In excitable cells, the molecular motions of ion channels are linked to the whole cell physiology via the transmembrane potential (Vm). To examine such events at the cellular level, we recently proposed a model that links the molecular dynamics (MD) and electrostatics of the slow delayed rectifier K+ channel (IKs) to the cell and tissue electrophysiology (Silva et al., 2009). Although the data for creating such a model are sparse and techniques to do a rigorous simulation are still being developed, the model provides a roadmap for future experiments and simulations that will provide a detailed understanding of the channel role in cardiac electrophysiology.
Here, we outline elements of the model that simulate processes at different time and spatial scales, and highlight recent and potential future advances that could improve the modeling at each level. These elements are:
Structure: Ion channel atomic structure, homology modeling, and MD. Finding energetically stable states of the channel based on atomic structure, and the energy imparted by the Vm.
Gating: Modeling the channel opening and closing in response to changing energetics.
Current: Incorporating the gating transitions of the molecular model into a kinetic scheme that simulates the channel current.
Action potential (AP) and higher levels: Studying the role of molecular interactions in the context of the whole cell AP. Simulating electrocardiographic waveforms that reflect the organ level electrophysiology.
Structure
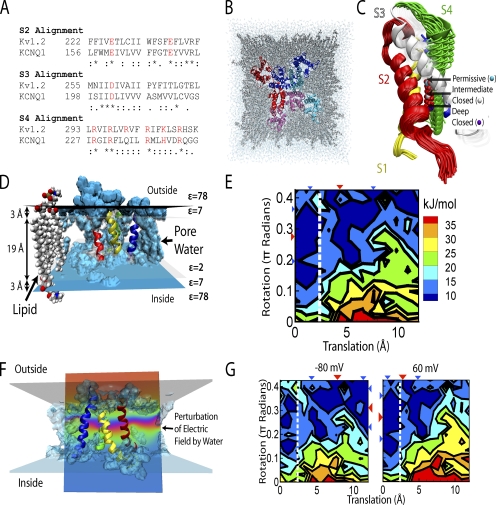
During the past decade, publication of crystal structures for several ion channels has enabled insight into their assembly, permeation, and gating. Several physiologically relevant channels are structurally homologous to the recently published rat Kv1.2 channel (Long et al., 2005a,b). In the absence of a crystal structure, this similarity has facilitated the creation of homology-based models of the IKs channel (Smith et al., 2007) and the bacterial sodium channel NaChBac (DeCaen et al., 2008), among others. Homology modeling is accomplished by aligning the sequence of the channel of interest with a known structure. An initial model can then be created by minimizing an objective function that includes the alignment, van der Waal’s forces, dihedral angles, bond lengths and torsions, and coulombic interactions. This task is accomplished with computer software, for example Modeller (Sali and Blundell, 1993). The alignment we used for KCNQ1 (the α subunit that together with KCNE1 forms IKs) with Kv1.2 serving as a template is shown in Fig. 1 A; it shows good agreement with respect to the positions of the charged residues. Note that four KCNQ1 subunits form a homomeric functioning channel, with KCNE1 modulating its conductance and kinetic properties (Silva and Rudy, 2005).
Figure 1.
KCNQ1 structure and gating energy landscape. (A) Kv1.2/KCNQ1 alignment. Symbols: *, identical; :, conserved; ., semi-conserved substitutions. Red residues are charged. (B) Top-down (extracellular) view of the all-atom system. Each of the four channel subunits (consisting of S1–S6 segments) is color coded. Pore regions (S5–S6) of one subunit interact with the adjacent subunit voltage-sensing region (S1–S4). (C) S4 translation and rotation. β carbon of R4 labeled with red beads shows motion. Arrows indicate stable configurations (labeled). Yellow, S1; red, S2; white, S3; green, S4; blue, S5. (D) Implicit membrane. Isocontours at dielectric constant ε = 78 (blue) and ε = 2 (transparent) show the transition region representing the lipid esters. Water (blue) can penetrate the protein and is represented by ε = 78. (E) Energy landscape at Vm = 0 mV and associated conformations. Increasing translation corresponds to movement toward the intracellular space; increasing rotation is counterclockwise as viewed from extracellular space. Left of dotted white line is permissive (open) state. (F) Vm. Water penetration causes significant perturbation of the electric field (ripples in the potential lines; arrow). (G) Energy landscape at positive (60 mV) and negative (−80 mV) Vm. Triangles are mean S4 position (red) and standard deviation (blue). Modified from Silva et al (2009).
Once a putative model is generated, it can be tested experimentally for accuracy using a variety of techniques, including metal bridges (Lainé et al., 2003; Webster et al., 2004; Campos et al., 2007; Lewis et al., 2008), disulphide bond formation (Liu et al., 2008; Chung et al., 2009), luminescence resonance energy transfer (Cha et al., 1999; Posson et al., 2005; Richardson et al., 2006), histidine scanning (Cymes et al., 2005), accessibility to methanethiosulphonate reagents (Liu et al., 1997; del Camino et al., 2000; del Camino and Yellen, 2001; Zhen et al., 2005) or fluorescent cysteine labeling (Zheng and Zagotta, 2000; Pathak et al., 2007), and paired amino acid substitutions (Tiwari-Woodruff et al., 1997; Chatelain et al., 2005; Kollewe et al., 2009). Because these experiments provide information regarding the spatial location of residues, they can be used effectively to define ion channel structures. Some examples that have been published recently include refinement of the Shaker channel open state (Lewis et al., 2008), sodium channel structure (DeCaen et al., 2008), the location of the KCNE1 β subunit in the IKs channel (Chung et al., 2009), and the structure of the K2P family of channels (Kollewe et al., 2009).
Additionally, MD simulations can be used to situate the model in a physiological environment, as we did for IKs with explicit lipid and water molecules surrounding the channel (Fig. 1 B). With this model in hand, we focused on the channel voltage-sensing regions, which include transmembrane segments S1–S4, and S5 of the neighboring subunit.
Channel gating
Transition rates between stable channel states are determined by the energy and availability of intermediate conformations. From a computational point of view, these transition rates can be estimated by performing many all-atom MD simulations in parallel for a given time duration. The proportion of these simulations that reach the target state, or a conformation with reasonable similarity, can then be used to estimate the transition rate (Voelz et al., 2010). This approach and others that accurately sample the conformational space pose a difficult challenge for ion channel simulations because of the long timescale of conformational transitions that is usually on the order of several milliseconds (or longer). Currently, long MD simulations typically represent no more than a microsecond in simulated time for a system of this size, and many of these simulations would be needed to probe a transition. It is also not guaranteed that any of the simulations would actually reach the target state. The orders of magnitude difference between possible MD-simulated time and protein transition time, even for fast channel transitions, makes proper sampling with current computational speeds and techniques extremely challenging.
On the horizon are several methodologies that could someday provide a means to alleviate this computational challenge. Most simply, computing speeds have been reliably improving over the last several decades. Also, specialized super computers for MD simulations, for example the Anton machine (Klepeis et al., 2009), have recently become available, allowing all-atom MD simulations on the order of milliseconds. New software approaches, with significant advancement, may also be of help. Some examples are accelerated MD (Hamelberg et al., 2004) and coarse-grained simulations (Scott et al., 2008) that are being developed to minimize the number of computing cycles necessary for long simulations. Finally, methodology has been recently published that uses the string method to find transition pathways in complex systems (Pan et al., 2008). It should be noted that even with substantial progress in these techniques, MD simulations will continue to be a significant limiting step in the modeling process.
In the absence of a full MD trajectory, the transition rates of the molecular motions that determine channel opening and closing can be inferred by calculating a multidimensional energy landscape (Sigg et al., 1999; Sigg and Bezanilla, 2003). This landscape is intended to reflect the stability of the protein as a function of reaction coordinates. Coordinates are chosen to track the progress of the channel between stable states. To describe gating of KCNQ1, we chose translation of S4 along its helical axis and rotation about it (Fig. 1 C). From a macroscopic perspective, possible degrees of freedom include tilting, horizontal translation, and bending of the helix in addition to the motion of other helices. Most simply, an estimate of the energy at a given point can be obtained from the electrostatic energy at each sampled point (Lecar et al., 2003), which is the approach we used by incorporating the voltage-sensing region into an implicit membrane (Fig. 1 D). The energy landscape is then constructed by assigning the electrostatic energy calculated from the Poisson–Boltzmann equation to each point along the reaction coordinates. Because we specified two degrees of freedom (translation and rotation; Fig. 1 C) a reduced (minimal) two-dimensional energy landscape resulted (Fig. 1 E). Future work will be necessary to determine how many degrees of freedom are required to accurately specify the voltage sensor movement.
The movement of positive charges on S4 across the membrane during channel opening and closing causes gating to depend on Vm. The change in the energy landscape, caused by the presence of Vm, can be computed using a modified Poisson–Boltzmann equation (Roux, 1997) (Fig. 1 F). During Vm application, the model correctly predicts favored occupancy in the open state at positive potentials and in the closed state at negative potentials (Fig. 1 G). The gating transitions associated with the movement of the S4 segment due to changes in Vm are then determined by the Smoluchowski equation, which specifies a transition rate reflecting the energy landscape in addition to a diffusive component (Sigg et al., 1999; Sigg and Bezanilla, 2003).
Current
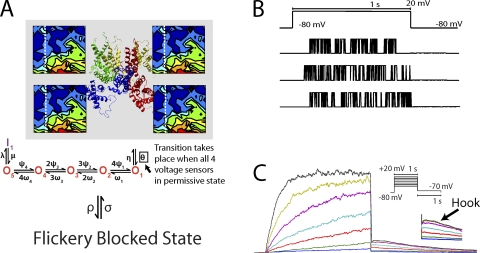
Once gating transitions have been successfully described, a model of a single KCNQ1 channel can be constructed by coupling the molecular transitions associated with the energy landscape to other known movements of the channel, such as inactivation (Silva et al., 2009). For simplicity, these transitions do not necessarily need to be described in as much detail. The main challenge at this level is representing the connectivity between transitions. For example, channel inactivation can be independent of or may occur preferentially from the open state of the channel. The final model (Fig. 2 A), which consists of the energy landscape and its linkage to additional channel states, can then be used to simulate the activity of a single channel via Monte Carlo simulation (Fig. 2 B).
Figure 2.
Markov model of KCNQ1 current. (A) KCNQ1 Markov model. Energy landscapes of four subunits determine channel residency in the permissive state, from which the channel can open. All four subunits in the permissive state (left of dotted white line) enable channel transitions to the open states (O1–O5). Inactivated state, I, is only accessible from O5. Flickery blocked state is available from any conformation. (B) Single-channel currents. Traces show random nature of opening. (C) Macroscopic KCNQ1 current is the sum of 1,000 single channels. A hook (arrow in inset that enlarges tail current), characteristic of KCNQ1, is observed upon repolarization and is caused by channel recovery from inactivation. Modified from Silva et al. (2009).
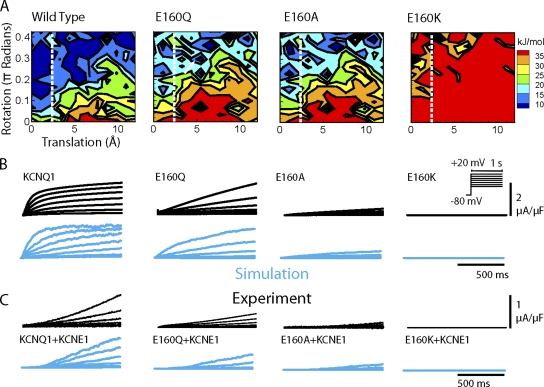
Using the single-channel model as a foundation, the macroscopic current can be obtained by computing the probability of residency in each channel state, or as the sum of activity in many (∼1,000) single channels (Fig. 2 C). Because the current through the channel links it to the other channels in a cell (via the changes the current causes in Vm), the model at this stage is ideal for validation. Experimentally, current kinetics can be readily observed in native cells with drugs that block the channel selectively, or in mammalian and frog oocyte expression systems. Ideally, a molecule-based model will be able to predict the effect of mutations on the current, providing insight into the details of the mutation effects. In our model, we compared several mutations that alter charge at residue E160 on S2 (Fig. 3). The wild-type channel carries a negative charge, glutamate (E). The naturally occurring mutation E160K, that causes the arrhythmogenic long QT syndrome type 1 (LQT1), replaces this negative charge with a fully positive charge. Intermediate to these are neutral residues E160Q and E160A. The model was able to predict slowing of current activation for E160Q and E160A, as observed experimentally (Fig. 3). For E160K, no current was detected in oocytes, so the conductance was set to 0 in the simulation.
Figure 3.
Mutation effects. (A) Energy landscapes for wild-type KCNQ1 and mutants E160Q, E160A, and E160K at Vm = 0 mV. (B) Comparison of macroscopic current between experiment (black) and simulation (cyan) for homomeric wild-type KCNQ1 and mutants (protocol in inset). (C) IKs (KCNQ1+KCNE1) is accurately simulated by slowing S4 movement. Mutations cause further slowing of activation in addition to the KCNE1 effect. Modified from Silva et al. (2009).
AP and beyond
As mentioned above, currents interact with each other at the cellular level through the changes they cause in Vm. This interrelationship is well characterized by the classical circuit model from Hodgkin and Huxley that represents the membrane as a capacitor in parallel with resistors that carry ionic current (Hodgkin and Huxley, 1952). To simulate cardiac ventricular cells, models based on the Luo–Rudy dynamic model (Luo and Rudy, 1994) are often adjusted to match the kinetics of a given species. Due to its similarity to the human AP, the canine ventricular AP has been extensively modeled. One of the most up to date models of the canine is the Hund–Rudy dynamic model (Hund and Rudy, 2004; Decker et al., 2009), which we used to compute the whole cell AP (Fig. 4 A).
Figure 4.
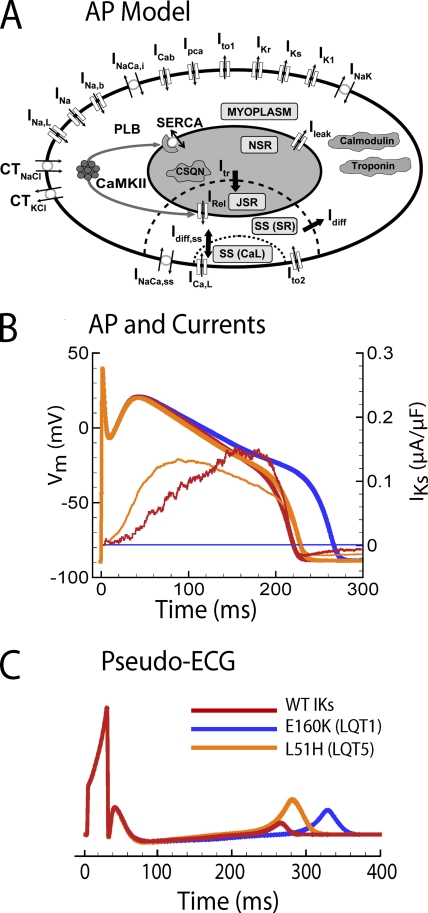
AP and electrocardiographic waveforms. Vm and IKs currents are color coded. Currents, with the exception of L51H, are in the presence of KCNE1. (A) Schematic of canine AP model (from Decker et al., 2009). (B) APs (thick lines) and currents (thin lines). Currents and APs are color coded: red, WT IKs; blue, E160K; orange, L51H. The mutations prolong the AP duration to varying degrees, depending on the severity of the mutation (degree of charge replacement at position 160 on S2). (C) Simulated pseudo ECGs. All mutations prolong the QT interval, the electrocardiographic marker of the long QT syndrome. For the naturally occurring mutations, the LQT1 mutation E160K (replacement of negative by positive charge) shows the greatest QT interval prolongation compared with mild prolongation by the LQT5 mutation L51H, consistent with the clinical phenotypes. Modified from Silva et al. (2009).
The primary benefit of incorporating a molecular model into AP simulations is to understand the role of the modeled kinetic properties in shaping the AP (Clancy and Rudy, 1999). As shown previously for IKs, the most pertinent parameters are its conductance and the rate of its activation relative to KCNQ1 alone (Silva and Rudy, 2005). Fig. 4 B shows that even though the conductance of KCNQ1 is only one fifth that of IKs, KCNQ1 still displays more current initially, due to its faster rate of activation (Fig. 3). The consequence of this interplay between conductance and activation rate can be seen in the difference in AP prolongation when comparing the LQT1 mutation E160K and the LQT5 mutation in KCNE1, L51H. E160K eliminates IKs current completely, whereas L51H prevents the conductance increase from KCNE1 and reduces conductance further by 20% to a total conductance that is 16% that of IKs. Still, because L51H mutant current activates faster, the consequence is only a moderately prolonged AP compared with a much greater prolongation of AP with E160K.
To extrapolate these findings to the body surface, we simulated pseudo-electrocardiographic waveforms. These simulations show QT interval prolongation with a more moderate phenotype for the LQT5 mutant compared with LQT1 (Fig. 4 C). Although this result is to be expected, the ability to simulate the electrocardiogram (ECG) starting from the molecular movement of ion channels shows promise for enabling interpretation of more complex waveforms for clinical diagnosis (Gima and Rudy, 2002). Akin to the accurate modeling of molecular transitions, the simulation of an accurate body-surface ECG will require substantial effort. Some current challenges are modeling accurately the spatial heterogeneity of cardiac APs in different regions of the heart, representing the distribution of non-excitable cells and gap junctions and the connectivity of cells, and the computational challenge of simulating the electrical activity of 5 × 109 myocytes (Adler and Costabel, 1975).
By formulating a comprehensive multi-scale model, we put forward testable hypotheses. For example, we assume that the folding of the KCNQ1 channel protein and the motion of its voltage sensor are similar to those of KV1.2, but the amount of charge moved per channel is less due to a missing charged residue. We also propose that the interaction with the KCNE1 β subunit slows the gating transitions, and this slowing counters an increase in the macroscopic conductance conferred by KCNE1. At the level of electrocardiographic waveforms, we hypothesize that the removal of negative charge at residue 160 on S2 will prolong the electrocardiographic QT interval. The model simulations can predict the physiological consequences of these hypotheses and propose experiments to test them. It is also hoped that by linking specific molecular changes to clinical phenotypes, the modeling approach will assist with individualized patient treatment.
Acknowledgments
We thank A. Kollewe for helpful comments on this manuscript.
This work was funded by National Institutes of Health National Heart Lung and Blood Institute Merit Award R37-HL33343, grant R01-HL49054 (to Y. Rudy), and fellowship F31-HL68318 (to J. Silva). Y. Rudy is the Fred Saigh Distinguished Professor at Washington University.
Footnotes
Abbreviations used in this paper:
- AP
- action potential
- ECG
- electrocardiogram
- IKs
- slow delayed rectifier K+ channel
- LQT1
- long QT syndrome type 1
- MD
- molecular dynamics
- Vm
- transmembrane potential
References
- Adler C.P., Costabel U. 1975. Cell number in human heart in atrophy, hypertrophy, and under the influence of cytostatics. Recent Adv. Stud. Cardiac Struct. Metab. 6:343–355 [PubMed] [Google Scholar]
- Campos F.V., Chanda B., Roux B., Bezanilla F. 2007. Two atomic constraints unambiguously position the S4 segment relative to S1 and S2 segments in the closed state of Shaker K channel. Proc. Natl. Acad. Sci. USA. 104:7904–7909 10.1073/pnas.0702638104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha A., Snyder G.E., Selvin P.R., Bezanilla F. 1999. Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature. 402:809–813 10.1038/45552 [DOI] [PubMed] [Google Scholar]
- Chatelain F.C., Alagem N., Xu Q., Pancaroglu R., Reuveny E., Minor D.L., Jr 2005. The pore helix dipole has a minor role in inward rectifier channel function. Neuron. 47:833–843 10.1016/j.neuron.2005.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D.Y., Chan P.J., Bankston J.R., Yang L., Liu G., Marx S.O., Karlin A., Kass R.S. 2009. Location of KCNE1 relative to KCNQ1 in the I(KS) potassium channel by disulfide cross-linking of substituted cysteines. Proc. Natl. Acad. Sci. USA. 106:743–748 10.1073/pnas.0811897106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy C.E., Rudy Y. 1999. Linking a genetic defect to its cellular phenotype in a cardiac arrhythmia. Nature. 400:566–569 10.1038/23034 [DOI] [PubMed] [Google Scholar]
- Cymes G.D., Ni Y., Grosman C. 2005. Probing ion-channel pores one proton at a time. Nature. 438:975–980 10.1038/nature04293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen P.G., Yarov-Yarovoy V., Zhao Y., Scheuer T., Catterall W.A. 2008. Disulfide locking a sodium channel voltage sensor reveals ion pair formation during activation. Proc. Natl. Acad. Sci. USA. 105:15142–15147 10.1073/pnas.0806486105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker K.F., Heijman J., Silva J.R., Hund T.J., Rudy Y. 2009. Properties and ionic mechanisms of action potential adaptation, restitution, and accommodation in canine epicardium. Am. J. Physiol. Heart Circ. Physiol. 296:H1017–H1026 10.1152/ajpheart.01216.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Camino D., Yellen G. 2001. Tight steric closure at the intracellular activation gate of a voltage-gated K(+) channel. Neuron. 32:649–656 10.1016/S0896-6273(01)00487-1 [DOI] [PubMed] [Google Scholar]
- del Camino D., Holmgren M., Liu Y., Yellen G. 2000. Blocker protection in the pore of a voltage-gated K+ channel and its structural implications. Nature. 403:321–325 10.1038/35002099 [DOI] [PubMed] [Google Scholar]
- Gima K., Rudy Y. 2002. Ionic current basis of electrocardiographic waveforms: a model study. Circ. Res. 90:889–896 10.1161/01.RES.0000016960.61087.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelberg D., Mongan J., McCammon J.A. 2004. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J. Chem. Phys. 120:11919–11929 10.1063/1.1755656 [DOI] [PubMed] [Google Scholar]
- Hodgkin A.L., Huxley A.F. 1952. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 117:500–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hund T.J., Rudy Y. 2004. Rate dependence and regulation of action potential and calcium transient in a canine cardiac ventricular cell model. Circulation. 110:3168–3174 10.1161/01.CIR.0000147231.69595.D3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepeis J.L., Lindorff-Larsen K., Dror R.O., Shaw D.E. 2009. Long-timescale molecular dynamics simulations of protein structure and function. Curr. Opin. Struct. Biol. 19:120–127 10.1016/j.sbi.2009.03.004 [DOI] [PubMed] [Google Scholar]
- Kollewe A., Lau A.Y., Sullivan A., Roux Benoît, Goldstein S.A. 2009. A structural model for K2P potassium channels based on 23 pairs of interacting sites and continuum electrostatics. J. Gen. Physiol. 134:53–68 10.1085/jgp.200910235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lainé M., Lin M.C., Bannister J.P., Silverman W.R., Mock A.F., Roux B., Papazian D.M. 2003. Atomic proximity between S4 segment and pore domain in Shaker potassium channels. Neuron. 39:467–481 10.1016/S0896-6273(03)00468-9 [DOI] [PubMed] [Google Scholar]
- Lecar H., Larsson H.P., Grabe M. 2003. Electrostatic model of S4 motion in voltage-gated ion channels. Biophys. J. 85:2854–2864 10.1016/S0006-3495(03)74708-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A., Jogini V., Blachowicz L., Lainé M., Roux B. 2008. Atomic constraints between the voltage sensor and the pore domain in a voltage-gated K+ channel of known structure. J. Gen. Physiol. 131:549–561 10.1085/jgp.200809962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Zakharov S.I., Yang L., Deng S.X., Landry D.W., Karlin A., Marx S.O. 2008. Position and role of the BK channel α subunit S0 helix inferred from disulfide crosslinking. J. Gen. Physiol. 131:537–548 10.1085/jgp.200809968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Holmgren M., Jurman M.E., Yellen G. 1997. Gated access to the pore of a voltage-dependent K+ channel. Neuron. 19:175–184 10.1016/S0896-6273(00)80357-8 [DOI] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B., Mackinnon R. 2005a. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903 10.1126/science.1116269 [DOI] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B., Mackinnon R. 2005b. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 309:903–908 10.1126/science.1116270 [DOI] [PubMed] [Google Scholar]
- Luo C.H., Rudy Y. 1994. A dynamic model of the cardiac ventricular action potential. I. Simulations of ionic currents and concentration changes. Circ. Res. 74:1071–1096 [DOI] [PubMed] [Google Scholar]
- Pan A.C., Sezer D., Roux B. 2008. Finding transition pathways using the string method with swarms of trajectories. J. Phys. Chem. B. 112:3432–3440 10.1021/jp0777059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak M.M., Yarov-Yarovoy V., Agarwal G., Roux B., Barth P., Kohout S., Tombola F., Isacoff E.Y. 2007. Closing in on the resting state of the Shaker K(+) channel. Neuron. 56:124–140 10.1016/j.neuron.2007.09.023 [DOI] [PubMed] [Google Scholar]
- Posson D.J., Ge P., Miller C., Bezanilla F., Selvin P.R. 2005. Small vertical movement of a K+ channel voltage sensor measured with luminescence energy transfer. Nature. 436:848–851 10.1038/nature03819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J., Blunck R., Ge P., Selvin P.R., Bezanilla F., Papazian D.M., Correa A.M. 2006. Distance measurements reveal a common topology of prokaryotic voltage-gated ion channels in the lipid bilayer. Proc. Natl. Acad. Sci. USA. 103:15865–15870 10.1073/pnas.0607532103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B. 1997. Influence of the membrane potential on the free energy of an intrinsic protein. Biophys. J. 73:2980–2989 10.1016/S0006-3495(97)78327-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A., Blundell T.L. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- Scott K.A., Bond P.J., Ivetac A., Chetwynd A.P., Khalid S., Sansom M.S. 2008. Coarse-grained MD simulations of membrane protein-bilayer self-assembly. Structure. 16:621–630 10.1016/j.str.2008.01.014 [DOI] [PubMed] [Google Scholar]
- Sigg D., Bezanilla F. 2003. A physical model of potassium channel activation: from energy landscape to gating kinetics. Biophys. J. 84:3703–3716 10.1016/S0006-3495(03)75099-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigg D., Qian H., Bezanilla F. 1999. Kramers’ diffusion theory applied to gating kinetics of voltage-dependent ion channels. Biophys. J. 76:782–803 10.1016/S0006-3495(99)77243-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Rudy Y. 2005. Subunit interaction determines IKs participation in cardiac repolarization and repolarization reserve. Circulation. 112:1384–1391 10.1161/CIRCULATIONAHA.105.543306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J.R., Pan H., Wu D., Nekouzadeh A., Decker K.F., Cui J., Baker N.A., Sept D., Rudy Y. 2009. A multiscale model linking ion-channel molecular dynamics and electrostatics to the cardiac action potential. Proc. Natl. Acad. Sci. USA. 106:11102–11106 10.1073/pnas.0904505106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.A., Vanoye C.G., George A.L., Jr., Meiler J., Sanders C.R. 2007. Structural models for the KCNQ1 voltage-gated potassium channel. Biochemistry. 46:14141–14152 10.1021/bi701597s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari-Woodruff S.K., Schulteis C.T., Mock A.F., Papazian D.M. 1997. Electrostatic interactions between transmembrane segments mediate folding of Shaker K+ channel subunits. Biophys. J. 72:1489–1500 10.1016/S0006-3495(97)78797-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelz V.A., Bowman G.R., Beauchamp K., Pande V.S. 2010. Molecular simulation of ab initio protein folding for a millisecond folder NTL9(1-39). J. Am. Chem. Soc. 132:1526–1528 10.1021/ja9090353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster S.M., Del Camino D., Dekker J.P., Yellen G. 2004. Intracellular gate opening in Shaker K+ channels defined by high-affinity metal bridges. Nature. 428:864–868 10.1038/nature02468 [DOI] [PubMed] [Google Scholar]
- Zhen X.G., Xie C., Fitzmaurice A., Schoonover C.E., Orenstein E.T., Yang J. 2005. Functional architecture of the inner pore of a voltage-gated Ca2+ channel. J. Gen. Physiol. 126:193–204 10.1085/jgp.200509292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Zagotta W.N. 2000. Gating rearrangements in cyclic nucleotide-gated channels revealed by patch-clamp fluorometry. Neuron. 28:369–374 10.1016/S0896-6273(00)00117-3 [DOI] [PubMed] [Google Scholar]