Abstract
The herpes simplex virus type 1 (HSV-1) latency-associated transcript (LAT) is abundantly expressed in latently infected sensory neurons. In small animal models of infection, expression of the first 1.5 kb of LAT coding sequences is necessary and sufficient for wild-type reactivation from latency. The ability of LAT to inhibit apoptosis is important for reactivation from latency. Within the first 1.5 kb of LAT coding sequences and LAT promoter sequences, additional transcripts have been identified. For example, the anti-sense to LAT transcript (AL) is expressed in the opposite direction to LAT from the 5′ end of LAT and LAT promoter sequences. In addition, the upstream of LAT (UOL) transcript is expressed in the LAT direction from sequences in the LAT promoter. Further examination of the first 1.5 kb of LAT coding sequences revealed two small ORFs that are anti-sense with respect to LAT (AL2 and AL3). A transcript spanning AL3 was detected in productively infected cells, mouse neuroblastoma cells stably expressing LAT and trigeminal ganglia (TG) of latently infected mice. Peptide-specific IgG directed against AL3 specifically recognized a protein migrating near 15 kDa in cells stably transfected with LAT, mouse neuroblastoma cells transfected with a plasmid containing the AL3 ORF and TG of latently infected mice. The inability to detect the AL3 protein during productive infection may have been because the 5′ terminus of the AL3 transcript was downstream of the first in-frame methionine of the AL3 ORF during productive infection.
INTRODUCTION
Most adults harbour latent herpes simplex virus type 1 (HSV-1) (Nahmias & Roizman, 1973; Whitley, 1997) in sensory neurons located in trigeminal ganglia (TG) or sacral dorsal root ganglia (Jones, 1998; Wagner & Bloom, 1997). Acute infection is initiated in the mucocutaneous epithelium. Despite a vigorous immune response during acute infection, HSV-1 establishes latency in sensory neurons. Recurrent ocular HSV-1 remains the leading cause of infectious corneal blindness in industrialized nations (Nesburn, 1983). HSV-1-induced encephalitis is a severe form of focal necrotizing encephalitis that affects at least 2000 individuals each year in the USA (Gesser & Koo, 1997; Lohr et al., 1990; Whitley, 1991, 1997).
The latency-associated transcript (LAT) is abundantly transcribed in latently infected neurons (Croen et al., 1987; Deatly et al., 1987, 1988; Krause et al., 1988; Mitchell et al., 1990; Rock et al., 1987; Stevens et al., 1987; Wagner et al., 1988a, b). The primary LAT transcript is approximately 8.3 kb (Deatly et al., 1988; Rock et al., 1987; Zwaagstra et al., 1990), and splicing yields a stable 2 kb LAT and an unstable 6.3 kb LAT. Expression of LAT enhances the latency–reactivation cycle in small animal models (reviewed by Jones, 1998, 2003). For example, the HSV-1 McKrae strain is frequently shed in tears of infected rabbits because of spontaneous reactivation (Perng et al., 1994, 1996a, b, c, 1999). A LAT deletion mutant (dLAT2903) has a low spontaneous reactivation phenotype in rabbits (Perng et al., 1994, 1996a) and a low explant-induced reactivation phenotype in mice (Perng et al., 2001). dLAT2903 contains a deletion (−161 to +1667) that prevents LAT expression at detectable levels (Perng et al., 1994, 1996a). The reactivation phenotype of dLAT2903 is restored to wild-type levels when the first 1.5 kb of LAT coding sequences (LAT nt 1–1499) and LAT promoter is inserted between UL37 and UL38 of dLAT2903 (Perng et al., 1996b). LAT interferes with apoptosis in transiently transfected cells (Ahmed et al., 2002; Henderson et al., 2002; Inman et al., 2001; Jin et al., 2003; Perng et al., 2000; Peng et al., 2003) and promotes neuronal survival in TG of infected rabbits (Perng et al., 2000) and mice (Ahmed et al., 2002; Branco & Fraser, 2005). Inhibiting apoptosis appears to be the most important function of LAT with respect to enhancing the reactivation phenotype, because three anti-apoptosis genes (Jin et al., 2005, 2008; Mott et al., 2003; Perng et al., 2002b) can each restore the wild-type reactivation phenotype to a LAT null mutant.
LAT coding sequences encode several transcripts in addition to LAT. For example, novel transcripts within the LAT promoter region have been reported (Singh & Wagner, 1993). More recently, a transcript and protein, upstream of LAT (UOL), was identified that is encoded within the LAT promoter regulatory region (Naito et al., 2005). Deletion of UOL does not affect the spontaneous reactivation phenotype in rabbits (Chan et al., 2006). Another transcript, antisense to LAT (AL), is expressed within the first 1.5 kb of LAT coding sequences and the start site of the LAT promoter, and appears to encode a protein (Perng et al., 2002a) (see Fig. 1b for location of UOL and AL).
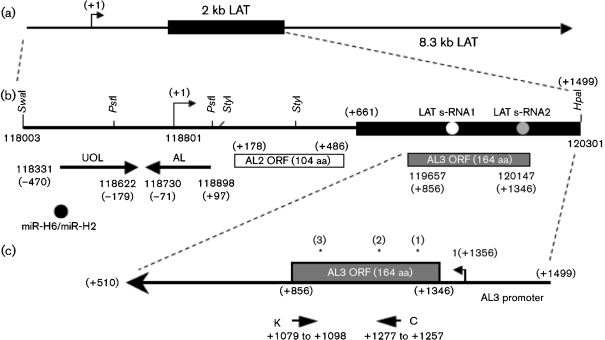
Fig. 1.
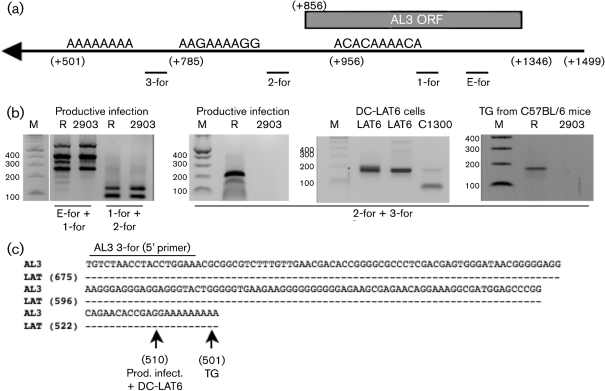
Schematic diagram of the HSV-1 genome and the LAT locus. (a) Schematic of the 8.3 kb primary LAT. The black rectangle represents the stable 2 kb LAT intron. The arrow at +1 denotes the start of LAT transcription. (b) Expanded view of the first 1.5 kb of the LAT coding sequence and the region upstream of this. The LAT promoter, the start of LAT transcription (arrow, +1; nt 118801) and the first 1.5 kb of the LAT coding sequence are shown. Restriction enzyme sites are shown for reference. The positions of the UOL transcript, AL transcript, the two LAT small RNAs (s-RNAs) located within the first 1.5 kb of the LAT coding sequences (Peng et al., 2008) and ORFs located on the opposite strand to LAT (AL2 and AL3) are shown. Nucleotide positions relative to the start of LAT transcription are shown in parentheses. Numbers not in parentheses represent HSV-1 nucleotide positions. (c) Schematic of the AL3 gene. The ORF encoding the putative AL3 protein is denoted by a grey rectangle. The putative AL3 promoter is between LAT nt +1356 and +1499. Primers K and C (Supplementary Table S1) were used for amplifying AL3 RNA. Asterisks above the AL3 ORF denote the positions of peptides used to generate AL3-specific antiserum.
In this study, we identified a novel transcript that is encoded within the first 1.5 kb of the LAT coding sequence (AL3). Peptide-specific IgG directed against the AL3 ORF recognized a protein in cells stably transfected with a large LAT fragment or a plasmid containing the AL3 ORF, and in trigeminal ganglionic neurons of infected mice. We suggest that AL3 protein expression was not detected during productive infection because the 5′ terminus of the AL3 transcript was downstream of the first initiating ATG of the AL3 ORF.
METHODS
Cells and viruses.
C1300 mouse neuroblastoma cells stably transfected with LAT sequences, DC-LAT6, DC-ΔLAT35 and DC-ΔLAT311, have been described previously (Carpenter et al., 2007). DC-LAT6 cells contain a NotI–NotI restriction fragment of HSV-1, which contains 361 nt of the LAT promoter followed by the first 3225 nt of LAT. DC-LAT6 cells express abundant levels of the 2 kb LAT stable intron (Carpenter et al., 2007). DC-ΔLAT35 and DC-ΔLAT311 cell lines contain the same NotI–NotI restriction fragment, but lack the LAT TATA box and start site of LAT transcription (−130 to +64). DC-ΔLAT35 and DC-ΔLAT311 cells express little or no 2 kb stable LAT .
C1300 cells, DC-LAT6, DC-ΔLAT35, DC-ΔLAT311, neuro-2A (mouse neuroblastoma) or rabbit skin (RS) cells were plated in 100 mm plastic dishes in Earl's modified Eagle's medium supplemented with 10 % fetal bovine serum, penicillin (10 U ml−1) and streptomycin (100 μg ml−1).
The HSV-1 McKrae strain, dLAT2903, dLAT2903R, LAT3.3A and LAT2.6A have been described previously (Perng et al., 1994, 1996a, 2000). RS cells were used for preparing virus stocks.
RNA preparation and RT-PCR.
Cultured cells or TG were lysed using lysis/binding solution (mirVana kit, Ambion) and total RNA was prepared using this kit. RNA samples were treated with amplification grade DNase I (Invitrogen). Reverse transcription (RT) was performed using SuperScript III reverse transcriptase (Invitrogen) and the designated primer (see Supplementary Table S1, available in JGV Online). PCR was performed using GoTaq DNA polymerase (Promega) and primers K and C. PCR was initiated at 90 °C for 10 min, followed by 30 cycles of 95 °C for 45 s, 56 °C for 45 s, and 72 °C for 45 s, and extension at 72 °C for 10 min. Southern blots were performed on RT-PCR products with oligonucleotide L or M (Supplementary Table S1).
ICP0 and ICP27 transcripts were detected as follows: DNase I-treated RNA samples were used to synthesize single-stranded cDNA using an Oligo-dT primer (Invitrogen). PCR was performed using GoTaq DNA Polymerase (Promega). Primers for ICP0 were: forward (5′–ACAGACCCCCAACACCTACA–3′), and reverse (5′–GCGTATGAGTCAGTGGGGA–3′). Primers for ICP27 were: forward (5′–CCCTTTCTCCAGTGCTACCTGAA–3′), and reverse (5′–GTGCGTGTGTAGGATTTCGAT–3′). PCR was initiated at 90 °C for 10 min, followed by 30 cycles of 95 °C for 30 s, 57 °C for 30 s, and 72 °C for 30 s. Extension was at 72 °C for 10 min.
5′- and 3′-random amplification of cDNA ends (RACE).
The 5′ and 3′ ends of AL3 RNA were identified using FirstChoice RLM-RACE kit (Ambion) according to the manufacturer's instructions. For the 5′-RACE protocol, cDNA was synthesized with primer K (Supplementary Table S1) and two rounds of PCR conducted with the supplied 5′-RACE outer and inner primers. AL3-specific primers (K and M) were used to amplify the 5′ terminus of AL3 mRNA. For the 3′-RACE protocol, cDNA synthesis was performed with the supplied 3′-RACE adaptor. PCR amplification was achieved using the 3′-RACE inner and outer primers. AL3-specific primers used for 3′-RACE were 2-for and 3-for primers (Supplementary Table S1). Amplified products were cloned using the TOPO cloning kit (Invitrogen) and the inserts were sequenced.
Generation of antiserum directed against the AL3 ORF.
Three antigenic peptides within the AL3 ORF were used to generate peptide-specific antibodies: (i) VTNPHPGMLGGMKEGGR (aa 5–21), (ii) GRGVGVQRHAHPRGQVGR (aa 29–46) and (iii) PPQAVRPKHREPG (aa 102–114). All three peptides were co-injected into two rabbits (0.5 mg per injection). Three additional injections (0.5 mg per injection) of all peptides were performed. Rabbits were bled 2 weeks after each antigen injection. Two weeks after the final antigen injection, rabbits were euthanized and a terminal bleed was performed. Serum from all bleeds (both rabbits) was pooled, IgG was purified and peptide-specific IgG was affinity purified. Peptides were prepared by ABR Affinity Bioreagents and peptide-specific antiserum was from Golden. This strategy was used to enhance the chances of obtaining a high-affinity IgG fraction directed against AL3 ORF.
Cloning and expression of AL3 ORF.
Sequences containing the AL3 ORF were synthesized by Integrated DNA Technology so the ORF was optimized for expression in mammals, and contained BamHI and HindIII restriction sites. The AL3 ORF was cloned into the unique BamHI–HindIII sites in pCMV-Tag2A, pCMV-Tag2B or pCMV-Tag2C (Stratagene) to obtain the respective AL3 sequences in all three reading frames. The pCMV-Tag vectors contain a FLAG epitope at the N terminus of an ORF inserted into the multiple cloning site. The pCMV-Tag2B construct contained the AL3 ORF in the correct reading frame.
Western blot analysis of AL3 protein expression.
Total cell lysate was prepared from the designated cell lines. Cells were scraped into PBS, pelleted at 3000 g for 10 min and lysed by adding CHAPS cell extract buffer (Cell Signalling catalogue no. 9852) to the cell pellet. Freeze–thawing was performed three times (−70 to 37 °C) to ensure lysis. Cell debris was removed by centrifugation at 18 000 g for 20 min in a microcentrifuge. A 500 μg aliquot of protein of the cell lysate was loaded in each lane of a 15 % SDS-PAGE gel and Western blot analysis was performed as described previously (Meyer et al., 2007a, b). The primary antibody was diluted from 10 to 1 mg ml−1 in double distilled water and then diluted to a final concentration of 1 : 2000 in 10 ml TBS [2.42 g Tris base, 8 g NaCl (l water)−1, pH 7.6] plus 0.1 % Tween-20 with 5 % non-fat dried milk (Cell Signalling catalogue no. 9999) (TBS/T). Membranes and primary antibody were incubated with gentle agitation overnight at 4 °C. Membranes were washed three times for 15 min in 1×TBS/T at room temperature and then incubated with horseradish peroxidase-conjugated secondary anti-rabbit antibody (1 : 2000) (Amersham NA 934) with gentle agitation for 1 h at room temperature. Membranes were finally washed three times for 15 min in 1× TBS/T, and antigen–antibody complexes were detected using the enhanced chemiluminescence detection kit (Amersham; RPN 2106). β-Actin antibody was used as a loading control (Santa Cruz).
Infection of mice and immunohistochemistry.
Eight- to ten-week-old Swiss Webster or C57BL/6 female mice (Jackson Laboratories) were infected with 2×105 p.f.u. per eye of dLAT2903 (LAT−) or dLAT2903R (LAT+). Infections were done without scarification, as described previously (Jones et al., 2005; Perng et al., 1994, 1996c, 2001). TG were removed, formalin fixed and embedded in paraffin and thin sections were cut. Latent TG were prepared at ≥30 days after infection. Immunohistochemistry was performed using the AL3-specific IgG as described previously (Meyer et al., 2007a, b; Winkler et al., 2000, 2002).
RESULTS
Detection of AL3 transcripts in cultured cells productively infected with HSV-1
Examination of the first 1.5 kb of LAT coding sequences revealed two additional small ORFs that, like AL, are anti-sense with respect to LAT (AL2 and AL3; Fig. 1b, c). We focused on AL3 because sequences spanning the putative AL3 ORF are crucial for the anti-apoptosis activity of LAT (Inman et al., 2001; Jin et al., 2003; Peng et al., 2004) and wild-type spontaneous reactivation from latency phenotype (Inman et al., 2001; Perng et al., 1996a, 2001).
To test whether a transcript spanning the AL3 ORF was expressed during productive infection, cDNA synthesis was primed using the K primer (for location of K, see Fig. 1c and Supplementary Table S1) and PCR was performed. RS cells were mock-infected or infected with dLAT2903 (LAT-null mutant) or its marker-rescued virus dLAT2903R, which expresses wild-type levels of LAT (Perng et al., 1996a). An amplified band of the expected size for the AL3 transcript was detected at 8 and 16 h after infection with dLAT2903R (Fig. 2a, lanes marked R). The AL3 amplified product was not detected after infection with dLAT2903 or in mock-infected cells (Fig. 2a). As judged by β-actin levels, similar levels of RNA were used for the RT-PCR.
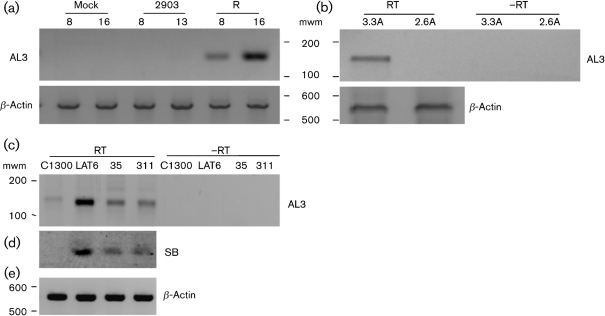
Fig. 2.
Analysis of AL3 transcription in cultured cells. (a) RS cells were infected with dLAT2903 (2903 lanes) or dLAT2903R rescued virus (R lanes) (m.o.i.=1). Cells were lysed at 8 or 16 h after infection and total RNA was prepared. DNase I-treated RNA samples (1 μg) were used to synthesize single-stranded cDNA using the AL3-specific primer K (see Fig. 1c and Supplementary Table S1 for position and sequence of K). PCR was performed using K as reverse primer and C as forward primer. (b) RS cells were infected with the LAT2.6A or LAT3.3A recombinant virus (m.o.i.=2). Total RNA was prepared 16 h after infection and RT-PCR was performed as in (a). −RT, Reverse transcriptase was not included in the reaction. (c, d) The indicated cells were lysed and total RNA was prepared. RT-PCR and PCR were performed as in (a). To confirm PCR results (c), a Southern blot was performed using internal Probe M (d). (e) β-Actin RT-PCR was used as a loading control for (a–d). Molecular mass markers (mwm) are from a 100 bp ladder (Invitrogen). Images of autoradiographs were scanned using a CanoScan 8600 F and the scanned image was saved as a JPEG file. Images were prepared from gels using a Molecular Imager FX (Bio-Rad) and these images were saved as JPEG files. The brightness of the respective images was adjusted to match the other panels.
To test whether the first 1.5 kb of LAT coding sequences was sufficient for AL3 transcription; RS cells were infected with LAT3.3A or LAT2.6A viruses. LAT3.3A contains the first 1.8 kb of the LAT promoter and the first 1.5 kb of LAT coding sequences between the UL37 and UL38 genes of dLAT2903 (Perng et al., 1996a). LAT3.3A makes no LAT RNA from the long repeats, but expresses a 1.5 kb LAT RNA from the ectopic insert. LAT2.6A contains the 1.8 kb LAT promoter and first 811 bp of LAT in the same location as dLAT2903 (Inman et al., 2001). LAT2.6A does not contain AL3 ORF sequences or the 5′ end of AL3 mRNA and should not express AL3 RNA. An AL3-specific cDNA was detected in RS cells infected with LAT3.3A for 16 h (Fig 2b, 3.3A lane) but not in RS cells infected with LAT2.6A (2.6A lane). This study indicated that the first 1.5 kb of LAT coding sequences contain the necessary sequences for expressing the AL3 transcript in productively infected cells.
AL3 is expressed in cells stably transfected with LAT
Mouse neuroblastoma cells (C1300) stably transfected with a NotI–NotI LAT restriction fragment express high levels of the stable 2 kb LAT fragment (DC-LAT6 cells), and these cells are resistant to cold-shock-induced apoptosis (Carpenter et al., 2007). Conversely, cell lines stably transfected with a PstI–PstI deletion in the LAT promoter (DC-ΔLAT311 and DC-ΔLAT35) express little detectable stable 2 kb LAT, and these cells undergo cold-shock-induced apoptosis with similar efficiency to the parental C1300 cells. An AL3-specific amplified cDNA was detected in DC-LAT6 cells, DC-LATΔ35 cells and DC-LATΔ311 cells (Fig. 2c). A faint, slightly larger amplified band was detected in parental C1300 cells, but was not detected following hybridization with an internal probe, oligonucleotide M (Fig. 2d). These results suggested that AL3 expression did not absolutely require LAT transcription or the intact LAT promoter.
Detection of AL3 transcription in TG of infected mice
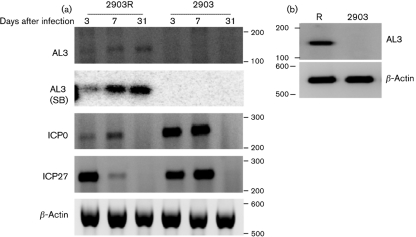
To test whether AL3 transcription occurred in TG of infected mice, female Swiss Webster mice were infected with HSV-1 (dLAT2903R or dLAT2903) and RT-PCR was performed. An AL3-specific cDNA amplified product was detected at 3 and 7 days after infection with dLAT2903R (Fig. 3a, AL3 panel, 2903R lanes). Southern blot analysis using an internal probe (Supplementary Table S1) confirmed the PCR results at 3 and 7 days after infection [Fig. 3a, AL3 (SB) panel]. As expected, AL3-specific amplified cDNA products were not observed when mice were infected with dLAT2903 (Fig. 3a, 2903 lanes). ICP0 and ICP27 transcripts (immediate-early expression) were detected at 3 and 7 days after infection. The intensities of the ICP0 and ICP27 amplified cDNA products were higher at 7 days after infection following infection with dLAT2903, which is consistent with previous studies (Chen et al., 1997; Garber et al., 1997).
Fig. 3.
Analysis of AL3 transcription in TG of infected mice. (a) Swiss Webster mice were infected with dLAT2903 (2903) or dLAT2903R (2903R). Total RNA was prepared from TG at 3, 7 or 31 days after infection. DNase I-treated RNA samples (1 μg) were used to synthesize single-stranded cDNA using the K primer. PCR was performed using K as the reverse primer and C as the forward primer. Southern blots were performed using internal primer L. ICP0 and ICP27 RNA expression was examined as described in Methods. β-Actin RT-PCR was used as a loading control. (b) C57BL/6 mice were infected with dLAT2903 (2903) or dLAT2903R (R) and AL3 expression was examined. β-Actin RT-PCR was used as a loading control. Molecular mass markers are from a 100 bp ladder (Invitrogen). Images were prepared as described in Fig. 2.
At 31 days after infection (latent infection), AL3, but not ICP0 or ICP27, was detected in total RNA prepared from TG of Swiss Webster (Fig. 3a, 2903R lanes) or C57BL/6 (Fig. 3b, lane R) mice infected with dLAT2903R. The AL3 transcript was not detected in TG of mice infected with dLAT2903 (Fig. 3b, 2903 lanes). Similar levels of β-actin were detected in all samples. These results indicate that relative to ICP0 and ICP27, the AL3 transcript was readily detected in TG of latently infected mice.
Mapping the 5′ terminus of the AL3 transcript
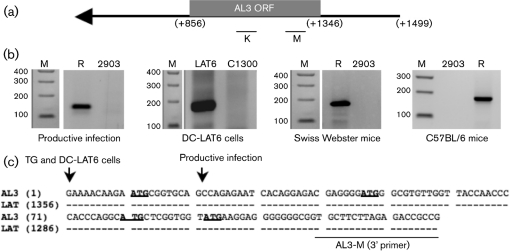
The 5′ end of the AL3 transcript was identified by 5′ RACE using the internal primer K to prime cDNA synthesis. Primer K was used for the primary PCR and then primer M was used for nested PCR (Fig. 4a and Supplementary Table S1). Total RNA was prepared from: (i) RS cells infected with dLAT2903R or dLAT2903 (m.o.i. of 2) for 16 h, (ii) DC-LAT6 cells or (iii) TG prepared from latently infected mice (31 days after infection). These primers have consistently yielded a single band from the respective cell lines or TG (Fig. 4b). The AL3-specific band was excised from the samples, cloned into a vector and then sequenced. At least five different clones were sequenced for each sample (productive infection, DC-LAT 6 cells or TG from latently infected mice). The 5′ end of the AL3 RNA prepared from productive infection was mapped to nt 1337 using the HSV-1 LAT numbering system (Fig. 4c). In contrast, the 5′ end of the AL3 RNA mapped to nt 1356 when RNA was prepared from DC-LAT6 cells or TG of latently infected mice. The 5′ end of the AL3 transcript in productively infected RS cells appeared to begin 20 nt downstream of the 5′ end of the AL3 transcript in latently infected TG (Fig. 4c). The initiating AL3 ORF ATG is located at LAT nt 1346. Thus, in latently infected TG and in DC-LAT6 cells, the AL3 transcript began 10 nt upstream of the first in-frame methionine of the AL3 ORF. However, in productively infected RS cells, the AL3 transcript began 10 nt downstream of the initiating ATG.
Fig. 4.
5′-RACE analysis of the AL3 transcript. (a) Schematic representation of the AL3 ORF and primers used for 5′-RACE. K and M were used in PCR and nested PCR to map the 5′ end of AL3. (b) Total RNA were prepared from RS cells infected with dLAT2903R (R), dLAT2903 (2903), cells stably transfected with LAT (DC-LAT6; LAT6) or TG extracted from infected mice (Swiss Webster mice or C57BL/6 mice) for 31 days. 5′ RACE was performed as described in Methods. Lane M is a 100 bp DNA Ladder (Invitrogen). (c) Summary of AL3 5′-RACE results. An arrow denotes the AL3 5′ terminus mapped by 5′ RACE. AL3 RNA indicates the DNA sequence corresponding to the AL3 RNA sequence. Dashes in the LAT sequence indicate identity to the AL3 sequence. ATG sequences in bold type are in-frame methionine residues within the AL3 ORF. AL3-M is the primer sequence used in the 5′-RACE analysis. Images were prepared as described in Fig. 2.
Mapping the 3′ end of AL3 RNA
To map the 3′ terminus of the AL3 transcript, 3′ RACE was performed. Two putative polyA addition sites are located downstream of the AL3 ORF and one is located within the AL3 ORF (Fig. 5a). Four primers (3-for, 2-for, 1-for and E-for) were tested. The 3-for primer gave reproducible results using samples prepared from TG of latently infected mice, productive infection and DC-LAT6 cells (Fig. 5b). The E-for and 1-for or 1-for and 2-for primers yielded bands in samples prepared from cells infected with dLAT2903, a mutant that lacks AL3 coding sequences (Fig. 5b, left panel). Thus, the E-for and 1-for or 1-for and 2-for primers were not useful because they annealed to cellular sequences or other viral transcripts. The 3′ RACE PCR products were excised, cloned into a PCR cloning vector and the inserts were sequenced (at least five separate clones were sequenced for each sample). These results consistently demonstrated that the 3′ terminus of the AL3 transcript in TG of latently infected mice was at position 501 using the numbering system for LAT (Fig. 5c). During productive infection and in DC-LAT6 cells, the 3′ terminus was located at position 510.
Fig. 5.
3′-RACE analysis of the AL3 transcript. (a) Schematic representation of the AL3 ORF, primers used in 3′ RACE (Supplementary Table S1) and positions of putative polyA addition signals. The position of the stop codon of the AL3 ORF is indicated by +856. (b) Total RNA was prepared from RS cells infected with the designated virus (R, dLAT2903R; 2903, dLAT2903), DC-LAT6 cells or TG of latently infected mice (C57/BL/6 mice). 3′-RACE was performed according to the manufacturer's recommendations (Ambion). Specific primers used for PCR and nested PCR are shown. Lane M is a 100 bp DNA Ladder (Invitrogen). (c) Summary of AL3 3′-RACE results. RT-PCR products were cloned and sequenced. AL3 indicates DNA sequences that correspond to the AL3 cDNA sequence. Dashes in the LAT sequence indicate identity to the AL3 sequence. The position of the AL3 3-for primer used for mapping the 3′ terminus of the AL3 transcript is indicated. Arrows denote the 3′ terminus of the AL3 transcript. Images were prepared from gels as described in Fig. 2.
Detection of an AL3 protein in cultured cells
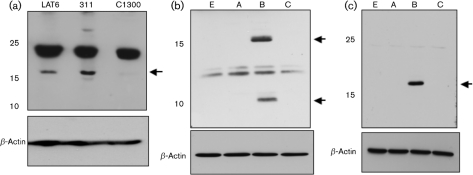
The affinity-purified peptide-specific antiserum directed against AL3 detected a protein migrating slightly higher than the 15 kDa marker in DC-LAT6 and DC-ΔLAT311 cells (Fig. 6a, LAT6 and 311 lanes) but not in parental C1300 cells. The predicted molecular mass of the AL3 ORF is 17.4 kDa, which was consistent with the AL3-specific band detected in the stably transfected cell lines. The AL3-specific antiserum also reacted non-specifically with a protein migrating near the 25 kDa marker (Fig. 6a). As expected, β-actin protein levels were similar for the three cell lines.
Fig. 6.
Detection of AL3 protein. (a) Cell lysate from DC-LAT6 (LAT6), DC-ΔLAT3-11 (311) or C1300 (C1300) cells was analysed by Western blot analysis using the AL3 antibody (top; 350 μg protein per lane) or β-actin antibody (bottom; 100 μg protein per lane). (b) Neuro-2A cells were transfected with the pCMV-Tag2 expression vectors containing AL3 ORF (5 μg plasmid DNA). Western blot analysis was performed using the AL3-specific antiserum (top; 350 μg protein per lane) or β-actin antibody (bottom; 100 μg protein per lane). Lane A was transfected with pCMV-Tag2A vector containing AL3 ORF. Lane B was transfected with pCMV-Tag2B vector containing AL3 ORF. Lane C was transfected with pCMV-Tag2C vector containing AL3 ORF. Lane E was transfected with empty pCMV-Tag2B vector. (c) Samples, as in (b), were examined using a FLAG-specific (top) or β-actin (bottom) antibody. Arrows on all panels denote AL3-specific bands. Images of autoradiographs were scanned using a CanoScan 8600 F and the scanned images were saved as JPEG files. The brightness of the respective images was adjusted to match the other panels.
To further analyse AL3 protein expression, CMV expression plasmids containing the AL3 ORF were used. Only pCMV-Tag2B should express the AL3 protein because the other two constructs do not contain AL3 in the proper reading frame. In mouse neuroblastoma cells (neuro-2a) transfected with pCMV-Tag2B, proteins migrating near the 15 and 10 kDa markers were detected by the AL3-specific peptide IgG (Fig. 6b, lane B). The predicted molecular mass of the AL3 ORF was 17.4 kDa, suggesting that the band migrating slightly higher than the 15 kDa marker was the AL3 protein. The 10 kDa band may be a stable degradation product or the result of protein synthesis initiating from an internal AL3 methionine residue. AL3-specific bands were not detected in neuro-2A cells transfected with pCMV-Tag2A, pCMV-Tag2C or the empty pCMV-Tag2B (Fig. 6b). The AL3 specific antiserum reacted non-specifically with proteins migrating between 12 and13 kDa (Fig. 6b,).
A FLAG-specific antibody recognized a protein migrating slightly higher than the 15 kDa marker when neuro-2A cells were transfected with pCMV-Tag2B containing the AL3 ORF, but not with pCMV-Tag2A, pCMV-Tag2C or the empty pCMV-Tag2B expression vector (Fig. 6c). Similar levels of β-actin were detected in the cell lysate regardless of the construct transfected into neuro-2A cells. Since the AL3 transcript appears to start downstream of the AL3 ATG in acutely infected cells, the AL3-specific protein was not detected following infection of RS cells with dLAT2903R, as expected (data not shown).
Analysis of AL3 protein expression in TG of infected mice
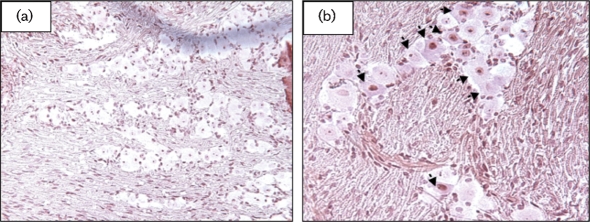
Immunohistochemistry was performed on formalin-fixed and paraffin-embedded TG sections using the AL3-specific IgG. Swiss Webster mice were infected with a LAT+ virus (dLAT2903R) or a LAT null mutant (dLAT2903) (Perng et al., 1996a) using 1×105 p.f.u. per eye as described previously (Jones et al., 2005; Peng et al., 2005). The AL3 peptide-specific IgG was then used to test whether AL3 protein expression occurred in TG during latency (30 days after infection). At 30 days after infection with dLAT2903R, TG neurons were stained with the AL3-specific IgG (Fig. 7b; arrows indicate positive neurons). Some neurons were stained in the nucleus, whereas others were clearly stained in the cytoplasm. Approximately 13 % of TG neurons were stained by the AL3-specific IgG at 30 days following infection with dLAT2903R. AL3-positive neurons were detected in approximately 3 % of neurons infected with dLAT2903 at 30 days after infection, which was similar to that in TG of mock-infected mice (Table 1). Commercially available normal rabbit IgG (Santa Cruz Biotechnology) also cross-reacted weakly with approximately 3 % of TG neurons, regardless of whether TG were prepared from infected or uninfected mice (data not shown). We suggest that IgG, in general, will non-specifically and weakly cross-react with a low percentage of neurons in formalin-fixed, paraffin-embedded TG sections. In summary, these studies indicated that the AL3 protein was detectable in TG neurons following infection.
Fig. 7.
Detection of an AL3 protein in TG of infected mice. Swiss Webster mice were infected with dLAT2903 (a) or dLAT2903R (b) and TG sections were prepared at 30 days after infection (latency). TG from mock-infected mice were used as a control. Arrows denote neurons stained by the AL3-specific IgG. Magnification is ×25.
Table 1.
Detection of an AL3 protein in TG of mice mock-infected or infected with dLAT2390R or dLAT2390
Neurons were stained by using AL3-specific IgG. The numbers of positive neurons versus total number of neurons counted are in parentheses.
| Neurons stained | Latency |
|---|---|
| 2903R | 13 % (21/165) |
| 2903 | 3.3 % (3/91) |
| Mock | 2.5 % (8/320) |
DISCUSSION
In this study, we identified a novel ORF and transcript (AL3) that was anti-sense to LAT. Relative to ICP0 and ICP27, AL3 transcription was readily detected in TG of latently infected mice. The AL3 transcript appeared to encode a protein that was detected in cells stably transfected with LAT or TG of latently infected mice. The recombinant virus LAT3.3A expressed the AL3 transcript during productive infection (Fig. 2b), suggesting that the minimal promoter for AL3 transcription was present within the first 1.5 kb of LAT coding sequences. A TATA box was not detected upstream of the 5′ terminus of the AL3 transcript. However, a perfect match for the mouse cytochrome C oxidase subunit Vb initiator (CGGAAG) was present 35 nt upstream of the start site for AL3 transcription (Yu et al., 1997). Initiators can direct transcription in the absence of a TATA box by interacting with a component of TFIID (Kaufmann & Smale, 1994). Sp1 binding sites within certain GC-rich TATA-less promoters can also specifically initiate transcription (Dynan & Tjian, 1983). Two Sp1 binding sites were identified in the putative AL3 promoter region, suggesting that the putative Sp1 binding sites and initiator element cooperate to activate AL3 transcription.
During productive infection, the 5′ terminus of the AL3 transcript was 20 bases downstream of the transcript expressed in DC-LAT6 cells or TG of latently infected mice (Fig. 4). This finding may be significant because during productive infection, the start site of AL3 transcription was downstream of the first in-frame methionine of the AL3 ORF, which may explain why we were unable to detect AL3 protein expression during productive infection (data not shown). Analysis of the AL3 ORF identified three in-frame methionine residues (LAT nt +1309, +1276 and +1264) that are downstream of the first in-frame methionine (LAT nt +1346) (Fig. 4c). If one of these downstream methionine residues acted as an initiating methionine, the AL3-specific IgG should have detected these smaller proteins because three peptides were used for generating the AL3-specific antiserum in rabbits (Fig. 1c). Since the AL3-specific IgG did not detect a viral-specific protein during productive infection, we suggest that a truncated AL3 protein was not expressed or was not stable. The minor differences in the 3′ terminus of the AL3 transcript do not appear to play a role in AL3 protein expression because DC-LAT6 cells contained the same 3′ terminus as in productive infection and AL3 protein expression was detected in DC-LAT6 cells.
DC-LAT6 cells appeared to express higher levels of AL3 RNA compared with DC-ΔLAT311 or DC-ΔLAT35 cells (Fig. 2c), suggesting that AL3 expression is partially dependent on LAT expression. DC-LAT6 cells were selected because they expressed high levels of the stable 2 kb LAT (Carpenter et al., 2007), which may be the result of DC-LAT6 cells containing more copies of the LAT expression plasmid stably integrated in these cells. Consequently, the levels of AL3 RNA may be higher because there are more copies of AL3 coding sequences. In contrast with AL3 RNA levels, AL3 protein expression levels appeared to be similar in DC-LAT6 and DC-ΔLAT311 cells (Fig. 6). Since DC-LAT6, but not DC-ΔLAT311, cells are resistant to cold-shock-induced apoptosis (Carpenter et al., 2007), it appears that AL3 protein expression does not directly inhibit apoptosis.
The AL3 ORF possessed 32 % identity to dynamin 1 (mouse, rat, bovine or human) and 30 % identity to a putative yeast phospholipid-transporting ATPase. Dynamin 1 is a GTPase that is crucial for synaptic vesicle endocytosis (Newton et al., 2006). Dynamin 1 retrieves molecular components of the vesicle from the synapse surface and then places these components into a new vesicle to be recycled after the vesicle has discharged its neurotransmitter (reviewed by Murthy & Camilli, 2003; Sudhof, 2004). Dynamin 1 is expressed exclusively in the brain and in neurons, dynamin 1 expression increases with synapse formation in parallel with levels of synaptic vesicle proteins. The functions of dynamin 1 are crucial during high levels of neuronal activity, but are not required to form functional synapses (Ferguson et al., 2007). These observations suggest that the AL3 protein mimics or regulates dynamin 1 functions following infection of neurons. It will be of interest to test whether AL3 protein expression plays a role in the latency–reactivation cycle.
Supplementary Material
Acknowledgments
C. J.'s laboratory is supported by a public health service (PHS) grant (R21AI069176), two USDA grants (2009- and 2006-01627) and a PHS grant 1P20RR15635 to the Nebraska Center for Virology. S. L. W.'s laboratory is supported by PHS grant EY13191, The Discovery Eye Foundation, The Henry L. Guenther Foundation and Research to Prevent Blindness (RPB). S. L. W. is an RPB Senior Scientific Investigator. G.-C. P.'s laboratory is supported by PHS grant NINDS/NS049556.
Footnotes
A supplementary table, listing AL3 primers used in this study, is available with the online version of this paper.
References
- Ahmed, M., Lock, M., Miller, C. G. & Fraser, N. W. (2002). Regions of the herpes simplex virus type 1 latency-associated transcript that protect cells from apoptosis in vitro and protect neuronal cells in vivo. J Virol 76, 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco, F. J. & Fraser, N. W. (2005). Herpes simplex virus type 1 latency-associated transcript expression protects trigeminal ganglion neurons from apoptosis. J Virol 79, 9019–9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, D., Hsiang, C., Jin, L., Osorio, N., BenMohamed, L., Jones, C. & Wechsler, S. L. (2007). Stable cell lines expressing high levels of the herpes simplex virus type 1 LAT are refractory to caspase 3 activation and DNA laddering following cold shock induced apoptosis. Virology 369, 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, D., Cohen, J., Naito, J., Mott, K. R., Osorio, N., Jin, L., Fraser, N. W., Jones, C., Wechsler, S. L. & Perng, G.-C. (2006). A mutant deleted for most of the herpes simplex virus type 1 (HSV-1) UOL gene does not affect the spontaneous reactivation phenotype in rabbits. J Neurovirol 12, 5–16. [DOI] [PubMed] [Google Scholar]
- Chen, S. H., Kramer, M. F., Schaffer, P. A. & Coen, D. M. (1997). A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J Virol 71, 5878–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croen, K. D., Ostrove, J. M., Dragovic, L. J., Smialek, J. E. & Straus, S. E. (1987). Latent herpes simplex virus in human trigeminal ganglia. Detection of an immediate early gene “anti-sense” transcript by in situ hybridization. N Engl J Med 317, 1427–1432. [DOI] [PubMed] [Google Scholar]
- Deatly, A. M., Spivack, J. G., Lavi, E. & Fraser, N. W. (1987). RNA from an immediate early region of the type 1 herpes simplex virus genome is present in the trigeminal ganglia of latently infected mice. Proc Natl Acad Sci U S A 84, 3204–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatly, A. M., Spivack, J. G., Lavi, E., O'Boyle, D. R. & Fraser, N. W. (1988). Latent herpes simplex virus type 1 transcripts in peripheral and central nervous system tissues of mice map to similar regions of the viral genome. J Virol 62, 749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan, W. S. & Tjian, R. (1983). The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell 35, 79–87. [DOI] [PubMed] [Google Scholar]
- Ferguson, S. M., Brasnjo, G., Hayashi, M., Wölfel, M, Collesi, C., Giovedi, S., Raimondi, A., Gong, L.-W., Ariel, P. & other authors (2007). A selective activity-dependent requirement for dynamin 1 in synaptic endocytosis. Science 316, 570–574. [DOI] [PubMed] [Google Scholar]
- Garber, D. A., Schaffer, P. A. & Knipe, D. M. (1997). A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J Virol 71, 5885–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesser, R. M. & Koo, S. C. (1997). Latent herpes simplex virus type 1 gene expression in ganglia innervating the human gastrointestinal tract. J Virol 71, 4103–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, G., Peng, W., Jin, L., Perng, G.-C., Nesburn, A. B., Wechsler, S. L. & Jones, C. (2002). Regulation of caspase 8- and caspase 9-induced apoptosis by the herpes simplex virus latency-associated transcript. J Neurovirol 8, 103–111. [DOI] [PubMed] [Google Scholar]
- Inman, M., Perng, G.-C., Henderson, G., Ghiasi, H., Nesburn, A. B., Wechsler, S. L. & Jones, C. (2001). Region of herpes simplex virus type 1 latency-associated transcript sufficient for wild-type spontaneous reactivation promotes cell survival in tissue culture. J Virol 75, 3636–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, L., Peng, W., Perng, G.-C., Nesburn, A. B., Jones, C. & Wechsler, S. L. (2003). Identification of herpes simplex virus type 1 (HSV-1) latency associated transcript (LAT) sequences that both inhibit apoptosis and enhance the spontaneous reactivation phenotype. J Virol 77, 6556–6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, L., Perng, G.-C., Mott, K. R., Osorio,, N., Naito, J., Brick, D. J., Carpenter, D., Jones, C. & Wechsler, S. L. (2005). A herpes simplex virus type 1 mutant expressing a baculovirus inhibitor of apoptosis gene in place of latency-associated transcript has a wild-type reactivation phenotype in the mouse. J Virol 79, 12286–12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, L., Carpenter, D., Moerdyk-Schauwecker, M., Vanarsdall, A. L., Osorio, N., Hsiang, C., Jones, C. & Wechsler, S. L. (2008). Cellular FLIP can substitute for the herpes simplex virus type 1 LAT gene to support a wild type virus reactivation phenotype in mice. J Neurovirol 14, 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, C. (1998). Alphaherpesvirus latency: its role in disease and survival of the virus in nature. Adv Virus Res 51, 81–133. [DOI] [PubMed] [Google Scholar]
- Jones, C. (2003). Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin Microbiol Rev 16, 79–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, C., Inman, M., Peng, W., Henderson, G., Doster, A., Perng, G.-C. & Kaenjak Angeletti, A. (2005). The herpes simplex virus type 1 (HSV-1) locus that encodes the latency-associated transcript (LAT) enhances the frequency of encephalitis in male BALB/C mice. J Virol 79, 14465–14469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, J. & Smale, S. T. (1994). Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev 8, 821–829. [DOI] [PubMed] [Google Scholar]
- Krause, P. R., Croen, K. D., Straus, S. E. & Ostrove, J. M. (1988). Detection and preliminary characterization of herpes simplex virus type 1 transcripts in latently infected human trigeminal ganglia. J Virol 62, 4819–4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr, J. M., Nelson, J. A. & Oldstone, M. B. (1990). Is herpes simplex virus associated with peptic ulcer disease? J Virol 64, 2168–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, F., Perez, S., Jiang, Y., Zhou, Y., Henderson, G. & Jones, C. (2007a). Identification of a novel protein encoded by the latency-related gene of bovine herpesvirus 1. J Neurovirol 13, 569–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, F., Perez, S., Geiser, V., Sintek, M., Inman, M. & Jones, C. (2007b). A protein encoded by the bovine herpes virus 1 latency-related gene interacts with specific cellular regulatory proteins, including CCAAT enhancer binding protein alpha. J Virol 81, 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, W. J., Lirette, R. P. & Fraser, N. W. (1990). Mapping of low abundance latency-associated RNA in the trigeminal ganglia of mice latently infected with herpes simplex virus type 1. J Gen Virol 71, 125–132. [DOI] [PubMed] [Google Scholar]
- Mott, K. R., Osorio, N., Jin, L., Brick, D. J., Naito, J., Cooper, J., Henderson, G., Inman, M., Jones, C. & other authors (2003). The bovine herpesvirus 1 LR ORF2 is critical for this gene's ability to restore the high wild-type reactivation phenotype to a Herpes simplex virus-1 LAT null mutant. J Gen Virol 84, 2975–2985. [DOI] [PubMed] [Google Scholar]
- Murthy, V. N. & Camilli, P. (2003). Cell biology of the presynaptic terminal. Annu Rev Neurosci 26, 701–728. [DOI] [PubMed] [Google Scholar]
- Nahmias, A. J. & Roizman, B. (1973). Infection with herpes-simplex viruses 1 and 2. 3. N Engl J Med 289, 781–789. [DOI] [PubMed] [Google Scholar]
- Naito, J., Mukerjee, R., Mott, K. R., Kang, W., Osorio, N., Fraser, N. W. & Perng, G.-C. (2005). Identification of a protein encoded in the herpes simplex virus type 1 latency associated transcript promoter region. Virus Res 108, 101–110. [DOI] [PubMed] [Google Scholar]
- Nesburn, A. B. (editor) (1983). Report of the Corneal Disease Panel: Vision Research – a National Plan, 1983–1987, part III. St Louis: CV Mosby.
- Newton, A. J., Kirchhusen, T. & Murthy, V. N. (2006). Inhibition of dynamin completely blocks compensatory synaptic vesicle endocytosis. Proc Natl Acad Sci U S A 103, 17955–17960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, W., Henderson, G., Perng, G.-C., Nesburn, A. B., Wechsler, S. L. & Jones, C. (2003). The gene that encodes the herpes simplex virus type 1 latency-associated transcript influences the accumulation of transcripts (Bcl-xL and Bcl-xS) that encode apoptotic regulatory proteins. J Virol 77, 10714–10718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, W., Jin, L., Henderson, G., Perng, G.-C., Brick, D. J., Nesburn, A. B., Wechsler, S. L. & Jones, C. (2004). Mapping herpes simplex virus type 1 (HSV-1) LAT sequences that protect from apoptosis mediated by a plasmid expressing caspase-8. J Neurovirol 10, 260–265. [DOI] [PubMed] [Google Scholar]
- Peng, W., Henderson, G., Inman, M., BenMohamed, L., Perng, G.-C., Wechsler, S. L. & Jones, C. (2005). The locus encompassing the latency-associated transcript (LAT) of herpes simplex virus type 1 interferes with and delays interferon expression in productively infected neuroblastoma cells and trigeminal ganglia of acutely infected mice. J Virol 79, 6162–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, W., Vitvitskaia, O., Carpenter, D., Wechsler, S. L. & Jones, C. (2008). Identification of two small RNAs within the first 1.5-kb of the herpes simplex virus type 1 (HSV-1) encoded latency-associated transcript (LAT). J Neurovirol 14, 41–52. [DOI] [PubMed] [Google Scholar]
- Perng, G.-C., Dunkel, E. C., Geary, P. A., Slanina, S. M., Ghiasi, H., Kaiwar, R., Nesburn, A. B. & Wechsler, S. L. (1994). The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J Virol 68, 8045–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng, G.-C., Ghiasi, H., Slanina, S. M., Nesburn, A. B. & Wechsler, S. L. (1996a). The spontaneous reactivation function of the herpes simplex virus type 1 LAT gene resides completely within the first 1.5 kilobases of the 8.3- kilobase primary transcript. J Virol 70, 976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng, G.-C., Chokephaibulkit, K., Thompson, R. L., Sawtell, N. M., Slanina, S. M., Ghiasi, H., Nesburn, A. B. & Wechsler, S. L. (1996b). The region of the herpes simplex virus type 1 LAT gene that is colinear with the ICP34.5 gene is not involved in spontaneous reactivation. J Virol 70, 282–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng, G.-C., Slanina, S. M., Ghiasi, H., Nesburn, A. B. & Wechsler, S. L. (1996c). A 371-nucleotide region between the herpes simplex virus type 1 (HSV-1) LAT promoter and the 2-kilobase LAT is not essential for efficient spontaneous reactivation of latent HSV-1. J Virol 70, 2014–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng, G.-C., Slanina, S. M., Yukht, A., Drolet, B. S., Keleher, W., Jr, Ghiasi, H., Nesburn, A. B. & Wechsler, S. L. (1999). A herpes simplex virus type 1 latency-associated transcript mutant with increased virulence and reduced spontaneous reactivation. J Virol 73, 920–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng, G.-C., Jones, C., Ciacci-Zanella, J., Stone, M., Henderson, G., Yukht, A., Slanina, S. M., Hoffman, F. M., Ghiasi, H. & other authors (2000). Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript (LAT). Science 287, 1500–1503. [DOI] [PubMed] [Google Scholar]
- Perng, G.-C., Esmaili, D., Slanina, S. M., Yukht, A., Ghiasi, H., Osorio, N., Mott, K. R., Maguen, B., Jin, L. & other authors (2001). Three herpes simplex virus type 1 latency-associated transcript mutants with distinct and asymmetric effects on virulence in mice compared with rabbits. J Virol 75, 9018–9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng, G.-C., Maguen, B., Jing, L., Mott, K. R., Kurylo, J., BenMohamed, O., Yukht, A., Osorio, N., Nesburn, A. B. & other authors (2002a). A novel herpes simplex virus type 1 transcript (AL-RNA) antisense to the 5′ end of LAT (latency associated transcript) produces a protein in infected rabbits. J Virol 76, 8003–8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng, G.-C., Maguen, B., Jin, L., Mott, K. R., Osorio, N., Slanina, S. M., Yukht, A., Ghiasi, H., Nesburn, A. B. & other authors (2002b). A gene capable of blocking apoptosis can substitute for the herpes simplex virus type 1 latency-associated transcript gene and restore wild-type reactivation levels. J Virol 76, 1224–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock, D. L., Nesburn, A. B., Ghiasi, H., Ong, J., Lewis, T. L., Lokensgard, J. R. & Wechsler, S. L. (1987). Detection of latency-related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus type 1. J Virol 61, 3820–3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, J. & Wagner, E. (1993). Transcriptional analysis of the herpes simplex virus type 1 region containing the TRL/UL junction. Virology 196, 220–231. [DOI] [PubMed] [Google Scholar]
- Stevens, J. G., Wagner, E. K., Devi-Rao, G. B., Cook, M. L. & Feldman, L. T. (1987). RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 235, 1056–1059. [DOI] [PubMed] [Google Scholar]
- Sudhof, T. C. (2004). The synaptic vesicle cycle. Annu Rev Neurosci 27, 509–547. [DOI] [PubMed] [Google Scholar]
- Wagner, E. K. & Bloom, D. C. (1997). Experimental investigation of herpes simplex virus latency. Clin Microbiol Rev 10, 419–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, E. K., Devi-Rao, G., Feldman, L. T., Dobson, A. T., Zhang, Y. F., Flanagan, W. M. & Stevens, J. G. (1988a). Physical characterization of the herpes simplex virus latency-associated transcript in neurons. J Virol 62, 1194–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, E. K., Flanagan, W. M., Devi-Rao, G., Zhang, Y. F., Hill, J. M., Anderson, K. P. & Stevens, J. G. (1988b). The herpes simplex virus latency-associated transcript is spliced during the latent phase of infection. J Virol 62, 4577–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley, R. J. (1991). Herpes simplex virus infections of the central nervous system. Encephalitis and neonatal herpes. Drugs 42, 406–427. [DOI] [PubMed] [Google Scholar]
- Whitley, R. J. (1997). Herpes Simplex Virus. Philadelphia, New York: Lippincott-Raven.
- Winkler, M. T., Schang, L. S., Doster, A., Holt, T. & Jones, C. (2000). Analysis of cyclins in trigeminal ganglia of calves infected with bovine herpesvirus-1. J Gen Virol 81, 2993–2998. [DOI] [PubMed] [Google Scholar]
- Winkler, M. T., Doster, A., Sur, J. H. & Jones, C. (2002). Analysis of bovine trigeminal ganglia following infection with bovine herpesvirus 1. Vet Microbiol 86, 139–155. [DOI] [PubMed] [Google Scholar]
- Yu, M., Yang, X.-Y., Schmidt, T., Chinenov, Y., Wang, R. & Martin, M. E. (1997). GA-binding protein-dependent transcription initiator elements. J Biol Chem 272, 29060–29067. [DOI] [PubMed] [Google Scholar]
- Zwaagstra, J. C., Ghiasi, H., Slanina, S. M., Nesburn, A. B., Wheatley, S. C., Lillycrop, K., Wood, J., Latchman, D. S., Patel, K. & Wechsler, S. L. (1990). Activity of herpes simplex virus type 1 latency-associated transcript (LAT) promoter in neuron-derived cells: evidence for neuron specificity and for a large LAT transcript. J Virol 64, 5019–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.