Abstract
GB virus B (GBV-B) causes acute hepatitis in experimentally infected tamarins. We compared evolutionary features in acute resolving and persistent GBV-B infection. We detected no evidence of evolution in four animals with clearance during weeks 9–12, whereas three animals with clearance during weeks 13–26 had several substitutions in their polyprotein sequence. A single tamarin had long-term GBV-B viraemia; analysis of virus recovered at weeks 2, 5, 12, 20, 26, 52 and 104 demonstrated that mutations accumulated over time. Overall, the amino acid substitution rate was 3.5×10−3 and 1.1×10−3 substitutions per site year−1 during weeks 1–52 and 53–104, respectively. Thus, there was a significant decrease in evolution over time, as found for hepatitis C virus. The rate of non-synonymous substitution per non-synonymous site compared with that of synonymous substitution per synonymous site decreased over time, suggesting reduction of positive selective pressure. These data demonstrate that prolonged GBV-B infection is associated with viral evolution.
Hepatitis C virus (HCV) is a virus for which there is urgent need to develop more effective therapeutic measures and a vaccine. However, small-animal models for HCV are not readily available; the only model available for studies of HCV pathogenesis is the chimpanzee. GB virus B (GBV-B), on the other hand, causes acute hepatitis in experimentally infected tamarins (Bukh et al., 1999; Simons et al., 1995). Furthermore, GBV-B is the virus genetically related most closely to HCV (Bukh et al., 1999; Muerhoff et al., 1995; Simons et al., 1995). Like HCV, it has a positive-sense, single-stranded RNA genome and a genomic organization similar to that of HCV, with highly structured 5′ and 3′ untranslated regions (UTRs) and a single long open reading frame (ORF) encoding structural (core, E1 and E2) and non-structural (p13, NS2, NS3, NS4A, NS4B, NS5A and NS5B) proteins. Thus, we and other investigators have proposed GBV-B infection of tamarins as a surrogate model of HCV infection in experimentally infected chimpanzees and in humans (Bukh, 2004). In the present study, we compared the molecular evolutionary features in acute resolving GBV-B infection with those in persistent GBV-B infection. The availability of the GBV-B model system in tamarins with prolonged or persistent infections could contribute to our understanding of how HCV persists in the majority of infected individuals, and might represent a relevant surrogate model for testing inhibitors of HCV that are under development.
The course of experimental GBV-B infection in tamarins following virus inoculation or intrahepatic transfection with RNA transcripts usually results in acute resolving infection (Bukh et al., 1999; Nam et al., 2004; Simons et al., 1995). In a recent study, we infected 17 Saguinus mystax tamarins with GBV-B following intrahepatic transfection with pGBB genomes (GenBank accession no. AF179612) containing mutations in the p13 gene (Takikawa et al., 2006). Animals were maintained under conditions that met or exceeded all requirements for their use in an approved facility. Weekly serum samples were tested for liver enzyme levels [isocitrate dehydrogenase (ICD)] and for GBV-B RNA by a real-time PCR as described previously (Nam et al., 2004). In previous studies, we found an excellent correlation between detection of GBV-B RNA in this assay and in a nested RT-PCR assay (Bukh et al., 2008; Nam et al., 2004). Two animals died before the outcome of infection could be determined, but 14 of the remaining 15 animals apparently developed a transient infection. We noticed that there were animals that cleared their infection rapidly without rebound in viraemia, as well as animals that had reappearance of viraemia after initial control. Therefore, our aim was to determine whether these differences were associated with virus evolution and to compare evolution in these animals with that in the animal with persistent GBV-B infection. We selected eight animals for analysis of virus evolution. Importantly, the animals selected were infected with mutants that did not appear to attenuate the GBV-B infection, as they had rapid appearance of viraemia with relatively high GBV-B RNA titres early in the acute phase (Fig. 1). However, it should be noted that infection in SM911 and SM912 might have been slightly attenuated initially, but the attenuating A681R mutant changed rapidly to 681C in these animals with subsequent robust infection (Takikawa et al., 2006).
Fig. 1.
Course of GBV-B infection in S. mystax tamarins selected for further analysis of GBV-B evolution. The animals became infected following intrahepatic transfection with RNA transcripts of pGBB-A669N (SM908 and SM910), pGBB-R654L-R704L (SM916 and SM917), pGBB-A681R (SM911 and SM912), pGBB-R704L (SM920) and pGBB-A680L (SM918) (Takikawa et al., 2006). Serum levels of ICD (shaded area) and the estimated GBV-B titre (◊), determined in a TaqMan quantitive PCR assay, were plotted against time. Samples found to be below the assay cut-off of approx. 103 genome equivalents ml−1 in the TaqMan assay are shown as not detected (nd). The complete ORF sequence of recovered GBV-B virus was determined at the weeks indicated by arrows.
We determined the consensus sequence of the genome corresponding to nt 36–9107 of pGBB (GenBank accession no. AF179612), which included the entire ORF of recovered GBV-B viruses, by using a long nested RT-PCR procedure as described in detail previously (Nam et al., 2004). As in prior GBV-B studies, we elected not to perform clonal analysis, but instead inspected the primary sequence data for the presence of minor sequences that are typically detected if a sequence is present in >10 % of the genomes, and at positions with dominant changes, this quasispecies was reported (Table 1). Viruses analysed included those recovered in the early acute phase and late in the acute infection and, in one animal, those recovered during persistent infection.
Table 1.
Nucleotide and amino acid changes in S. mystax animals with acute resolving (SM911, SM920 and SM918) and persistent (SM912) GBV-B infection
The consensus sequence corresponding to nt 36–9107 of pGBB, including the entire ORF of recovered GBV-B viruses, was determined for each sample, using a long nested RT-PCR procedure. Positions with differences from pGBB are shown for each animal (mutations in p13 originally introduced into these viruses are not shown). Only dominant changes observed at one or more time point in an individual animal are included. Dominant sequences are shown in capital letters and minor sequences are shown in lower-case letters.
| Genome region [nt (aa)]* | pGBB | SM912 | SM911 | SM920 | SM918 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| wk 2 | wk 5 | wk 12 | wk 20 | wk 26 | wk 52 | wk 104 | wk 5 | wk 12 | wk 2 | wk 9 | wk 20 | wk 2 | wk 19 | wk 25 | ||
| 5′ UTR (1–445) | ||||||||||||||||
| 237 | A | A | A | A | A | A | G | G | ||||||||
| C (446–913) | ||||||||||||||||
| 710 (89) | A (I) | A (I) | A (I) | A (I) | A (I) | G/a (V/i) | A (I) | A (I) | A (I) | G (V) | A (I) | |||||
| 846 (134) | C (A) | C (A) | C (A) | T (V) | ||||||||||||
| E1 (914–1489) | ||||||||||||||||
| 982 | C | A | A | |||||||||||||
| E2 (1490–2284) | ||||||||||||||||
| 1528 | T | T | T | T | T/C | T | C | C | ||||||||
| 1550 (369) | A (I) | A (I) | A (I) | A (I) | A (I) | A (I) | G (V) | G (V) | ||||||||
| 1842 (466) | A (H) | A (H) | A (H) | A (H) | A/g (H/r) | A/G (H/R) | A (H) | A (H) | ||||||||
| 1895 (484) | A (M) | A (M) | A (M) | A (M) | A (M) | A (M) | T (L) | T (L) | ||||||||
| 1976 (511) | G (V) | G (V) | G (V) | G (V) | G/A (V/M) | G (V) | A (M) | A (M) | ||||||||
| 2053 | A | A | A | A | A | A | G | G | ||||||||
| 2267 (608) | T (F) | T (F) | T (F) | T (F) | T (F) | T (F) | G (V) | G (V) | ||||||||
| 2276 (611) | G (A) | G (A) | G (A) | G (A) | G (A) | G (A) | G (A) | A (T) | ||||||||
| p13 (2285–2641) | ||||||||||||||||
| 2304 (620) | T (L) | T (L) | T (L) | T (L) | T (L) | C/t (P/l) | T (L) | T (L) | ||||||||
| 2552 (703) | A (N) | A (N) | A/G (N/D) | G (D) | G (D) | G (D) | G (D) | G (D) | A (N) | G (D) | A (N) | G (D) | G (D) | |||
| NS2 (2642–3265) | ||||||||||||||||
| 3136 | A | A | A | G | ||||||||||||
| 3212 (923) | G (G) | G (G) | G (G) | A (S) | A (S) | A (S) | A (S) | A (S) | G (G) | A/g (S/g) | G (G) | A (S) | A (S) | |||
| NS3 (3266–5125) | ||||||||||||||||
| 3367 | C | C | C | C | C | C | T | T | ||||||||
| 3463 | T | T | C | T | ||||||||||||
| 4198 | A | A | A | G | G | G | G | G | ||||||||
| 4246 | C | C | T | C | ||||||||||||
| 4285 | C | C | C | C | C | C | C | T | ||||||||
| 4915 | T | G | G | |||||||||||||
| NS4A (5126–5290) | ||||||||||||||||
| 5200 | T | T | T | A | ||||||||||||
| 5272 | A | A | A | A | A | A/G | A | A | ||||||||
| NS4B (5291–6034) | ||||||||||||||||
| 5353 | C | C | C | C | C | C | C | T | ||||||||
| 5386 | T | T | T | T | T | T | T | C | ||||||||
| 5674 | C | C | C | T | ||||||||||||
| 5794 | C | C | C | T | ||||||||||||
| 5910 (1822) | A (N) | A (N) | G (S) | A (N) | ||||||||||||
| 5914 (1823) | G (E) | G (E) | G (E) | G (E) | G (E) | C/g (D/e) | G (E) | G (E) | G (E) | G (E) | T (D) | |||||
| NS5A (6035–7267) | ||||||||||||||||
| 6131 (1896) | A (N) | A (N) | G (D) | A (N) | ||||||||||||
| 6472 | G | G | G | G | G | G | G | A | ||||||||
| 6690 (2082) | C (T) | C (T) | T (I) | C (T) | ||||||||||||
| 6849 (2135) | T (F) | T (F) | T (F) | T (F) | T (F) | T (F) | C (S) | C (S) | ||||||||
| 6956 (2171) | A (N) | A (N) | A (N) | A (N) | A (N) | A (N) | A (N) | G (D) | ||||||||
| 7031 (2196) | T (F) | T (F) | T (F) | T (F) | C/t (L/f) | T (F) | C (L) | C (L) | ||||||||
| 7072 | C | C | C | C | T/c | C | T | T/c | ||||||||
| 7105 | G | G | G | A | ||||||||||||
| 7248 (2268) | G (G) | G (G) | G (G) | G (G) | A/g (E/g) | G/a (G/e) | A (E) | A (E) | ||||||||
| NS5B (7268–9037) | ||||||||||||||||
| 7440 (2332) | T (L) | T (L) | T (L) | T (L) | T (L) | T (L) | A (Q) | A (Q) | ||||||||
| 7994 (2517) | A (S) | A (S) | G (G) | |||||||||||||
| 8001 (2519) | A (Q) | A (Q) | A (Q) | G/a (R/q) | ||||||||||||
| 8122 | A | A | A | A | A | A | G | G | ||||||||
| 8314 | T | T | T | C | ||||||||||||
| 8336 | T | T | T | C | T | T | T | T | ||||||||
| 8472 (2676) | G (S) | G (S) | G (S) | A (N) | G (S) | G (S) | G (S) | G (S) | ||||||||
| 8653 | T | T | C/t | |||||||||||||
| 8663 (2740) | A (I) | A (I) | A (I) | G (V) | ||||||||||||
| 8727 (2761) | C (T) | C (T) | C (T) | C (T) | C (T) | C (T) | C (T) | T (I) | ||||||||
| 8766 (2774) | A (K) | A (K) | A (K) | G (R) | ||||||||||||
| 8860 | A | A | A | A | A | A | A | G | ||||||||
| 8938 | G | G | A/g | |||||||||||||
| 8942 (2833) | A (I) | A (I) | G (V) | A (I) | ||||||||||||
| 8971 | C | C | C | C | C | C | C | T | ||||||||
| 3′ UTR (9038–9399) | ||||||||||||||||
| 9046 | C | C | T/c | |||||||||||||
*Nucleotide and amino acid positions corresponding to pGBB (GenBank accession no. AF179612).
In seven animals with resolved GBV-B infection, we compared the genome sequence of virus recovered from a sample taken 1–3 weeks prior to resolution with the sequence found in the early acute phase (Fig. 1). We found no evidence of evolution in four animals (SM908, SM910, SM916 and SM917) that had clearance during weeks 9–12 (Fig. 1). However, we recognize that, since we did not perform clonal analysis, some quasispecies might not have been recognized. In fact, such quasispecies were recently reported in tamarins as early as 17 days post-infection (McGarvey et al., 2008).
In the three animals with prolonged acute resolving infection, we did however detect evidence of virus evolution. In SM911, with clearance at week 13, we observed six nucleotide changes during weeks 5–12 (Table 1), which resulted in three amino acid changes (N703D, G923S and S2517G) (Table 1). In SM920, with clearance at week 21, the week 2 and 9 samples had identical sequences. In this animal, GBV-B RNA was not detected during weeks 11–18, but the virus reappeared transiently during weeks 19 and 20 (Fig. 1); the viruses recovered at week 20 had six nucleotide changes (Table 1), which resulted in three amino acid changes (A134V, Q2519R and I2740V) (Table 1). Finally, in SM918, with clearance at week 26, GBV-B RNA was not detected during weeks 14–16, 21 or 22. In this animal, two different virus populations were detected at weeks 19 and 25: we observed nine and seven nucleotide changes at weeks 19 and 25, respectively (Table 1), which resulted in seven (I89V, N703D, G923S, N1822S, N1896D, T2082I and I2833V) and four (N703D, G923S, E1823D and K2774R) amino acid changes, respectively, compared with the week 2 sequence (Table 1). Thus, prolonged infection or transient reappearance following control of viraemia could have resulted from emergence of minor variants that temporarily escaped immune control.
Persistent GBV-B infections have been reported (Martin et al., 2003; Nam et al., 2004; Takikawa et al., 2006). After transfection with wild-type GBV-B, a Saguinus oedipus tamarin remained viraemic for 2 years, but subsequently cleared the virus (Martin et al., 2003). After transfection with a poly(U) tract deletion mutant, an S. mystax tamarin remained infected until its death at week 90 (Nam et al., 2004). In the present study, one (SM912) of two S. mystax tamarins transfected with the pGBB-A681R mutant (Takikawa et al., 2006) remained viraemic for >2 years of follow-up with fluctuating liver enzyme levels (Fig. 1), whereas the other animal (SM911) transfected with this mutant cleared the virus during week 13 (see above). It is possible that SM912 eventually cleared the virus, as GBV-B RNA was not detected during weeks 120–156 (end of follow-up).
Unlike the two previous reports, the persistently infected animal identified in the present study had relatively low titres of GBV-B and exhibited only marginally elevated liver enzymes, although enzyme flares were observed in association with spikes in the GBV-B RNA titre throughout follow-up (Fig. 1). We determined the genome sequence (corresponding to nt 36–9107 of pGBB) of virus recovered at weeks 2, 5, 12, 20, 26, 52 and 104 (Table 1). Generally, mutations accumulated over time in the recovered viruses. At week 5, we found the amino acid change N703D (in p13), a mutation that was also observed in SM911 and SM918 with resolving infection (see above) and in SM890, another animal with persistent infection (Nam et al., 2004), suggesting that this site might be under host immune pressure. At week 12, we found the amino acid change G923S; this NS2 mutation was also detected in SM911 and SM918. Interestingly, two changes (F2196L and G2268E) observed in NS5A during the first year of follow-up were observed previously in the other two animals that developed persistent GBV-B infection (Martin et al., 2003; Nam et al., 2004).
The viruses recovered from SM912 at week 52 had a total of 19 nucleotide changes (Table 1), encoding 10 amino acid changes (I369V, M484L, V511M, F608V, N703D, G923S, F2135S, F2196L, G2268E and L2332Q) in the polyprotein sequence (Table 1). However, during the second year of follow-up, we observed only nine additional nucleotide changes (Table 1), encoding three amino acid changes (A611T, N2171D and T2761I) (Table 1). The nucleotide mutation rate in SM912 was 1.9×10−3 and 1.0×10−3 changes per site year−1 during years 1 and 2 of follow-up, respectively, and the amino acid substitution rate was 3.5×10−3 and 1.1×10−3 substitutions per site year−1, respectively. Thus, there was a dramatic decrease in evolution rate during the second year of follow-up; a similar decrease in mutation rates was found in chronic HCV infections (Fernandez et al., 2004; Major et al., 1999; J. Bukh, unpublished data).
Non-synonymous substitutions per non-synonymous site (dN) and synonymous substitutions per synonymous site (dS) were determined during weeks 0–26, 0–52 (first year) and 53–104 (second year), respectively (Sakai et al., 2007). In contrast to the dS rate, the dN rate decreased over time and, during the second year, the dS rate was higher than the dN rate (Fig. 2), suggesting that significant reduction of immune pressure took place in SM912. Furthermore, comparing the synonymous mutation rate with the non-synonymous mutation rate at different time points revealed that the dN/dS ratio decreased over time, indicating less immune pressure in the chronic phase. Thus, the non-synonymous substitution rate/synonymous substitution rate ratio during weeks 0–26, 0–52 and 53–104 was 1.67, 1.12 and 0.83, respectively. Similar findings have been reported for HCV (Fernandez et al., 2004). Thus, the evolutionary features of GBV-B are consistent with the molecular evolution of HCV.
Fig. 2.
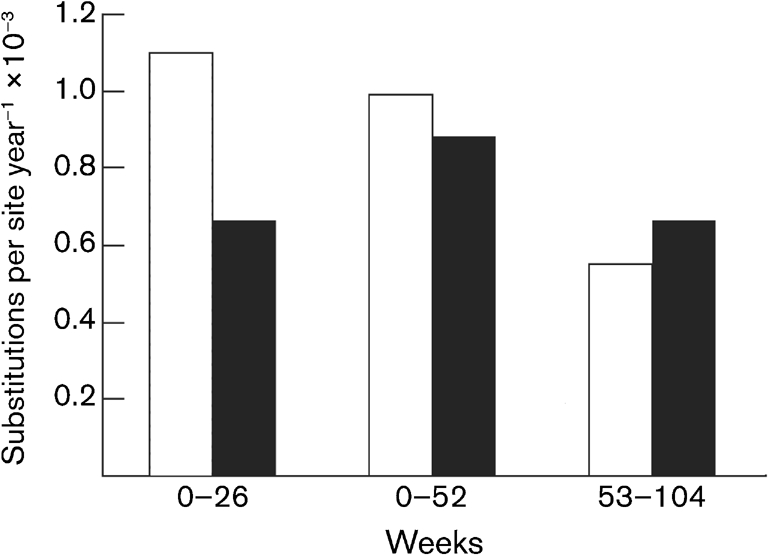
Mutation rates of GBV-B in SM912 with persistent infection. A comparison of non-synonymous (empty bars) and synonymous (filled bars) mutation rates during weeks 0–26, 0–52 and 53–104 was performed by using GBV-B consensus sequences of the complete ORF at weeks 26, 52 and 104, respectively.
HCV has a hypervariable region (HVR1) at the N-terminal end of the E2 protein that evolves rapidly over time in an infected individual (Ogata et al., 1991) and contains a neutralization epitope (Farci et al., 1996); this region was believed to be involved in viral persistence through immune evasion. However, a study of experimentally infected chimpanzees with recombinant monoclonal infection indicated that establishment of persistent HCV infection does not necessarily require mutations in HVR1 (Major et al., 1999). During 60 weeks observation, HVR1 did not accumulate mutations at a high rate compared with the rest of the viral genome. Moreover, a chimpanzee that was inoculated with an RNA transcript lacking HVR1 became persistently infected (Forns et al., 2000). According to these studies, it appears that HCV infection can progress to persistence without amino acid changes or diversity in HVR1. As GBV-B infection usually results in acute resolving infection (typically within 20 weeks) in tamarins, it is not clear whether this virus has a highly variable region, like HCV. Comparison of the polyprotein sequences over 104 weeks in SM912 showed that most changes occurred in E2 and NS5A, with five amino acid changes in each deduced protein (Table 1), but with no apparent evidence of a hypervariable region in E2. The absence of a hypervariable region in GBV-B E2 was also noted by McGarvey et al. (2008) in a clonal analysis of E2.
Interestingly, among the mutations observed in NS5A, F2196L and G2286E were also detected in two other persistently infected tamarins (Martin et al., 2003; Nam et al., 2004). These mutations occurred between weeks 7 and 15 in SM890 (Nam et al., 2004), between weeks 12 and 20 in SM912 and between weeks 40 and 48 in a third tamarin with persistent infection (Martin et al., 2003). In SM890, these mutations were, however, not present at weeks 60 or 90, and thus were not present in chronic phase passaged virus, which resulted in acute resolving infection (Nam et al., 2004). In contrast, the NS5A mutations persisted until week 104 in SM912. Kyuregyan et al. (2005) reported that these mutations were observed in two marmosets that resolved the acute GBV-B infection by weeks 14 and 17. Thus, it remains unclear whether these mutations are involved with GBV-B persistence in tamarins. HCV NS5A has been shown to possess an ability to induce interleukin-8, a chemokine that inhibits the antiviral function of interferon (Polyak et al., 2001). This is one of the possible mechanisms of evasion from innate immune responses.
The GBV-B/tamarin infection model could play a role in efforts to develop antiviral reagents against HCV and for analysis of the immune-clearance mechanisms and pathogenesis of HCV. It is possible that detailed analysis of virus evolution and associated immune responses may add information on how GBV-B persists in a tamarin. In this context, observation of an additional animal with persistent GBV-B, as well as animals with prolonged viraemia, and detailed evolutionary analysis are valuable.
Acknowledgments
J. B. is the recipient of a professorship at the University of Copenhagen with external funding from the Lundbeck Foundation. This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
References
- Bukh, J. (2004). A critical role for the chimpanzee model in the study of hepatitis C. Hepatology 39, 1469–1475. [DOI] [PubMed] [Google Scholar]
- Bukh, J., Apgar, C. L. & Yanagi, M. (1999). Toward a surrogate model for hepatitis C virus: an infectious molecular clone of the GB virus-B hepatitis agent. Virology 262, 470–478. [DOI] [PubMed] [Google Scholar]
- Bukh, J., Engle, R. E., Govindarajan, S. & Purcell, R. H. (2008). Immunity against the GBV-B hepatitis virus in tamarins can prevent productive infection following rechallenge and is long-lived. J Med Virol 80, 87–94. [DOI] [PubMed] [Google Scholar]
- Farci, P., Shimoda, A., Wong, D., Cabezon, T., De, G. D., Strazzera, A., Shimizu, Y., Shapiro, M., Alter, H. J. & Purcell, R. H. (1996). Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci U S A 93, 15394–15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, J., Taylor, D., Morhardt, D. R., Mihalik, K., Puig, M., Rice, C. M., Feinstone, S. M. & Major, M. E. (2004). Long-term persistence of infection in chimpanzees inoculated with an infectious hepatitis C virus clone is associated with a decrease in the viral amino acid substitution rate and low levels of heterogeneity. J Virol 78, 9782–9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forns, X., Thimme, R., Govindarajan, S., Emerson, S. U., Purcell, R. H., Chisari, F. V. & Bukh, J. (2000). Hepatitis C virus lacking the hypervariable region 1 of the second envelope protein is infectious and causes acute resolving or persistent infection in chimpanzees. Proc Natl Acad Sci U S A 97, 13318–13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyuregyan, K. K., Poleschuk, V. F., Zamyatina, N. A., Isaeva, O. V., Michailov, M. I., Ross, S., Bukh, J., Roggendorf, M. & Viazov, S. (2005). Acute GB virus B infection of marmosets is accompanied by mutations in the NS5A protein. Virus Res 114, 154–157. [DOI] [PubMed] [Google Scholar]
- Major, M. E., Mihalik, K., Fernandez, J., Seidman, J., Kleiner, D., Kolykhalov, A. A., Rice, C. M. & Feinstone, S. M. (1999). Long-term follow-up of chimpanzees inoculated with the first infectious clone for hepatitis C virus. J Virol 73, 3317–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, A., Bodola, F., Sangar, D. V., Goettge, K., Popov, V., Rijnbrand, R., Lanford, R. E. & Lemon, S. M. (2003). Chronic hepatitis associated with GB virus B persistence in a tamarin after intrahepatic inoculation of synthetic viral RNA. Proc Natl Acad Sci U S A 100, 9962–9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey, M. J., Iqbal, M., Nastos, T. & Karayiannis, P. (2008). Restricted quasispecies variation following infection with the GB virus B. Virus Res 135, 181–186. [DOI] [PubMed] [Google Scholar]
- Muerhoff, A. S., Leary, T. P., Simons, J. N., Pilot-Matias, T. J., Dawson, G. J., Erker, J. C., Chalmers, M. L., Schlauder, G. G., Desai, S. M. & Mushahwar, I. K. (1995). Genomic organization of GB viruses A and B: two new members of the Flaviviridae associated with GB agent hepatitis. J Virol 69, 5621–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, J. H., Faulk, K., Engle, R. E., Govindarajan, S., St.Claire, M. & Bukh, J. (2004). In vivo analysis of the 3′ untranslated region of GB virus B after in vitro mutagenesis of an infectious cDNA clone: persistent infection in a transfected tamarin. J Virol 78, 9389–9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata, N., Alter, H. J., Miller, R. H. & Purcell, R. H. (1991). Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci U S A 88, 3392–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak, S. J., Khabar, K. S., Rezeiq, M. & Gretch, D. R. (2001). Elevated levels of interleukin-8 in serum are associated with hepatitis C virus infection and resistance to interferon therapy. J Virol 75, 6209–6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, A., Takikawa, S., Thimme, R., Meunier, J. C., Spangenberg, H. C., Govindarajan, S., Farci, P., Emerson, S. U., Chisari, F. V. & other authors (2007). In vivo study of the HC-TN strain of hepatitis C virus recovered from a patient with fulminant hepatitis: RNA transcripts of a molecular clone (pHC-TN) are infectious in chimpanzees but not in Huh7.5 cells. J Virol 81, 7208–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, J. N., Pilot-Matias, T. J., Leary, T. P., Dawson, G. J., Desai, S. M., Schlauder, G. G., Muerhoff, A. S., Erker, J. C., Buijk, S. L. & Chalmers, M. L. (1995). Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc Natl Acad Sci U S A 92, 3401–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takikawa, S., Engle, R. E., Emerson, S. U., Purcell, R. H., St. Claire, M. & Bukh, J. (2006). Functional analyses of GB virus B p13 protein: development of a recombinant GB virus B hepatitis virus with a p7 protein. Proc Natl Acad Sci U S A 103, 3345–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]



