS (Table 1). The average errors between the theoretical and detected masses of two peptides were approximately 0.010% and 0.015% respectively, which supports this attribution.
3.1.6. CHH precursor-related peptide (CPRP)
The peptides RSTQGYGRMDRILAALKTSPMEPSAALAVEHGTTHPLE and RSTPGYGRMDRILAALKTSPMEPSAALAVEHGTTHPLE, two known isoforms of C. maenas CPRP (Dircksen et al., 2001), were sequenced via ESI-Q-TOF MS/MS and direct tissue MALDI-FTMS analysis from SG (Table 1). In addition to these full-length peptides, seven putative CPRP truncations, RSTPGYGRMDRIL, RSTPGYGRMDRILAA, RSTQGYGRMDPIL, RSTQGYGRMDPILAA, PSAALAVEHGTTHPLE, SPMEPSAALAVEHGTTHPLE and TSPMEPSAALAVEHGTTHPLE, were de novo sequenced from the SG via ESI-Q-TOF MS/MS (Table 1).
3.1.7. FMRFamide-like peptides
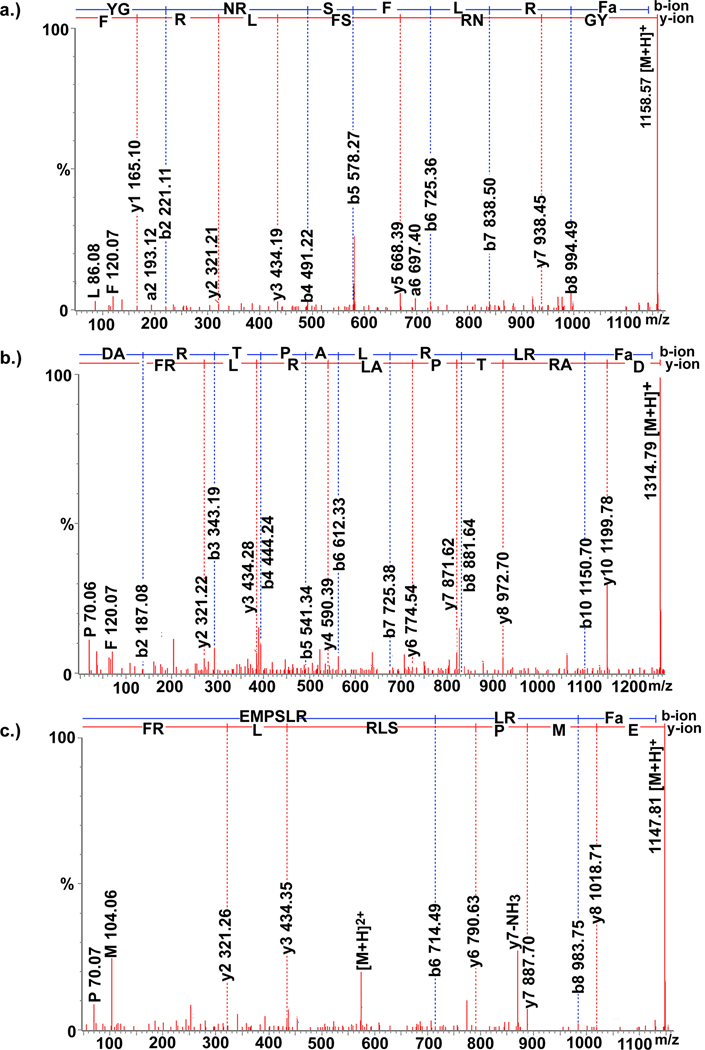
The FMRFamide-like peptides (FLPs) are a large and diverse family of peptides found in both invertebrates and vertebrates (Zajac and Mollereau, 2006). Many subfamilies have been identified in arthropods, including the myosuppressins and the short neuropeptide Fs (sNPFs) (Brown et al., 1999; Garczynski et al., 2006; Nichols, 2003). In our study, 25 FMRFamide-like peptides were identified in C. maenas neural tissues using a combination of ESI-Q-TOF MS/MS sequencing and direct tissue/off-line HPLC fractionation coupled to MALDI-FTMS analysis (Table 1). Two of these peptides, QDLDHVFLRFamide and pQDLDHVFLRFamide, were identified by both MALDI-FTMS and ESI-Q-TOF MS/MS in the brain and the thoracic ganglia, with pQDLDHVFLRFamide also identified in the PO. These two peptides each possess the C-terminal motif -HVFLRFamide, implicates them as members of the myosuppressin subfamily (Table 1). Both QDLDHVFLRFamide and pQDLDHVFLRFamide are known isoforms in the lobster Homarus americanus (Ma et al., 2008), with the latter being a known C. maenas neuropeptide (Stemmler et al., 2007). Eight other peptides, PSLRLRFamide, PSMRLRFamide, PSM(O)RLRFamide (where M(O) represents an oxidized methionine residue), SMPSLRLRFamide, SM(O)PSLRLRFamide, EMPSLRLRFamide, EM(O)PSLRLRFamide and DARTPALRLRFamide, exhibit -RXRFamide C-termini (where X represents a variable residue), the identifying characteristic of the sNPF subfamily. The sNPFs have been proposed to be the invertebrate homolog of the vertebrate neuropeptide Ys (McVeigh et al., 2005). Three of the C. maenas sNPFs, EMPSLRLRFamide (Figure 1(c)), EM(O)PSLRLRFamide and DARTPALRLRFamide (Figure 1(b)), were de novo sequenced here for the first time. The remaining sNPF isoforms were previously identified in the crab Cancer borealis and/or the lobster Homarus americanus (Fu and Li, 2005; Ma et al., 2008), but were newly discovered in C. maenas.
Figure 1.
Collision-induced dissociation spectra of three de novo sequenced peptides. ESI-Q-TOF MS/MS sequencing of three FLPs: (a.) YGNRSFLRFamide (579.792+), (b.) DARTPALRLRFamide (657.902+), and (c.) EMPSLRLRFamide(574.412+).
Of the remaining 15 FMRFamide-like peptides identified here, 12 possess the C-terminal motif -FLRFamide, and three exhibit -YLRFamide C-termini. Eight of the -FLRFamide isoforms, RNFLRFamide, NRSFLRFamide, NRNFLRFamide, DRNFLRFamide, APRNFLRFamide, GNRNFLRFamide, APQRNFLRFamide and SENRNFLRFamide and one – YLRFamide isoform GAHKNYLRFamide, were identified previously from the PO of the crabs Cancer productus and/or C. borealis (Fu et al., 2005a; Fu and Li, 2005b), but are described here for the first time in C. maenas. The other four -FLRFamide-containing peptides, pQGNFLRFamide, APQGNFLRFamide, DGNRNFLRFamide and YGNRSFLRFamide (Figure 1(a)), as well as the -YLRFamide isoforms SRNYLRFamide and GLSRNYLRFamide, were de novo sequenced for the first time.
3.1.8. HIGSLYRamide
HIGSLYRamide, a previously described C. maenas neuropeptide (Christie et al., 2008), was identified via both MALDI-FTMS and ESI-Q-TOF MS/MS from the brain, SG and PO.
3.1.9. Orcokinin
Six full-length orcokinins, NFDEIDRSGFGFA, NFDEIDRSGFGFV, DFDEIDRSGFGFV, NFDEIDRSGFGFN, NFDEIDRSSFGFV and NFDEIDRSSFGFN, and five putative orcokinin truncations, EIDRSGFGFA, NFDEIDRSGFG, NFDEIDRSGFA, NFDEIDRSSFA, NFDEIDRSSFG and NFDEIDRSGFGF, were characterized from C. maenas neural tissues via ESI-Q-TOF MS/MS and/or direct tissue/off-line HPLC fractionation coupled to MALDI-FTMS analysis. Each of these peptides has been described previously from crustacean neural tissues, and the full-length peptides NFDEIDRSGFGFA, NFDEIDRSGFGFV, NFDEIDRSGFGFN and NFDEIDRSSFGFN were identified previously in C. maenas (Bungart et al., 1995). In addition, two amidated truncations, NFDEIDRSGFamide and NFDEIDRSSFamide (Figure 2(a), (b)) were identified via ESI-Q-TOF MS/MS from the brain (Table 1). NFDEIDRSGFamide was previously identified in H. americanus (Ma et al., 2008), but is new to C. maenas; FDEIDRSGFGFA and NFDEIDRSSFamide were de novo sequenced for the first time in this study.
Figure 2.
Collision-induced dissociation spectra of three de novo sequenced peptides. ESI-Q-TOF MS/MS sequencing of two orcokinins: (a.) NFDEIDRSGFamide (599.782+), (b.) NFDEIDRSSFamide (614.702+), and (c.) the putative cryptocyanin truncation KIFEPLRDKN (630.312+).
3.1.10. Orcomyotropin
The orcomyotropin-related peptide FDAFTTGFGHS, a known C. maenas neuropeptide (Stemmler et al., 2007), was detected by both MALDI-FTMS and ESI-Q-TOF MS/MS from brain, thoracic ganglia, PO and SG.
3.1.11. Pigment dispersing hormone
The peptide NSELINSILGLPKVMNDAamide, commonly known as β-pigment dispersing hormone (β-PDH) and a known C. maenas neuropeptide (Lohr et al., 1993), was identified by both MALDI-FTMS and ESI-Q-TOF MS/MS from the SG (Table 1).
3.1.12. Proctolin
The pentapeptide RYLPT, also known as proctolin (Brown, 1975; Starratt and Brown, 1975) and a previously described C. maenas neuropeptide (Stangier et al., 1986), was identified in the PO via MALDI-FTMS (Table 1).
3.1.13. Pyrokinin
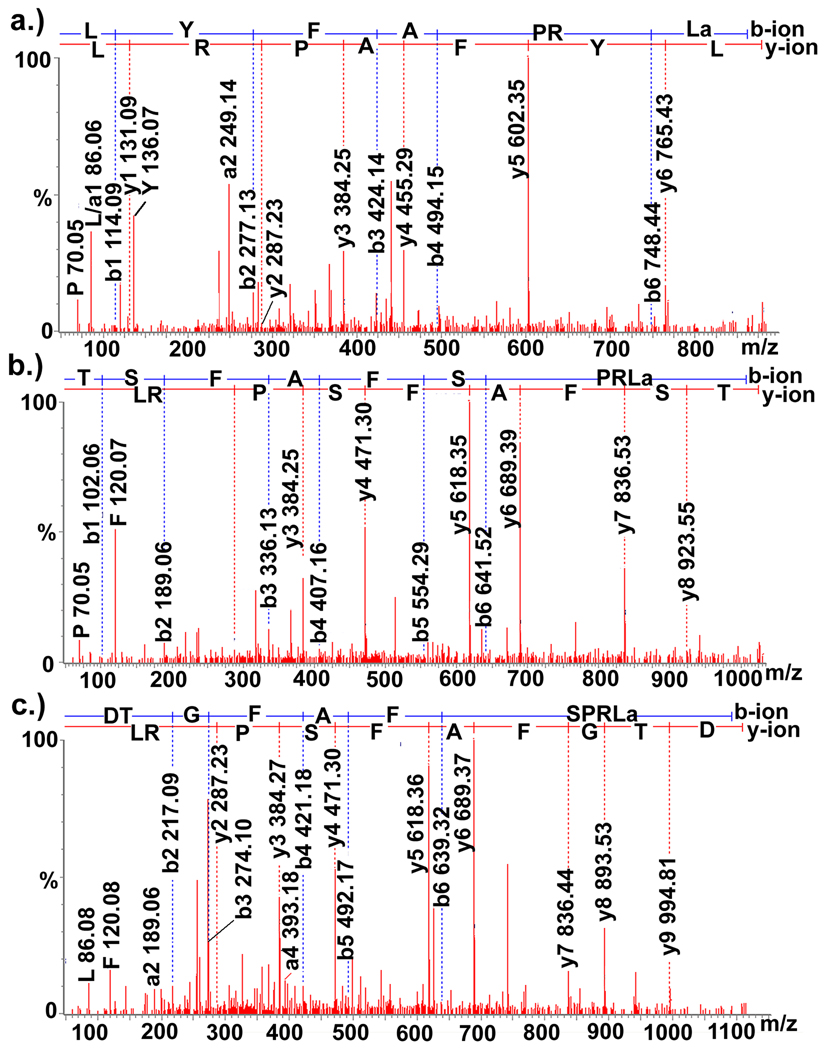
Recently, peptides possessing the C-terminal motif -FXPRLamide (where X is a variable amino acid) were identified from the penaeid shrimp Litopenaeus vannamei and the crab Cancer borealis (Saideman et al., 2007; Torfs et al., 2001). This sequence is characteristic of members of the pyrokinin/periviscerokinin/pheromone biosynthesis activating neuropeptide family of peptides (Torfs et al., 2001). Here, two novel pyrokinins, TSFAFSPRLamide and DTGFAFSPRLamide (Figure 3(a), (b)), were de novo sequenced using ESI-Q-TOF MS/MS from brain tissue (Table 1). In addition, a third pyrokinin, LYFAPRLamide, a known C. borealis variant (Ma et al.), was also identified in the brain via ESI-Q-TOF MS/MS.
Figure 3.
Collision-induced dissociation spectra of three de novo sequenced peptides. ESI-Q-TOF MS/MS sequencing of three pyrokinins: (a.) LYAFPRLamide (439.762+), (b.) TSFAFSPRLamide (512.782+), and (c.) DTGFAFSPRLamide(555.292+).
3.1.14. Red pigment concentrating hormone
The peptide pQLNFSPGWamide, a known C. maenas neuropeptide (Fernlund, 1974) commonly known as red pigment concentrating hormone (RPCH), was sequenced via ESI-Q-TOF MS/MS from the SG (Table 1).
3.1.15. RYamide
Nine peptides containing a RYamide C-terminus were characterized via ESI-Q-TOF MS/MS sequencing from the C. maenas PO. Six RYamide isoforms, FVGGSRYamide, FYANRYamide, FYSQRYamide, SGFYANRYamide, pEGFYSQRYamide and SSRFVGGSRYamide, are identical in structure to peptides identified from the POs of the crabs C. borealis and C. productus (Fu et al., 2005a; Li et al., 2003). The existence of these isoforms in C. maenas is described for the first time in this study. The remaining RYamide peptides, SGFYAPRYamide, SGFYADRY and (X)YANRYamide (m/z 2091 Da, with X representing several uncharacterized amino acid residues), were de novo sequenced here for the first time, though the latter peptide remains to be fully characterized.
3.1.16. SIFamide
Three peptides possessing the C-terminal sequence SIFamide, GYRKPPFNGSIFamide, VYRKPPFNGSIFamide and RKPPFNGSIFamide, were identified from C. maenas via ESI-Q-TOF MS/MS and/or direct tissue/offline HPLC fractionation coupled to MALDI-FTMS analysis (Table 1). GYRKPPFNGSIFamide (Gly1-SIFamide), a known C. maenas isoform (Stemmler et al., 2007), was identified in brain, thoracic ganglia and SG. The second peptide, VYRKPPFNGSIFamide (Val1-SIFamide), was identified only in the thoracic ganglia. This peptide is a known SIFamide variant, but prior to our study it was thought to only exist in a member of the Astacidean genus Homarus (Dickinson et al., 2008). Lastly, RKPPFNGSIFamide was identified solely in the brain. This peptide, likely a truncation of one of the full-length isoforms and previously identified in both H. americanus and C. borealis (Ma et al., 2008; Ma et al.), is another new peptide identified in C. maenas.
3.1.17. Tachykinin-related peptides
Two full-length tachykinin-related peptides (TRPs), APSGFLGMRamide and TPSGFLGMRamide, their methionine oxidized forms APSGFLGM(O)Ramide and TPSGFLGM(O)Ramide, and two putative truncations, PSGFLGMRamide and SGFLGMRamide, were sequenced via ESI-Q-TOF MS/MS and MALDI-FTMS from the brain and thoracic ganglia of C. maenas (Table 1). In addition, the putative precursor of APSGFLGMRamide, APSGFLGMRG, was sequenced from brain tissue via ESI-Q-TOF MS/MS (Table 1). Both full-length TRPs were also sequenced from the SG via ESI-Q-TOF MS/MS (Table 1). While each of these peptides is a known TRP isoform, this is their first description in C. maenas, with the exception of APSGFLGMRamide (Stemmler et al., 2007).
3.1.18. Other peptides
Four peptides possessing the N-terminal motif KIFEPL- were de novo sequenced via ESI-Q-TOF MS/MS from the thoracic ganglia (Table 1): KIFEPLR, KIFEPLVA, KIFEPLRDKN (Figure 2(c)) and KIFEPLRDKNL. The similarity of these unique peptide sequences suggests that they may represent members of a common and previously unknown peptide family.
3.2. blastp analyses of KIFEPL-containing sequences
As stated in 3.1.18, four peptides possessing the N-terminal motif KIFEPL were de novo sequenced for the first time in our study. The consensus motif exhibited by each peptide did not immediately place this set of isoforms into a known peptide family. To assess if they might be truncations of larger, known proteins, homology searches of the crustacean protein database at NCBI were conducted using the sequence of the longest of the KIFEPL peptides as a query. The blastp analysis revealed this particular peptide to have significant homology to members of the cryptocyanin family, with the top blast hit being cryptocyanin 1 from the blue swimmer crab Portunus pelagicus (accession no. ABM54471 [BLAST score, 35.8; E-value, 4e-04](Kuballa et al., 2007)). Based on this finding, it is likely that the five KIFEPL-containing peptides are truncations of as of yet uncharacterized C. maenas cryptocyanins, rather than representing full-length isoforms of a novel peptide family.
3.3. Peptide identification using functional genomics
Using the sequences of known decapod crustacean and insect peptide precursors as queries, the C. maenas EST database at NCBI was searched for putative peptide-encoding prepro-hormone transcripts. The sequences used in this search encode the following peptide precursors: A-type allatostatin, B-type allatostatin, C-type allatostatin, allatotropin, bursicon (both the α and β subunit peptides), corazonin, CCAP, members of the CHH superfamily (including the CHH, molt-inhibiting hormone (MIH) and mandibular organ-inhibiting hormone (MOIH) subfamilies), diuretic hormone (both the calcitonin- and corticotropin-releasing factorlike families), ecdysis-triggering hormone (ETH), eclosion hormone, members of the FLP superfamily (including the myosuppressin, neuropeptide F, short NPF [sNPF], and sulfakinin subfamilies), HIGSLYRamide, insect kinin, intocin, neuroparsin, orcokinin, PDH, proctolin, members of the pyrokinin/periviscerokinin/pheromone biosynthesis activating neuropeptide (PBAN) family, SIFamide, and TRP. Those searches that identified putative precursors are described here, with the data presented in alphabetical order based on peptide family name.
3.3.1. Bursicon
In insects the bursicon has long been implicated in the tanning of the cuticle following molting (Fraenkel and Hsiao, 1962; Fraenkel and Hsiao, 1963; Fraenkel and Hsiao, 1965; Fraenkel and Hsiao, 1966). This hormone has recently been shown to be a heterodimer composed of two cystine knot proteins, namely bursicon α and bursicon β (Luo et al., 2005; Mendive et al., 2005). In this study, using the amino acid sequence of a D. melanogaster prepro-bursicon β (accession no. Q9VJS7; (Luo et al., 2005; Mendive et al., 2005)) as a query, a single C. maenas EST (accession no. DY656914 [BLAST score, 144; E-value, 7e-36]) was determined to encode a putative bursicon β precursor (Figure 4A). Translation of this transcript yielded a 138 amino acid, putative full-length precursor protein, the first 23 residues of which were predicted by SignalP to form a signal sequence (cleavage suggested to occur between Ala23 and Arg24). Based on homology to Drosophila bursicon β, no internal prohormone convertase processing is predicted for the C. maenas prohormone. Thus the enzymatic action of signal peptidase is hypothesized to produce the following bursicon β-like peptide from the deduced prepro-hormone: RSYGVECETLPSTIHISKEEYDDTGRLVRVCEEDVAVNKCEGACVSKVQPSVNTPSGFLK DCRCCREVHLRARDITLTHCYDGDGARLSGAKATQHVKLREPADCQCFKCGDSTR.
Figure 4.

Deduced amino acid sequences of C. maenas bursicon β, HIGSLYRamide and neuroparsin encoding precursors. (A) The putative full-length prepro-bursicon β. (B) A putative partial pro-hormone encoding HIGSLYRamide. (C) The putative full-length prepro-neuroparsin. The predicted signal peptides, when present, are shown in italics (gray in the online version), with all predicted dibasic prohormone convertase cleavage sites underlined (black in the online version). In the online version, the putative isoform(s) of the named peptide family (i.e. bursicon β, HIGSLYRamide, or neuroparsin) are shown in red, while all other precursor-related peptides are shown in blue. Asterisks indicate the presence of a stop codon.
3.3.2. HIGSLYRamide
As stated earlier in 3.1.8, the peptide HIGSLYRamide was identified via mass spectral analyses from the brain, SG and PO of C. maenas. The assignments of the position 2 Ile and the position 5 Leu in this peptide are based on a putative C. maenas prepro-hormone deduced from EST DV111329, the identification of which was described previously (Christie et al., 2008). In addition to HIGSLYRamide, the precursor deduced from DV111329 is also predicted to generate several other peptides, including the peptide HIEALYRamide, likely a member of the same peptide family as HIGSLYRamide. Another 9 peptides generated appear to be isoforms of a common family, with X representing the possibility of additional amino acids at the N-terminus: XRNSQNLSED, DEDNSQNLSED, DEDNTNNLSED, DEDNSPNLSED, DGGNTSTFSED, DDDTNSLSED, DEDNANDLSED, DEDNGNSISED and DEDNAHDLSED. Three more peptides may be generated from the predicted precursor: HQQDFPDLIQDEEAIE, LHELPRQRYFASLL and NRPSAFGX, where X represents the possibility of additional amino acids added at the C-terminus. The peptide products generated from the initial precursor and their organization within that precursor are summarized in Figure 4B.
3.3.3. Neuroparsin
The neuroparsins are a family of pleiotropic neuropeptides originally identified in insects (Badisco et al., 2007). Using the sequence of a Schistocerca gregaria prepro-neuroparsin 1 (accession no. CAC38869) (Janssen et al., 2001) as a query, six C. maenas ESTs (accession nos. DN635465 [BLAST score, 53.9; E-value, 5e-09], DN634851 [BLAST score, 53.9; E-value, 5e-09], DN634570 [BLAST score, 53.9; E-value, 5e-09], DN551462 [BLAST score, 53.9; E-value, 5e-09], DV466984 [BLAST score, 53.1; E-value, 1e-08], and DV467101 [BLAST score, 52.4; E-value, 2e-08]) were determined to encode putative neuroparsin precursor(s). Translation of these transcripts revealed that all encode an identical 101 amino acid, putative full-length prepro-hormone, the first 27 amino acids of which were predicted by SignalP to form a signal peptide. As no internal prohormone convertase cleavage sites were identified, signal peptidase processing of the deduced prepro-hormone is hypothesized to liberate a single isoform of neuroparsin: APRCDRHDEEAPKNCKYGTTQDWCKNGVCAKGPGETCGGYRWSEGKCGEGTFCSCGI CGGCSPFDGKCGPTSIC (Figure 4C).
4. Discussion
4.1. Peptide discovery in C. maenas using a strategy combining mass spectrometry and functional genomics
Due to its extreme adaptability to environmental challenges, the European green crab C. maenas has become arguably one of the most significant marine invasive species known. The physiological flexibility of this animal is a key factor in its ability to successfully establish itself in new habitats. This is undoubtedly under paracrine/hormonal control, and peptides are likely the largest single contributor to this unique physiology. Thus, a thorough understanding of the peptides used by C. maenas is of critical importance for understanding the physiological and behavioral adaptability of this species. In this study, a combination of mass spectrometry and functional genomics was used to elucidate the C. maenas neuropeptidome.
For the mass spectrometry aspect of our investigation, we combined MALDI-based high resolution mass profiling and tandem mass spectrometric sequencing to identify the native peptides present in the C. maenas nervous system. Specifically, the highly sensitive and accurate mass measurements provided by internally calibrated MALDI-FTMS (both direct tissue and offline analysis of HPLC fractions, including some peptide fragmentations using SORI-CID) was used to identify known peptides based on predicted mass-to-charge ratios (m/z). The mass measurement accuracy is within 5 ppm for the peptides identified by MALDI-FTMS. Figure 5 shows representative mass spectra via direct tissue analysis of brain, SG and PO with MALDI-FTMS. With the high mass measurement accuracy of MALDI-FTMS, numerous neuropeptides from a variety of families were identified based on accurate mass matching. Off-line HPLC separation removes interfering molecules such as salts, lipids and proteins and thus greatly simplifies the chemical complexity of the tissue extracts, significantly improving the neuropeptide detection (Figure 6).
Figure 5.
Direct tissue peptide profiling of C. maenas brain (a), SG (b), (c) and PO (d) by MALDI-FTMS. Signals correspond to the protonated molecular ions, [M+H]+, where M is the molecular weight of each peptide.
Figure 6.
Peptide profiles of off-line collected HPLC fractions of C. maenas brain extract detected by MALDI-FTMS. Three fractions show different peptide profiles as (a), (b) and (c). Signals correspond to the protonated molecular ions, [M+H]+, where M is the molecular weight of each peptide.
In the second part of this strategy, we utilized the power of nanoLC-ESI-Q-TOF MS/MS to de novo sequence novel peptides. Formaldehyde labeling was used to solve ambiguities during the de novo sequencing. It is especially useful for the de novo sequencing of the singly charged neuropeptides such as A-type ASTs because it enhances the a1 ion and simplifies the MS/MS fragmentation pattern. Figure 7 shows an example of using formaldehyde labeling to facilitate de novo sequencing of three A-type AST peptides: EAYAFGLamide, NPYAFGLamide and GGPYAFGLamide. As shown in Figure 7(a), the initial MS/MS spectrum of the native peptide EAYAFGLamide is very complex due to extensive internal fragmentations. Furthermore, it is difficult to resolve the ambiguity of the N-terminus sequence EA/AE due to the absence of cleavage between the first two amino acid residues. In contrast, after the reductive methylation, a- and b-ion series are enhanced while internal fragmentations are suppressed, yielding a much cleaner MS/MS spectrum (Figure 7(b)). An additional benefit is that the enhanced a1 ion helps resolve the ambiguities of the N-terminal residue, which suggests that the N-terminal sequence is EA. Figure 7(c) and 7(d) show two additional examples of improved de novo sequencing of A-type ASTs via formaldehyde labeling. Upon reaction, two A-type ASTs with the same molecular masses (m/z = 780.4) were identified as NPYAFGLamide and GGPYAFGLamide; the reaction differentiated the isobaric GG and N at the N-termini of the two peptides.
Figure 7.
ESI-Q-TOF MS/MS spectra of an A-type AST peptide EAYAFGLamide (769.391+) before (a) and after (b) formaldehyde labeling, and two A-type AST peptides NPYAFGLamide (780.401+, c.) and GGPYAFGLamide (780.401+, d.) after formaldehyde labeling. Asterisks in the peptide sequences indicate sites of dimethylation. In panels b), c) and d), the a-, b-, and y-type fragment ion series are labeled according to the MS/MS of (CH3)2EAYAFGLamide (870.461+), (CH3)2NPYAFGLamide (780.401+) and (CH3)2GGPYAFGLamide (780.401+).
Via this combined mass spectral strategy, 122 peptides from 14 peptide families were identified. This included 31 previously known C. maenas peptides (e.g., crustacean cardioactive peptide (CCAP) (Stangier et al., 1987), β-PDH (Fu et al., 2005a), the orcokinins NFDEIDRSGFGFA, NFDEIDRSGFGFV and NFDEIDRSSFGFN (Bungart et al., 1995), proctolin (Stangier et al., 1986) and RPCH (Linck et al., 1993)) and 49 peptides known from other species, but newly described in the European green crab (e.g. Arg7-corazonin (Li et al., 2003) and the RYamide isoforms FYANRYamide, FYSQRYamide, SGFYANRYamide, pEGFYSQRYamide and SSRFVGGSRYamide (Fu et al., 2005a; Li et al., 2003)). Additionally, 42 peptides were de novo sequenced for the first time in this study (e.g. the B-type ASTs AWSNLGQAWamide, AGWNKFQGSWamide, VTWGKFQGSWamide, GSNWSNLRGAWamide and GVNWSNLRGAWamide and the pyrokinins TSFAFSPRLamide and DTGFAFSPRLamide). Of particular note was the identification of 25 distinct FMRFamide-like peptides (including 9 novel ones), one of the most diverse peptide families found in the animal kingdom. In addition, functional genomic analysis leveraging ESTs derived from a normalized C. maenas cDNA library enabled identification of 15 additional peptides, including a putative bursicon β subunit peptide and a putative isoform of neuroparsin. Collectively, the data presented here not only dramatically increases the number of known C. maenas neuropeptides, but also provides a foundation for future investigations of the physiological roles played by these molecules in this highly adaptable marine invasive species.
4.2. Most known C. maenas neuropeptides were detected using mass spectral analyses
Prior to our study, 31 peptides had been characterized from the nervous system of C. maenas (Bungart et al., 1995; Duve et al., 1997a; Kegel et al., 1989; Lohr et al., 1993; Stangier et al., 1986; Stangier et al., 1987). We sought to apply our identification strategy to this same source of peptides, and thereby gain insight into the benefits and weaknesses of the different methods. Here, the vast majority of the initial set of peptides, particularly those with m/z less than 4000 Da, was identified using a strategy combining MALDI-FTMS and ESI-Q-TOF MS/MS. However, there were peptides that required further examination, particularly the isoforms of A-type AST.
Previously, Duve et al. (1997a) identified 21 A-type ASTs from pooled samples of C. maenas brain and thoracic ganglia; approximately 500 animals used. Of these peptides, thirteen isoforms were again identified in our study: EAYAFGLamide, GGPYAFGLamide, NPYAFGLamide, DPYAFGLamide, AGPYAFGLamide, EPYAFGLamide, AGPYSFGLamide, ASPYAFGLamide, SDMYSFGLamide, ATGQYAFGLamide, APGPYAFGLamide, EYDDMYTEKRPKVYAFGLamide, and GYEDEDEDRPFYALGLGKRPRTYSFGLamide. However, 7 of the peptides characterized by Duve et al. (1997a) were not detected in our mass spectral analyses: YAFGLamide, SPYAFGLamide, YSFGLamide, GGPYSYGLamide, PDMYAFGLamide, SGQYSFGLamide and APTDMYSFGLamide. Conversely, 12 C. maenas A-type isoforms not identified by Duve et al. (1997a) were sequenced here: AAPYAFGLamide, GKPYAFGLamide, EPYEFGLamide, RGPYAFGLamide, ARPYAFGLamide, FSGASPYGLamide, ARPYSFGLamide, AASPYSFGLamide, LKAYDFGLamide, TRPYSFGLamide, KLPYSFGLamide and PADLYEFGLamide. The origin of the differences between our A-type AST identifications and those Duve et al. (1997) remains unknown. The most plausible cause is individual- and/or regional-specific A-type AST variants. Specifically, all of the animals used by Duve et al. (1997) were obtained from European waters, while all of those used here were obtained from the greater Mount Desert Island area of Maine. The presence of isoform heterogeneity in neuropeptides has recently been demonstrated for the CHH precursor-related peptides of Cancer crabs (Stemmler et al., 2007). A similar situation could be at play here if the European animals possessed individual- and/or regional-specific A-type AST variants not present in the Maine population. Clearly, additional experiments will be needed to determine if this is indeed true. However, it is important to remember that, while extensive, the catalog of C. maenas peptides described in our study undoubtedly represents only a subset of the total peptidome of C. maenas as a species. Peptides may be missing from this study due to factors such as not readily ionizable structures, existence in very minute abundance, and the presence of isoforms that may be population and/or individual specific.
4.3. Val1-SIFamide: an individual-specific variant?
As described in 3.1.16, three isoforms of SIFamide were sequenced from the nervous system of C. maenas: GYRKPPFNGSIFamide, VYRKPPFNGSIFamide and RKPPFNGSIFamide. The identification of Gly1-SIFamide was anticipated, as it is a previously described C. maenas isoform (Stemmler et al., 2007). Likewise, the detection of RKPPFNGSIFamide was expected, as it is a known truncation product of the Gly1-variant (Ma et al., 2008). In contrast, our detection of VYRKPPFNGSIFamide (Val1-SIFamide) was surprising, as this SIFamide variant was thought to exist only in the Astacidean genus Homarus (Christie et al., 2006; Stemmler et al., 2007) and had never been described in C. maenas, despite this species being included in a recent mass spectral investigation in which this peptide family was specifically targeted and only Gly1-SIFamide was identified (Stemmler et al., 2007).
While it is not possible to unambiguously determine the origin of the VYRKPPFNGPIFamide identified and characterized from C. maenas in this study, it is likely the result of an individual, or a small population of animals, possessing a mutation in a gene encoding a C. maenas SIFamide precursor. This hypothesis is supported by the fact that, in our study, Val1-SIFamide was identified only via ESI-Q-TOF MS/MS from pooled tissue extract and not in any direct tissue studies, a finding supported by the individual tissue MALDI-FTMS data of Stemmler et al. (2007). In addition, other individual-specific SIFamide variants have been described, specifically a GYRKPPFNGPIFamide from the hermit crab Pagurus pollicarus (Stemmler et al., 2008). Regardless of origin, the finding of Val1-SIFamide in C. maenas should give pause before the assumption is made that all peptides described in our study are ubiquitously present in all individuals of this species, as this data is obtained from pooled tissue extracts where individual-specific variants may bias the detection of a given peptide.
Acknowledgements
We thank the University of Wisconsin School of Pharmacy Analytical Instrumentation Center for access to the MALDI-FTMS instrument. We wish to thank Dr. Peter O’Connor from Boston University for the use of BUDA software to make FTMS figures. Financial support for this study was provided in part by the University of Wisconsin School of Pharmacy, Wisconsin Alumni Research Foundation, National Science Foundation CAREER Award CHE-0449991, National Institutes of Health through grant 1R01DK071801 and a research fellowship from the Alfred P. Sloan Foundation (to L.L.). Furthermore, grants from the National Center for Research Resources’ Maine INBRE Program (NIH P20 RR-016463; to Mount Desert Island Biological Laboratory [MDIBL]), the National Science Foundation’s Research Experience for Undergraduates Program (NSF DBI-0453391; to the MDIBL REU site) and the National Institute of Environmental Health Sciences STEER program (NIEHS R25 ES016254), as well as the MDIBL High School Fellows Research Program, a MDIBL New Investigator Award (from the Salisbury Cove Research Fund provided through the Thomas H. Maren Foundation [to A.E.C.]) and institutional funds provided by MDIBL (to A.E.C) are gratefully acknowledged. R.P.H. was supported by NSF IBN 02-30005 and NSF EPS 04-47675.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Badisco L, Claeys I, Van Loy T, Van Hiel M, Franssens V, Simonet G, Broeck JV. Neuroparsins, a family of conserved arthropod neuropeptides. Gen. Comp. Endocrinol. 2007;153:64–71. doi: 10.1016/j.ygcen.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Brown BE. Proctolin: a peptide transmitter candidate in insects. Life Sci. 1975;17:1241–1252. doi: 10.1016/0024-3205(75)90133-2. [DOI] [PubMed] [Google Scholar]
- Bungart D, Hilbich C, Dircksen H, Keller R. Occurrence of analogues of the myotropic neuropeptide orcokinin in the shore crab, Carcinus maenas: Evidence for a novel neuropeptide family. Peptides. 1995;16:67–72. doi: 10.1016/0196-9781(94)00145-v. [DOI] [PubMed] [Google Scholar]
- Carlton JT, Cohen AN. Episodic global dispersal in shallow water marine organisms: the case history of the European shore crabs Carcinus maenas and C. aestuarii. J. Biogeography. 2003;30:1809–1820. [Google Scholar]
- Christie AE, Stemmler EA, Peguero B, Messinger DI, Provencher HL, Scheerlinck P, Hsu YW, Guiney ME, de la Iglesia HO, Dickinson PS. Identification, physiological actions, and distribution of VYRKPPFNGSIFamide (Val1)-SIFamide) in the stomatogastric nervous system of the American lobster Homarus americanus. J. Comp. Neurol. 2006;496:406–421. doi: 10.1002/cne.20932. [DOI] [PubMed] [Google Scholar]
- Cohen AN, Carlton TJ. Introduction, dispersal and potential impacts of the green crab Carcinus maenas in San Francisco Bay, California. Mar. Biol. 1995;122:225–237. [Google Scholar]
- Dircksen H, Bocking D, Heyn U, Mandel C, Chung JS, Baggerman G, Verhaert P, Daufeldt S, Plosch T, Jaros PP, Waelkens E, Keller R, Webster SG. Crustacean hyperglycaemic hormone (CHH)-like peptides and CHH-precursor-related peptides from pericardial organ neurosecretory cells in the shore crab, Carcinus maenas, are putatively spliced and modified products of multiple genes. Biochem. J. 2001;356:159–170. doi: 10.1042/0264-6021:3560159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell JA, VanderHeyden W, Li L. Rat neuropeptidomics by LC-MS/MS and MALDI-FTMS: enhanced dissection and extraction techniques coupled with 2D RP-RP HPLC. J. Proteome Res. 2006;5:3368–3375. doi: 10.1021/pr0603452. [DOI] [PubMed] [Google Scholar]
- Duve H, Johnsen AH, Maestro J-L, Scott AG, Jaros PP, Thorpe A. Isolation and Identification of multiple neuropeptides of the allatostatin superfamily in the shore crab Carcinus maenas. Eur. J. Biochem. 1997a;250:727–734. doi: 10.1111/j.1432-1033.1997.00727.x. [DOI] [PubMed] [Google Scholar]
- Duve H, Johnsen AH, Maestro J-L, Scott AG, Winstanley D, Davey M, East PD, Thorpe A. Lepidopteran peptides of the allatostatin superfamily. Peptides. 1997b;18:1301–1309. doi: 10.1016/s0196-9781(97)00188-5. [DOI] [PubMed] [Google Scholar]
- Dyrlov Bendtsen J, Nielsen H, von Heijne G, Brunak S. Improved Prediction of Signal Peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Fernlund P. Structure of the red-pigment-concentrating hormone of the shrimp, Pandalus borealis. Biochim. Biophys. Acta. 1974;371:304–311. doi: 10.1016/0005-2795(74)90027-0. [DOI] [PubMed] [Google Scholar]
- Fu Q, Kutz KK, Schmidt JJ, Hsu YW, Messinger DI, Cain SD, de la Iglesia HO, Christie AE, Li L. Hormone complement of the Cancer productus sinus gland and pericardial organ: an anatomical and mass spectrometric investigation. J. Comp. Neurol. 2005a;493:607–626. doi: 10.1002/cne.20773. [DOI] [PubMed] [Google Scholar]
- Fu Q, Li L. De novo sequencing of neuropeptides using reductive isotopic methylation and investigation of ESI QTOF MS/MS fragmentation pattern of neuropeptides with N-terminal dimethylation. Anal. Chem. 2005b;77:7783–7795. doi: 10.1021/ac051324e. [DOI] [PubMed] [Google Scholar]
- Grosholz ED. Recent biological invasion may hasten invasional meltdown by accelerating historical introductions. Proc. Natl. Acad. Sci. 2005;102:1088–1091. doi: 10.1073/pnas.0308547102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosholz ED, Ruiz GM. Predicting the impact of introduced marine species: Lessons from the multiple invasions of the European green crab Carcinus maenas. Biol. Conserv. 1996;78:59–66. [Google Scholar]
- Hidalgo FJ, Barón PJ, Orensanz JM. A prediction come true: the green crab invades the Patagonian coast. Biol. Invasions. 2005;7:547–552. [Google Scholar]
- Huybrechts J, Nusbaum MP, Bosch LV, Baggerman G, Loof AD, Schoofs L. Neuropeptidomic analysis of the brain and thoracic ganglion from the Jonah crab, Cancer borealis. Biochem. Biophys. Res. Commun. 2003;308:535–544. doi: 10.1016/s0006-291x(03)01426-8. [DOI] [PubMed] [Google Scholar]
- Janssen T, Claeys I, Simonet G, De Loof A, Girardie J, Vanden Broeck J. cDNA cloning and transcript distribution of two different neuroparsin precursors in the desert locust, Schistocerca gregaria. Insect Mol. Biol. 2001;10:183–189. doi: 10.1046/j.1365-2583.2001.00257.x. [DOI] [PubMed] [Google Scholar]
- Kegel G, Reichwein B, Weese S, Gaus G, Peter-Katalinic J, Keller R. Amino acid sequence of the crustacean hyperglycemic hormone (CHH) from the shore crab, Carcinus maenas. FEBS Letters. 1989;255:10–14. doi: 10.1016/0014-5793(89)81051-8. [DOI] [PubMed] [Google Scholar]
- Kuballa AV, Merritt DJ, Elizur A. Gene expression profiling of cuticular proteins across the moult cycle of the crab Portunus pelagicus. BMC Biol. 2007;5:45. doi: 10.1186/1741-7007-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafferty KD, Kuris KD. Biological control of marine pests. Ecology. 1996;77:1989–2000. [Google Scholar]
- Li L, Kelley WP, Billimoria CP, Christie AE, Pulver SR, Sweedler JV, Marder E. Mass spectrometric investigation of the neuropeptide complement and release in the pericardial organs of the crab, Cancer borealis. J. Neurochem. 2003;87:642–656. doi: 10.1046/j.1471-4159.2003.02031.x. [DOI] [PubMed] [Google Scholar]
- Li L, Ma M, Chen R, Christie AE. Peptidomic analysis of the central nervous systems of the american lobster Homarus americanus and the European green crab Carcinus maenas. 37th annual meeting of the Society for Neuroscience. 2007 [Google Scholar]
- Linck B, Klein JM, Mangerich S, Keller R, Weidemann WM. Molecular cloning of crustacean red pigment concentrating hormone precursor. Biochem. Biophys. Res. Commun. 1993;195:807–813. doi: 10.1006/bbrc.1993.2117. [DOI] [PubMed] [Google Scholar]
- Lohr J, Klein J, Webster SG, Dircksen H. Quantification, immunoaffinity purification and sequence analysis of a pigment-dispersing hormone of the shore crab, Carcinus maenas. Comp. Biochem. Physiolo. B. 1993;104:699–706. doi: 10.1016/0305-0491(93)90200-o. [DOI] [PubMed] [Google Scholar]
- Ma M, Chen R, Sousa GL, Bors EK, Kwiatkowski MA, Goiney CC, Goy MF, Christie AE, Li L. Mass spectral characterization of peptide transmitters/hormones in the nervous system and neuroendocrine organs of the American lobster Homarus americanus. Gen. Comp. Endocrinol. 2008;156:395–409. doi: 10.1016/j.ygcen.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Wang J, Chen R, Li L. Expanding the crustacean neuropeptidome using a multifacetecd mass spectrometric approach. Submitted. doi: 10.1021/pr801047v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen SS. The role of cortisol and growth hormone in seawater adaptation and development of hypoosmoregulatory mechanisms in sea trout parr (Salmo trutta trutta) Gen. Comp. Endocrinol. 1990;79:1–11. doi: 10.1016/0016-6480(90)90082-w. [DOI] [PubMed] [Google Scholar]
- McVeigh P, Kimber MJ, Novozhilova E, Day TA. Neuropeptide signalling systems in flatworms. Parasitology. 2005;131:S41–S55. doi: 10.1017/S0031182005008851. [DOI] [PubMed] [Google Scholar]
- Monigatti F, Gasteiger E, Bairoch A, Jung E. The Sulfinator: predicting tyrosine sulfation sites in protein sequences. Bioinformatics. 2002;18:769–770. doi: 10.1093/bioinformatics/18.5.769. [DOI] [PubMed] [Google Scholar]
- Morris S, Airriess CN. Integration of physiological responses of crustaceans to environmental challenge. South African J. Zool. 1998;33:87–106. [Google Scholar]
- Stangier J, Dircksen H, Keller R. Identification and immunocytochemical localization of proctolin in pericardial organs of the shore crab, Carcinus maenas. Peptides. 1986;7:67–72. doi: 10.1016/0196-9781(86)90063-x. [DOI] [PubMed] [Google Scholar]
- Stangier J, Hilbich C, Beyreuther K, Keller R. Unusual cardioactive peptide (CCAP) from pericardial organs of the shore crab Carcinus maenas. Proc. Natl. Acad. Sci. 1987;84:575–579. doi: 10.1073/pnas.84.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starratt AN, Brown BE. Structure of the pentapeptide proctolin, a proposed neurotransmitter in insects. Life Sci. 1975;17:1252–1256. doi: 10.1016/0024-3205(75)90134-4. [DOI] [PubMed] [Google Scholar]
- Stay B, Tobe SS. The role of allatostatins in juvenile hormone synthesis in insects and crustaceans. Annu. Rev. Entomol. 2007;52:277–299. doi: 10.1146/annurev.ento.51.110104.151050. [DOI] [PubMed] [Google Scholar]
- Stemmler EA, Cashman CR, Messinger DI, Gardner NP, Dickinson PS, Christie AE. High-mass-resolution direct-tissue MALDI-FTMS reveals broad conservation of three neuropeptides (APSGFLGMRamide, GYRKPPFNGSIFamide and pQDLDHVFLRFamide) across members of seven decapod crustaean infraorders. Peptides. 2007;28:2104–2115. doi: 10.1016/j.peptides.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Stemmler EA, Dickinson PS, Christie AE. Identifiction of a putative individual-specific variant of Gly1-SIFamide via direct tissue Matrix assisted laser desorption/ionization-Fourier transform mass spectromentry (MALDI-FTMS) and sustained off resonance irradiation-collision induced dissociation (SORI-CID) 2008 In preparation. [Google Scholar]
- Strand FL. Neuropeptides: regulators of physiological processes. Cambridge, MA; London, UK: MIT Press; 1999. [Google Scholar]
- Torfs P, Nieto J, Cerstiaens A, Boon D, Baggerman G, Poulos C, Waelkens E, Derua R, Calderon J, De Loof A, Schoofs L. Pyrokinin neuropeptides in a crustacean. Eur. J. Biochem. 2001;268:149–154. doi: 10.1046/j.1432-1327.2001.01858.x. [DOI] [PubMed] [Google Scholar]
- Veenstra JA. Isolation and structure of corazonin, a cardioactive peptide from the American cockroach. FEBS Letters. 1989;250:231–234. doi: 10.1016/0014-5793(89)80727-6. [DOI] [PubMed] [Google Scholar]
- Yamada SB, Hauck L. Field identification of the European green crab species: Carcinus maenas and Carcinus aestuarii. J. Shell Res. 2001;20:905–912. [Google Scholar]
- Zajac J-M, Mollereau C. Introduction. Peptides. 2006;27:941–942. doi: 10.1016/j.peptides.2005.12.005. [DOI] [PubMed] [Google Scholar]