Abstract
P-TEFb functions to induce the elongation step of RNA polymerase II transcription by phosphorylating the carboxyl-terminal domain of the largest subunit of RNA polymerase II. Core P-TEFb is comprised of Cdk9 and a cyclin regulatory subunit, with Cyclin T1 being the predominant Cdk9-associated cyclin. The kinase activity of P-TEFb is dependent on phosphorylation of the Thr186 residue located within the T-loop domain of the Cdk9 subunit. Here, we used immunofluorescence deconvolution microscopy to examine the subcellular distribution of phospho-Thr186 Cdk9/Cyclin T1 P-TEFb heterodimers. We found that phospho-Thr186 Cdk9 displays a punctate distribution throughout the non-nucleolar nucleoplasm and it co-localizes with Cyclin T1 almost exclusively within nuclear speckle domains. Phospho-Thr186 Cdk9 predominantly co-localized with the hyperphosphorylated forms of RNA polymerase II. Transient expression of kinase-defective Cdk9 mutants revealed that neither is Thr186 phosphorylation or kinase activity required for Cdk9 speckle localization. Lastly, both the Brd4 and HEXIM1 proteins interact with P-TEFb at or very near speckle domains and treatment of cells with the Cdk9 inhibitor flavopiridol alters this distribution. These results indicate that the active form of P-TEFb resides in nuclear speckles and raises the possibility that speckles are sites of P-TEFb function and exchange between negative and positive P-TEFb regulatory complexes.
RNA polymerase II (RNAPII) transcription is a highly regulated process involving several different stages that include pre-initiation, initiation, promoter clearance, elongation, and termination (Sims et al., 2004). The elongation step is of particular interest, as it has become increasingly clear that this stage is coordinately regulated for both the production of full-length transcripts and mRNA processing events (Sims et al., 2004; Bentley, 2005). Moreover, recent studies have shown that RNAPII elongation is the rate limiting step for expression of a large portions of cellular genes (Guenther et al., 2007; Hargreaves et al., 2009). The carboxyl-terminal domain (CTD) of the largest subunit of RNAPII is critical for the transition from transcriptional initiation to elongation and the integration of mRNA processing (Phatnani and Greenleaf, 2006). The CTD is an evolutionarily conserved domain and is comprised of multiple tandem copies of the consensus repeat heptad YSPTSPS (Corden, 1990). The phosphorylation pattern of the heptad repeats within the CTD change as the polymerase progresses through the stages of transcription and this appears to orchestrate the association of different cellular factors involved in transcription and co-transcriptional mRNA processing (Buratowski, 2003; Sims et al., 2004; Bentley, 2005; Phatnani and Greenleaf, 2006).
The transcription cycle begins with the formation of pre-initiation complexes in which the hypophosphorylated CTD form of RNAPII is recruited to gene promoters (Dahmus, 1995). Soon after the initiation of transcription, Ser5 residues in the heptad repeats of the CTD are heavily phosphorylated by Cdk7, a component of the basal transcription factor TFIIH (Orphanides et al., 1996). Progression into the elongation phase of transcription for most class II genes requires the action of positive elongation factor b (P-TEFb), a cellular kinase complex that phosphorylates the CTD at Ser2 positions and antagonizes negative factors associated with the initiating RNAPII complex such as negative elongation factor (NELF) and DRB sensitivity-inducing factor (DSIF) (Peterlin and Price, 2006; Zhou and Yik, 2006).
Core P-TEFb is a heterodimeric complex composed of the cellular kinase Cdk9 in association with a regulatory cyclin subunit being either, Cyclins T1, T2, or K (Peng et al., 1998; Fu et al., 1999). Although all of these complexes appear to function to a similar extent, Cyclin T1 is the predominant regulatory partner for Cdk9 in cells examined to date. The Cyclin T1/Cdk9 complex has been extensively studied, as the HIV-1 Tat protein specifically targets these P-TEFb molecules for efficient transcription of the proviral genome, an essential process for replication of the virus (Rice and Herrmann, 2003).
Given the importance of P-TEFb to the transcriptional activity of the cell, it is subject to considerable functional regulation. An initial level of P-TEFb regulation can be the differential expression of Cdk9 and the cyclin proteins within cells. In resting CD4+ T-cells and monocytes, the level of Cyclin T1 is substantially lower than those of Cdk9 and Cyclin T2. Upon T-cell activation or monocyte differentiation, the expression of Cyclin T1 is highly up-regulated while Cdk9 and Cyclin T2 protein levels remain fairly constant (Garriga et al., 1998; Herrmann et al., 1998; Ghose et al., 2001; Liou et al., 2002, 2006; Sung and Rice, 2006). A second mode of control over P-TEFb relies on the activation state of the Cdk9 subunit. The kinase activity of Cdk9 is dependent upon phosphorylation of the Thr186 residue within a region of the protein called the T-loop (Chen et al., 2004). Phosphorylation of the T-loop in Cdk family members induces a large conformation change in the protein that allows substrates to access the catalytic core of the enzyme (Morgan, 1995). A recent structural study has shown that the Cdk9 T-loop can be autophosphorylated (Baumli et al., 2008), a mechanism of auto-activation that has been seen in a number of kinases (Lochhead, 2009). Phosphatases specific for the Cdk9 T-loop have been identified recently (Chen et al., 2008; Wang et al., 2008). We refer to the phosphorylated-Thr186 Cdk9 (p-Cdk9) in association with Cyclin T1 as the “active” form of P-TEFb throughout this manuscript. It is currently believed that the active form of P-TEFb is regulated through reversible association between positive and negative regulators, namely the Brd4 protein and the 7SK snRNP, respectively (Nguyen et al., 2001; Yang et al., 2001; Byers et al., 2005).
In HeLa, cells roughly half of the active P-TEFb molecules are bound in the so-called “large” P-TEFb complex containing 7SK snRNA, HEXIM1 or HEXIM2, MePCE (BCDIN3), and PIP7S (LARP7) (Michels and Bensaude, 2008), while the other half of P-TEFb are referred to as the “small” P-TEFb complexes and consist of Cdk9, a cyclin partner, and the Brd4 protein. The 7SK snRNP complexes are repressed for kinase activity (Michels et al., 2003; Yik et al., 2003). In contrast, the P-TEFb/Brd4 complexes are capable of RNAPII-CTD phosphorylation and stimulation of transcriptional elongation (Jang et al., 2005; Yang et al., 2005). Brd4 is a bromodomain containing protein that directs the small P-TEFb complex to active chromatin regions that are marked by acetylated histones (Hargreaves et al., 2009). The dynamic equilibrium between the large and small P-TEFb complexes is dependent upon the transcriptional activity of the cell and this may play a crucial role in the global regulation of transcription (Zhou and Yik, 2006).
Given the multifaceted degree of interacting partners that P-TEFb has within the cell, and the effect that these interactions have on P-TEFb function, we were interested in determining the spatial organization and localization of the active T-loop phosphorylated form of P-TEFb at the cellular level. In the present study, we used a phospho-Thr186 Cdk9 specific antiserum to examine the subnuclear distribution of T-loop phosphorylated Cdk9 and the localization of both the large and small P-TEFb complexes by indirect immunofluorescence microscopy. We found that p-Cdk9 is present at numerous foci scattered throughout the non-nucleolar nucleoplasm and concentrates in larger speckle-like clusters that co-localize with nuclear speckles. Furthermore, in both HeLa cells and primary activated CD4+ T lymphocytes, the population of p-Cdk9 that co-localizes with Cyclin T1 and represents the active form of P-TEFb is almost entirely located within the speckle domains. We also show that neither is Thr186 phosphorylation or kinase activity required for Cdk9 localization within nuclear speckles. By dual immunolabeling HeLa cells for p-Cdk9 and the various phosphoforms of RNAPII, we found that T-loop phosphorylated Cdk9 co-localizes mostly with the hyperphosphorylated forms of RNAPII. In HeLa cells immunolabeled for Cyclin T1, SC35, and either Brd4 or HEXIM1, we observed that both the Brd4/Cyclin T1 and the HEXIM1/Cyclin T1 complexes co-localize with nuclear speckles. Lastly, we found that the Cdk9 inhibitor flavopiridol causes increased co-localization of the small P-TEFb complex with nuclear speckles and an abatement of the large complex from these domains. Our data further support the view that nuclear speckles are sites of storage/assembly of proteins involved in transcription and RNA processing and suggest that they may be sites of active P-TEFb function and exchange between the Brd4 and 7SK/HEXIM1 complexes.
Materials and Methods
Cell culture, immunofluorescence, and immunoblots
HeLa cells were grown in DMEM supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA) at 37°C with 5% CO2. Resting CD4+ T lymphocytes were isolated from healthy blood donors (Gulf Coast Regional Blood Center, Houston, TX) using the MACS CD4+ cell isolation kit II (Miltenyi Biotec, Bergisch Gladbach, Germany) and anti-CD30 microbeads to deplete activated cells as described previously (Ramakrishnan et al., 2009). The purity and resting phenotype of CD4+ T cells was determined by flow cytometry using a Beckman-Coulter XL-MCL cytometer with FITC-CD4, PE-CD3, PE-CD69, and FITC-CD25 antibodies. Cells were cytospun onto glass coverslips as described (Ramakrishnan et al., 2009). For inhibition of Cdk9, cells were incubated with 500 nM flavopiridol for 2h. Indirect immunofluorescence was conducted as previously described (Herrmann and Mancini, 2001) except in the case of the RNAPII antibodies in which 4% BSA (Sigma, St. Louis, MO) diluted in PBS was used as the blocking solution. Coverslips were mounted onto slides using ProLong Gold Antifade reagent (Invitrogen, Carlsbad, CA). Appropriate controls were conducted to demonstrate the specificity of both primary and secondary antibody labeling under all conditions. Deconvolution microscopy was conducted as described previously (Herrmann and Mancini, 2001) using a Carl Zeiss AxioVert S100 TV microscope and a DeltaVision Restoration Microscopy System. Multiple images containing several cells (minimum of 3 cells per field) taken in a Z-series of at least 10 focal planes were collected and examined. All images shown are representative 0.2 µm single Z-sections, are deconvolved, and were processed for presentation using Adobe Photoshop. Immunoblots were conducted as described previously (Wang et al., 2008).
Antibodies and competition assay
The phospho-Thr186-Cdk9 antibody (Cell Signaling Technology, Danvers, MA) was produced in rabbits and used at a dilution of 1:50 for immunofluorescence and 1:500 for immunoblotting. The competition assay to confirm specificity of the p-Cdk9 antibody was conducted by combining a working dilution of the antibody with the generating peptide at a 1:10 molar ratio. The antibody/peptide mixtures were then incubated with gentle shaking for 2 h at room temperature prior to use for primary staining in immunofluorescence. The Cyclin T1 antibody (T-18; Santa Cruz Biotechnology, Santa Cruz, CA) was used at a 1:200 dilution for immunofluorescence and at 1:1,000 for immunoblotting. The pan-Cdk9 antibody (C-20; Santa Cruz Biotechnology) was used at the dilutions of 1:500 and 1:5,000 for immunofluorescence and immunoblotting, respectively. The RNAPII antibodies (8WG16, H14, and H5) were purchased from Covance and used at a 1:500 dilution. The rabbit anti-HEXIM1 antibody was a kind gift from Jiemin Wong (Baylor College of Medicine) and used at a 1:5,000 dilution for immunofluorescence and a 1:20,000 dilution for immunoblotting. The rabbit anti-Brd4 antibody was a kind gift from Cheng-Ming Chiang (UT Southwestern) and was diluted 1:2,000 for immunofluorescence and 1:1,000 for immunoblotting. The SC35 and anti-Flag antibodies were both purchased from Sigma and used at a 1:10,000 dilution. Primary antibody detection was conducted using appropriate secondary antibodies conjugated to Alexa-488, Alexa-594, Alexa-647, or Texas Red and used at a 1:500 dilution. All Alexa conjugates were purchased from Invitrogen and the mouse-lgM specific Texas Red conjugate was purchased from Jackson ImmunoResearch Laboratories, West Grove, PA.
Plasmid transfection
HeLa cells were grown on glass coverslips in six-well dishes to 50% confluency prior to transfections. pBABE-Flag-Cdk9-IRES-eGFP, pBABE-Flag-Cdk9T186A-IRES-eGFP, and pBABE-Flag-Cdk9D167N-IRES-eGFP (500 ng) were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Cells were fixed at 24 h post-transfection and processed for immunofluorescence.
Results
Evaluation of phospho-Thr186-Cdk9 specific immunofluorescence
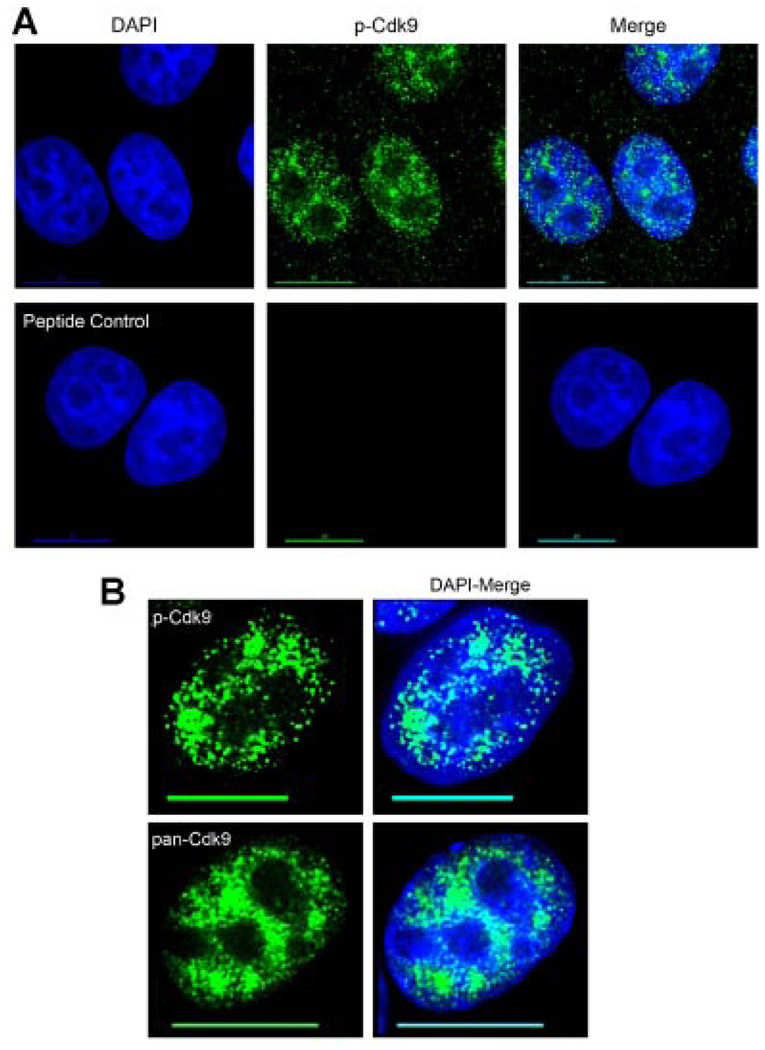
To determine the subcellular localization of T-loop activated Cdk9, we took advantage of a recently developed antiserum specific for the phosphorylated threonine-186 residue in the T-loop domain of Cdk9. We have shown previously with immunoblots that this antiserum recognizes wild-type Cdk9, but not a mutant Cdk9 protein in which the threonine-186 position has been changed to alanine (Wang et al., 2008). To determine if this antiserum is useful for immunofluorescence, we conducted a binding competition assay in which the generating peptide was incubated with the antiserum prior to the immunofluorescence procedure. HeLa cells labeled with either the p-Cdk9 antiserum alone or the pre-incubated peptide/antiserum mixture were then compared. As shown in Figure 1A, the phospho-Thr186 peptide completely abolished the p-Cdk9 signal demonstrating that the antiserum is specific for T-loop activated Cdk9.
Fig. 1.
The p-Cdk9 antibody is specific for T-loop activated Cdk9. Indirect immunofluorescence was conducted on HeLa cells to evaluate the specificity and staining pattern of the phospho-T186 antiserum (p-Cdk9). A: (upper parts) Cells were labeled using a 1:50 dilution of the p-Cdk9 antibody (determined as the optimum dilution by titration, data not shown) which was detected with an Alexa-488 (green) conjugated secondary antibody and the DNA was counterstained with DAPI. A: (lower parts) HeLa cells were labeled with a working dilution (1:50) of the p-Cdk9 antibody pre-incubated with a 10-fold molar excess of the peptide used to generate it. As can be seen, the cognate peptide effectively competes out the p-Cdk9 signal, demonstrating the specificity of this antiserum for use in indirect immunofluorescence. B: A comparison of the subnuclear distribution of T-loop activated Cdk9 (p-Cdk9) versus total Cdk9 (pan-Cdk9) shows that a more defined punctate, non-nucleolar staining pattern is seen with the p-Cdk9 signal with large clusters reminiscent of nuclear speckles. Bar, 10 µm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
As shown in Figure 1B, the p-Cdk9 antibody showed a more defined punctate nuclear staining pattern throughout the non-nucleolar space than did an antiserum that recognizes total Cdk9 (this antiserum is termed pan-Cdk9). Particularly evident was the enriched signal of large p-Cdk9 clusters in what looked to be nuclear speckle domains as compared to the total Cdk9 pattern (Fig. 1B). We note that although most of the labeling seen with the p-Cdk9 antibody was nuclear, we also saw a cytoplasmic population of p-Cdk9. This cytoplasmic signal is also seen with the pan-Cdk9 antibody (data not shown) and appears to be specific, as the generating peptide blocked the p-Cdk9 signal throughout the entire cell (Fig. 1A). A cytoplasmic and polysome-associated fraction of Cdk9 has been previously reported and we speculate that the T-loop activated Cdk9 found in the cytoplasm may be associated with polysomes (Napolitano et al., 2002; Rother and Strasser, 2007).
Active P-TEFb is located within nuclear speckles
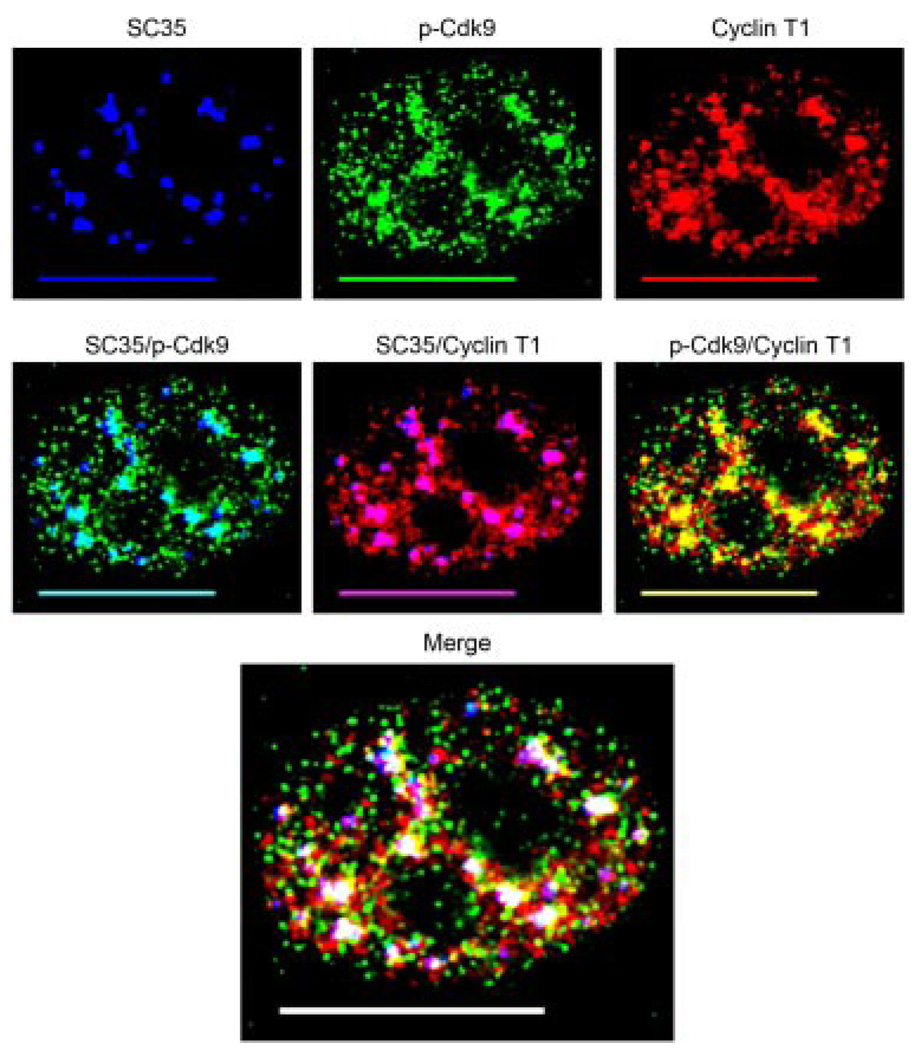
Previous work has shown that the P-TEFb subunits Cdk9 and CyclinT1 co-localize with nuclear speckles (Herrmann and Mancini, 2001). Nuclear speckles are a highly dynamic subdomain of the nucleus enriched in pre-mRNA splicing components and transcription factors (Lamond and Spector, 2003). To determine the extent to which T-loop activated Cdk9 co-localizes with Cyclin T1 and nuclear speckles, we triple immunolabeled HeLa cells for p-Cdk9, Cyclin T1, and the commonly used speckle marker SC35. As can be seen in Figure 2, the large clusters of p-Cdk9 mentioned above showed extensive co-localization with SC35 indicating they were indeed located within nuclear speckles (indicated by aqua, middle parts). Consistent with the findings of Herrmann and Mancini, we found considerable co-localization between Cyclin T1 and nuclear speckles, indicated by the purple color in the SC35 overlay (Fig. 2, middle parts) and as we expected, the p-Cdk9 signal co-localized with Cyclin T1 (Fig. 2, indicated by yellow). To our surprise, we found that almost all of the T-loop activated Cdk9 associated with Cyclin T1 was localized to the nuclear speckle domains (Fig. 2, indicated by white in the merge) with only a small fraction present outside these areas (Fig. 2, indicated by yellow in the merge). Collectively these data show that the active form of P-TEFb is located in nuclear speckles.
Fig. 2.
The active form of P-TEFb is located within nuclear speckles. HeLa cells were triple immunolabeled for SC35 (blue), p-Cdk9 (green), and Cyclin T1 (red) as shown in the upper parts. Co-localization of p-Cdk9 with SC35 (aqua), Cyclin T1 with SC35 (purple), and p-Cdk9 with Cyclin T1 (yellow) are shown in the middle parts. As can be seen, the large clusters of p-Cdk9 co-localize with nuclear speckles and to a high degree with Cyclin T1 (middle parts). The merge of all three channels shows that almost all of the active form of P-TEFb (p-Cdk9/Cyclin T1) is found within nuclear speckles. In the merge part areas of co-localization between p-Cdk9 and Cyclin T1 with nuclear speckles (SC35) appear as white, whereas those outside the speckle domains are indicated by yellow. Bar, 10 µm.
Co-localization of T-loop activated Cdk9 with RNAPII
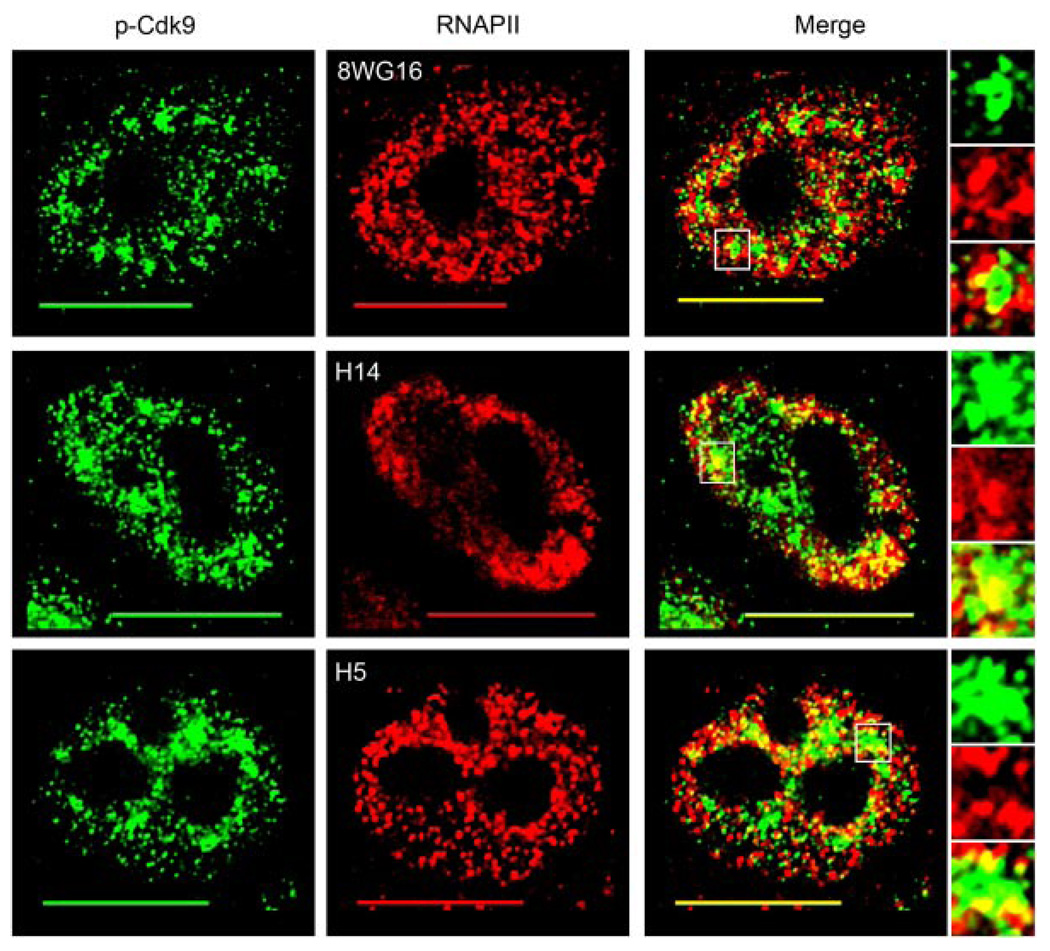
To find the degree to which T-loop activated Cdk9 co-localizes with RNAPII in cells, we conducted dual-label immunofluorescence using the p-Cdk9 antibody and antibodies specific for the various phosphoforms of the RNAPII CTD. RNAPII exists in cells primarily in two major forms – one are those with an unphosphorylated CTD and known as the hypophosphorylated form, while the second population are those that have been extensively phosphorylated, mainly at the Ser2 and/or Ser5 positions of the CTD and are known as the hyperphosphorylated forms (Phatnani and Greenleaf, 2006). The CTD of RNAPII is a major target of P-TEFb kinase activity and the different phosphorylation states of the CTD are associated with different functionality. The hypophosphorylated form is found in pre-initiation complexes and is recognized by the 8WG16 antibody (Thompson et al., 1989; Dahmus, 1994). The monoclonal antibody H14 recognizes phosphorylated Ser5 residues within the heptad repeats of the CTD, which occur via TFIIH activity and are required for transcription initiation (Bregman et al., 1994; Dahmus, 1994). The transition from initiating to elongating RNAPII complexes occurs when P-TEFb phosphorylates Ser2 residues within the CTD heptads and the monoclonal antibody H5 recognizes these phosphorylated epitopes (Warren et al., 1992; Marshall et al., 1996).
The results of our immunofluorescence analysis revealed that although there was some co-localization between the hypophosphorylated form of RNAPII and p-Cdk9, a large portion of T-loop activated Cdk9 co-localized with the hyperphosphorylated forms of RNAPII (Fig. 3). Moreover, the areas of co-localization between p-Cdk9 and all of the three forms of RNAPII predominantly occurred at or near the large clusters of p-Cdk9 determined in Figure 2 to be localized in nuclear speckles (Fig. 3 merge part insets). As can be seen in the enlarged insets, the 8WG16 antibody co-localizes with p-Cdk9 mostly around the periphery of these speckle-like regions whereas the H14 antibody co-localizes deeper into these domains (Fig. 3 compare upper merge part insets to middle). Upon analyzing the co-localization pattern seen with the H5 antibody, we found that the p-Cdk9 and H5 signals co-localized not only at the periphery of the speckle-like clusters, but also at well-defined punctae surrounding them (Fig. 3 lower merge part and insets). These data indicate that T-loop activated Cdk9 associates to a considerable extent with hyperphosphorylated RNAPII complexes within cells and that these interactions most likely take place at or near nuclear speckles.
Fig. 3.
T-loop activated Cdk9 co-localizes mostly with the hyperphosphorylated forms of RNAPII. HeLa cells were dual immunolabeled for p-Cdk9 (green) and the various phosphoforms of RNAPII (red). Co-localization between p-Cdk9 and hypophosphorylated RNAPII (8WG16), Ser5-phosphorylated RNAPII (H14), and Ser2-phosphorylated RNAPII (H5) is shown in the upper, middle, and lower merge parts, respectively. Areas of co-localization are indicated by the yellow color in the merge parts and an enlargement of a speckle-like cluster of p-Cdk9 (boxed region in merge) from each cell is shown in the insets. Bar, 10 µm.
Cdk9 does not require Thr186 phosphorylation or kinase activity for nuclear speckle localization
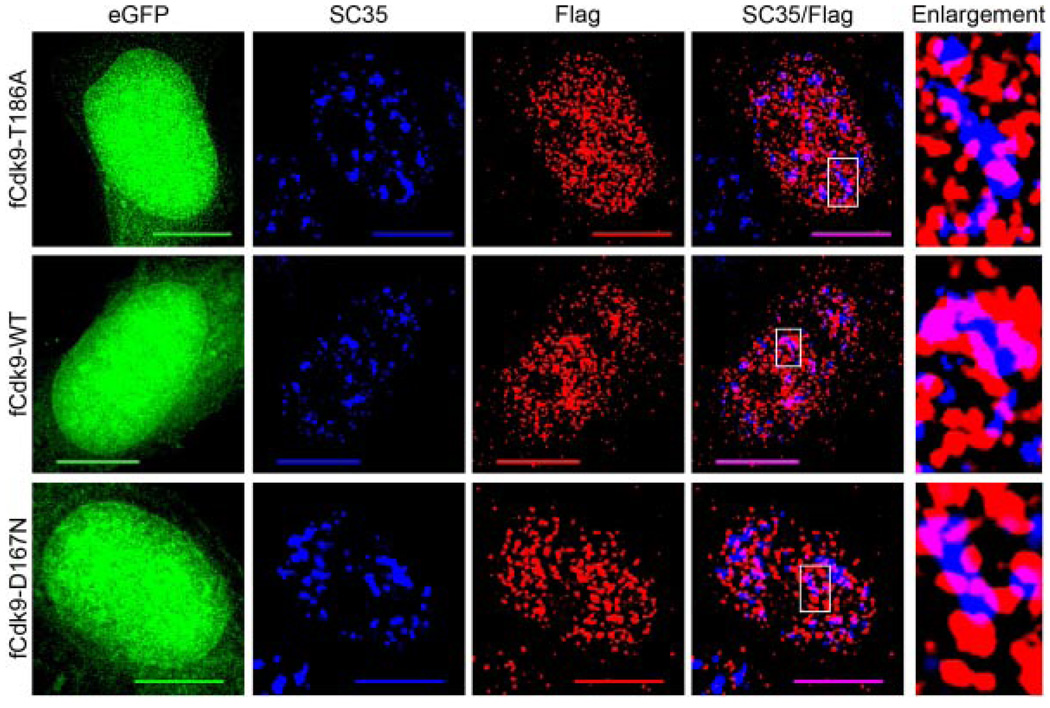
Upon finding that active P-TEFb (p-Cdk9/Cyclin T1) localizes predominantly within nuclear speckles, we were interested in determining if phosphorylation of Cdk9 at Thr186 was required for the association with the speckle domains, as phosphorylation/dephosphorylation has been found to act as a toggle for exit/entry of other speckle-associated proteins (Misteli et al., 1998). To test this, we transfected HeLa cells with a plasmid vector that expresses a Flag epitope-tagged Cdk9 mutant in which the Thr186 position was changed to Ala (fCdk9-T186A). We transfected another culture of HeLa cells with a plasmid vector that expresses the wild-type epitope-tagged Cdk9 (fCdk9-WT). The expression plasmids expressed eGFP from an IRES located at the 3′ end of the Cdk9 cDNA sequences. Transfected cells were then immunolabeled for Flag and SC35 and cells expressing similar levels of the eGFP internal control protein were then examined (Fig. 4). We observed no significant difference between the co-localization of SC35 with the fCdk9-T186A protein relative to that with the fCdk9-WT protein (indicated by purple in merged images, compare upper row to middle), suggesting that the phosphorylation of Thr186 in Cdk9 is not required for nuclear speckle localization.
Fig. 4.
Cdk9 does not require Thr186 phosphorylation or kinaseactivity for nuclear speckle localization. HeLa cells grown on glass coverslips were transfected with 500 ng of either: pBABE-Flag-Cdk9T186A-IRES-eGFP (fCdk9-T186A, upper parts), pBABE-Flag-Cdk9-IRES-eGFP (fCdk9-WT, middle parts), or pBABE-Flag-Cdk9D167N-IRES-eGFP (fCdk9-D167N, bottom parts) and fixed at 24 h post-transfection. The fixed cells were then immunolabeled for Flag (red) and SC35 (blue). The eGFP (green) was used as an internal control in conjunction with the Flag signal for assessing cells with similar expression levels. Immunoblots were also conducted to confirm that the expression levels of eGFP and the epitope-tagged Cdk9 proteins were similar in all cultures examined (data not shown). As can be seen, all three of the Flag-tagged Cdk9 proteins co-localize with SC35 (purple) to a similar extent, and show a similar non-nucleolar distribution throughout the nucleoplasm. An enlargement of a nuclear speckle from each of the three cells (boxed region on SC35/Flag parts) is shown on the right. Bar, 10 µm.
We also determined if the kinase activity of Cdk9 was a determinant of speckle localization. To do this, we transfected HeLa cells with a plasmid vector that expresses a kinase-defective Cdk9 mutant (fCdk9-D167N), and again compared the localization pattern of SC35 with this mutant Cdk9 protein and wild-type epitope tagged Cdk9 (Fig. 4). Similar to the Thr186 mutant, we found that the kinase-defective Cdk9 mutant showed no significant difference in co-localization with SC35 when compared to wild-type Cdk9 (Fig. 4, indicated by purple, compare bottom row to middle). These data suggest that neither Cdk9 kinase activity nor the phosphorylation of Thr186 is required for localization of Cdk9 within nuclear speckles.
Co-localization of Cyclin T1 with the Brd4 and HEXIM1 proteins
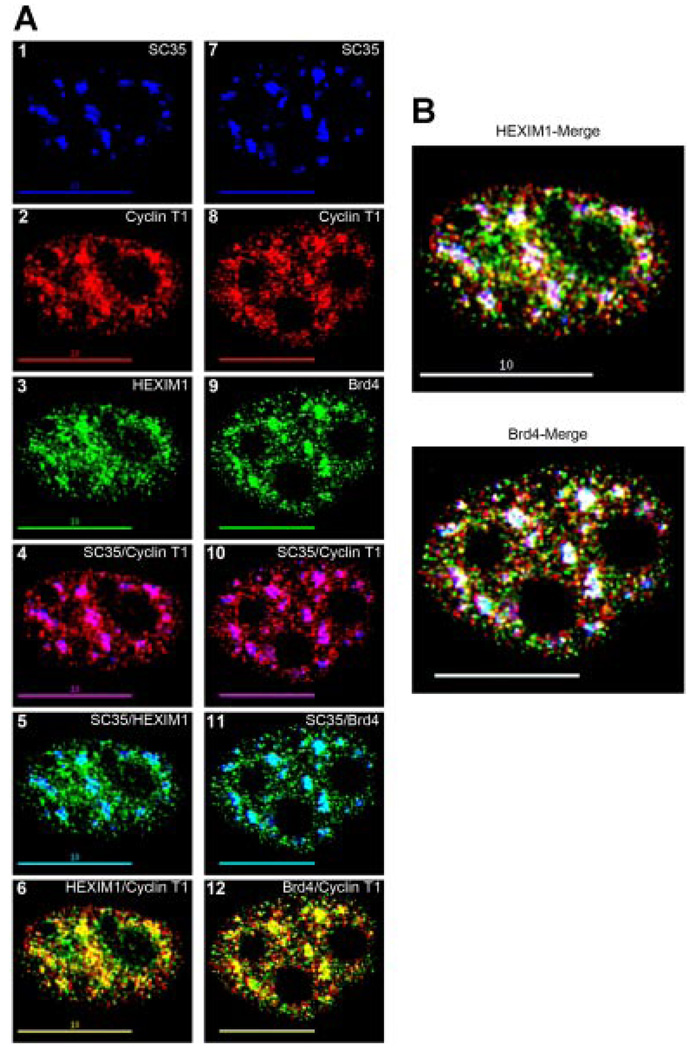
To further determine the potential function of active P-TEFb localization within nuclear speckles, we sought to examine the spatial organization of the small and large P-TEFb complexes in relation to the speckle domains. Although we could not observe the co-localization of p-Cdk9 with Brd4 or the HEXIM1 proteins because the antibodies suitable for immunofluorescence were all generated in rabbits, we were able to multi-label cells using an antiserum specific for Cyclin T1. Both the Brd4 and HEXIM1 proteins interact directly with Cyclin T1 (Jang et al., 2005; Dames et al., 2007), and we hypothesized that due to the functional differences between the association of Brd4 and HEXIM1 with P-TEFb, these interactions may take place in different regions of the nucleus.
We observed that both the Brd4 and HEXIM1 proteins were present throughout the nucleoplasm with exception of the nucleoli (Fig. 5A, parts 3 and 9). While co-localization with nuclear speckles was apparent with both Brd4 and HEXlM1 (indicated by aqua), in most cells examined the extent of direct overlap between Brd4 and SC35 was somewhat greater than that observed between HEXIM1 and SC35 (Fig. 5A, compare parts 5–11). Most often Brd4 was present throughout the majority of the SC35 area whereas the HEXIM1 protein quite often localized to the outer edges of the speckles.
Fig. 5.
Co-localization of the small and large P-TEFb complexes with nuclear speckles. A: HeLa cells were triple immunolabeled for SC35 (blue), Cyclin T1 (red), and HEXIM1 or Brd4 (green) as shown in parts 1–3 and 7–9, respectively. The co-localization of Cyclin T1 with SC35 (purple), HEXIM1 with SC35 (aqua), and Cyclin T1 with HEXIM1 (yellow) are shown in parts 4–6. The areas of co-localization between Cyclin T1 and SC35 (purple), Brd4 and SC35 (aqua), and Cyclin T1 with Brd4 (yellow) are shown in parts 10–12. As can be seen, both the HEXIM1 and Brd4 proteins co-localize with nuclear speckles (parts 5 and 11) and both HEXIM1 and Brd4 co-localize with Cyclin T1 (parts 6 and 12). B: Merge parts from the cells in A, in which co-localization of the large complex (HEXIM1/Cyclin T1) with nuclear speckles (SC35) is indicated by white and shown in the upper part, and co-localization of the small complex (Brd4/Cyclin T1) with nuclear speckles (SC35) is shown in the bottom part (also indicated by white). Areas of localization of the large and small complexes outside of the speckles regions are indicated by yellow in these parts. As can be seen, while both the large and small P-TEFb complexes co-localize with nuclear speckles (white), the small complex has a very tight co-localization pattern within the speckles, whereas the large complex localizes not only in the speckles, but also at many areas surrounding the outside of these regions (yellow). Bar, 10 µm.
As shown in Figure 5A, both Brd4 and HEXIM1 co-localized with Cyclin T1 at a number of nucleoplasmic sites (parts 6 and 12, indicated by yellow). We found that Cyclin T1 and Brd4 co-localized tightly within nuclear speckles (Fig. 5B, indicated by white in the Brd4-merge) with very few foci per focal plane of Brd4 associated Cyclin T1 located outside these regions (Fig. 5B, indicated by yellow in the Brd4 merge). While there was co-localization between Cyclin T1 and HEXIM1 in speckles, we noted that in the majority of the speckles we observed the co-localization was running along the edges of these domains (Fig. 5B, indicated by white in the HEXIM1-merge). Along with the speckle localized Cyclin T1/HEXIM1 fraction there was another population of HEXIM1 associated Cyclin T1 consistently seen outside the speckle regions (Fig. 5B, indicated by yellow in the HEXIM1-merge). Most often this was observed as a concentric gradient of yellow trending to white from outside to into the SC35 signal (Fig. 5B, HEXIM1-merge). These results imply that nuclear speckles are important sites of P-TEFb interaction with the Brd4 and HEXIM1 proteins.
Effects of flavopiridol on p-Cdk9, Brd4, and HEXIM1 co-localization with speckles
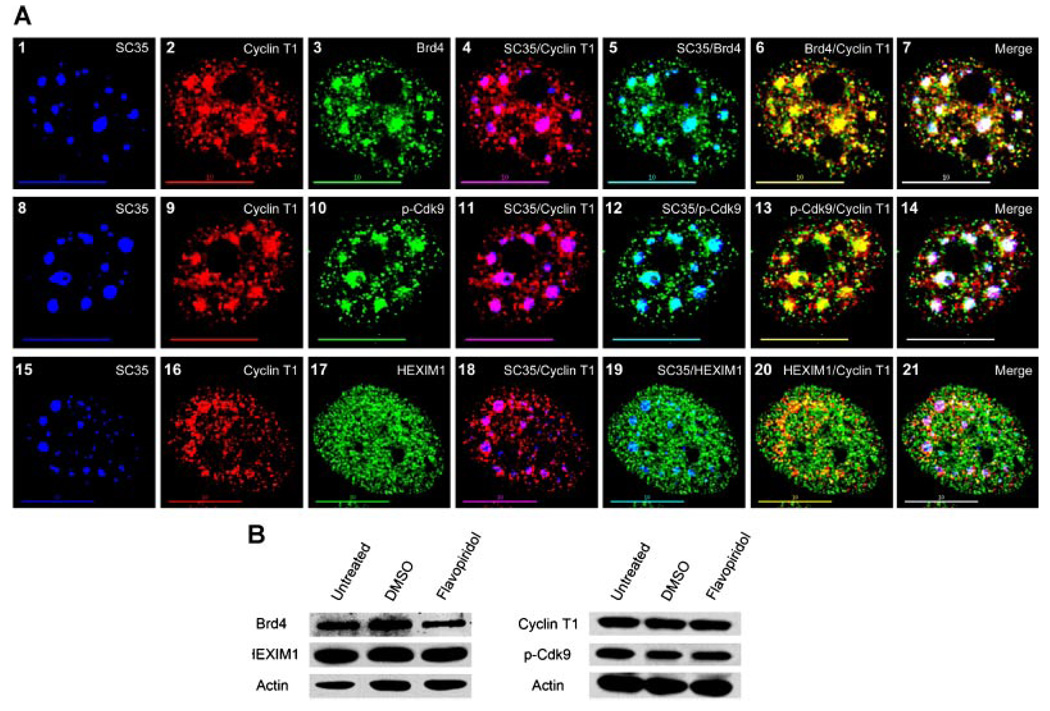
To address the question of why both negative (HEXlM1) and positive (Brd4) regulators of P-TEFb would localize to the same subnuclear site, we hypothesized that nuclear speckles may be the sites where active P-TEFb is being exchanged between these regulatory complexes. To test this idea, we treated HeLa cells with the Cdk9 inhibitor flavopiridol (Biglione et al., 2007; Baumli et al., 2008) and examined the subnuclear localization of active P-TEFb and both the Brd4 and HEXlM1 complexes under conditions where P-TEFb activity was inhibited. Previous work has determined that treatment of cells with the transcription inhibitors Actinomycin D and DRB causes almost complete dissociation of P-TEFb from the 7SK/HEXlM1 complex which is quantitatively shifted to the Brd4 complex (Yang et al., 2005). We reasoned that if speckles were indeed the sites where this exchange were to take place, then treatment with flavopiridol should cause an increased co-localization of the Brd4/P-TEFb complexes within the speckle domains and conversely a reduced co-localization of the 7SK/HEXlM1 complexes from these sites.
Figure 6A presents representative cells treated for 2 h with 500 nM flavopiridol and then triple-labeled for SC35 (blue), Cyclin T1 (red), and either Brd4 (green, upper parts), p-Cdk9 (green, middle parts), or HEXlM1 (green, bottom parts). The effect of transcription inhibitors in cells can be seen when speckles immunolabeled with SC35 become enlarged and rounded in appearance due to the concomitant reduction in splicing activity (O’Keefe et al., 1994). This effect on the SC35 signal is seen in parts 1, 8, and 15 (Fig. 6A) demonstrating that the flavopiridol is indeed inhibiting transcription in these cells. As we expected from previous work (Herrmann and Mancini, 2001), the flavopiridol treatment caused Cyclin T1 to also coalesce into enlarged puncta (parts 2, 9, and 16) which co-localized with SC35 as indicated by the purple color in parts 4, 11, and 18 (Fig. 6A). A similar effect occurred with both Brd4 and p-Cdk9 in the presence of flavopiridol as shown in parts 3 and 10, causing the proteins to co-localize tightly with SC35 indicated by the aqua color in parts 5 and 12 (Fig. 6A). However, the flavopiridol had the opposite result on the HEXlM1 distribution which became diffusely dispersed throughout the nucleoplasm (part 17) and greatly reduced its co-localization with SC35, which can be seen as the almost non-existent areas of aqua color in part 19 (Fig. 6A). Furthermore, the co-localization patterns of Brd4, p-Cdk9, and HEXlM1 with SC35 were analogous to the co-localization of these proteins with Cyclin T1 in the presence of flavopiridol (Fig. 6A, indicated by yellow in parts 6, 13, and 20). A merge of all three channels for each cell shows that flavopiridol does indeed cause increased co-localization of the small P-TEFb complex (Brd4/Cyclin T1) and active P-TEFb (p-Cdk9/Cyclin T1) with SC35 (indicated by white in parts 7 and 14), and conversely greatly reduces the co-localization of the large complex (HEXlM1/Cyclin T1) with SC35 (indicated by white in part 21).
Fig. 6.
Effects of flavopiridol on the localization of active P-TEFb and the small and large P-TEFb complexes. A: HeLa cells were treated with 500 nM flavopiridol for 2 h prior to immunofluorescence labeling for SC35 (blue), Cyclin T1 (red), and either Brd4 (green, upper parts), p-Cdk9 (green, middle parts), or HEXIM1 (green, bottom parts). As can be seen, flavopiridol causes the speckles to round up (parts 1, 8, and 15) and Cyclin T1 to co-localizegreatly with the nuclearspeckles (indicated by purple in parts4, 11, and 18). Similarly, both Brd4 and p-Cdk9 also greatly co-localize with speckles (indicated by aqua in parts 5 and 12) and with Cyclin T1 (indicated by yellow in parts 6 and 13). In contrast, flavopiridol causes HEXIM1 to become more diffusely scattered throughout the nucleoplasm (part 17) and abatement of its co-localization with speckles (part 19) and Cyclin T1 (part 20). A merge part for each cell specifies areas of co-localization between Brd4/Cyclin T1, p-Cdk9/Cyclin T1, and HEXIM1/Cyclin T1 complexes with nuclear speckles (indicated by white in parts 7, 14, and 21, respectively) and outside of speckles (indicated by yellow in parts 7, 14, and 21 respectively). Bar, 10 µm. B: HeLa cells were treated with 500 nM flavopiridol or DMSO (solvent control) for 2 h prior to preparation of cell lysates. The expression levels of the indicated proteins were examined in immunoblots. No significant differences in levels of Brd4, HEXIM1, Cyclin T1, or p-Cdk9 were observed in lysates of flavopiridol-treated cells versus control cells. This suggests that the dynamic shift of active P-TEFb from the large to the small complex may occur in nuclear speckles.
To confirm that the differences in the subnuclear distribution noted for active P-TEFb and the small and large complexes was due to the effect of flavopiridol on Cdk9 activity and not changes in the cellular concentration of the constituent proteins or levels of p-Cdk9, we conducted an immunoblot analysis of flavopiridol-treated and DMSO (solvent control) treated cells. We observed no changes in the levels of Brd4, HEXlM1, Cyclin T1 or p-Cdk9 in flavopiridol-treated cells relative to control cells (Fig. 6B). Taken together the data in Figure 6 imply that the exchange of active P-TEFb between the large and small P-TEFb complexes is likely to take place in nuclear speckles.
Active P-TEFb localizes to nuclear speckles in primary activated CD4+ T lymphocytes
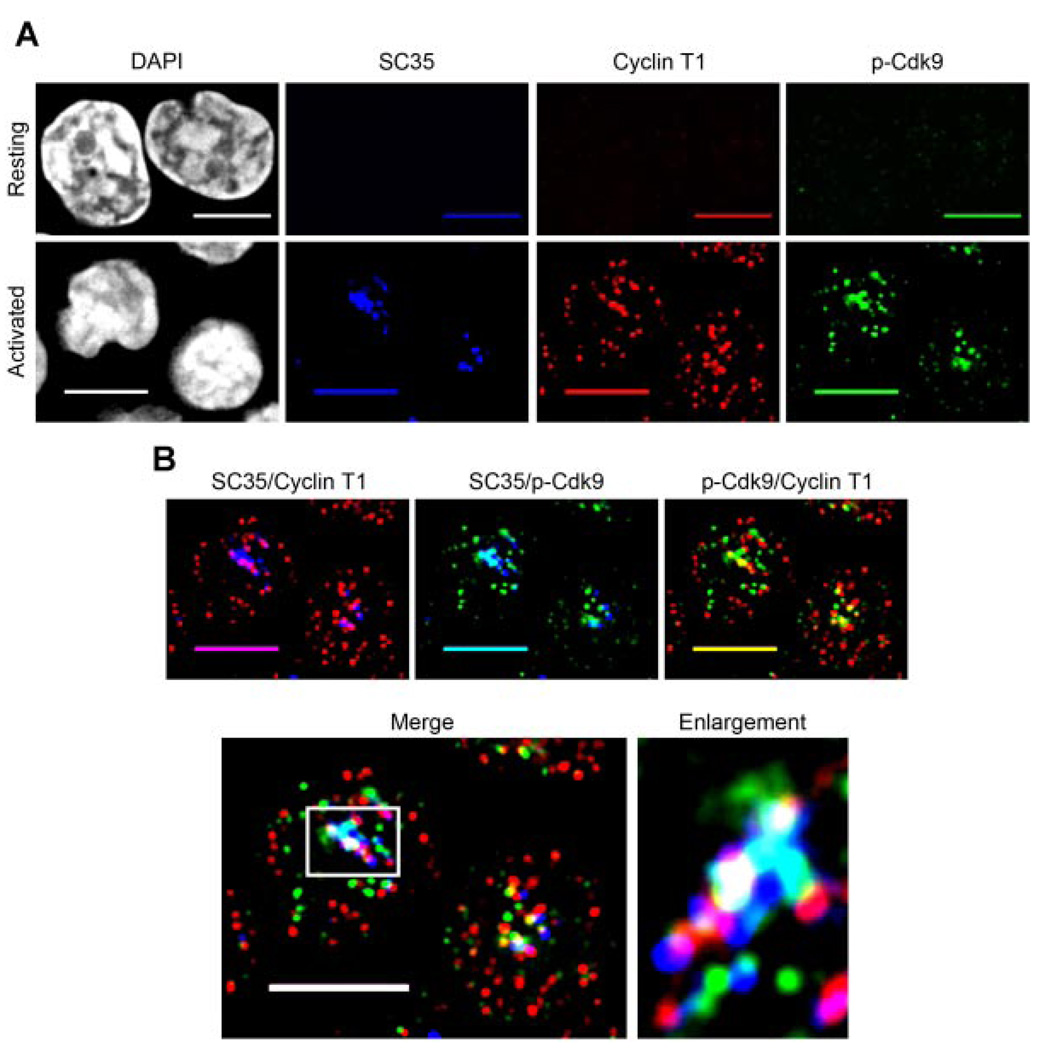
To determine if the localization pattern of active P-TEFb seen in HeLa cells is similar in a primary cell type in which P-TEFb function is highly regulated, we examined primary human CD4+ T lymphocytes. Cdk9 T-loop phosphorylation is low-to-undetectable in resting CD4+ T lymphocytes, and upon activation there is a rapid (< 1 h) increase in T-loop phosphorylation that does not require protein synthesis (Ramakrishnan et al., 2009). Cyclin T1 levels are also up-regulated in resting primary CD4+ T lymphocytes (Sung and Rice, 2006).
Resting CD4+ T lymphocytes were isolated from healthy blood donors and either left resting or activated for 1h with phorbol 12-myristate 13-acetate (PMA) + ionomycin prior to fixing and immunolabeling. We triple immunolabeled cells for Cyclin T1, p-Cdk9, and SC35. Interestingly, we observed that the expression pattern of SC35 follows that of Cyclin T1 in resting and activated CD4+ T lymphocytes (Fig. 7A). Consistent with our findings in HeLa cells, we found that indeed, active P-TEFb (Cyclin T1/p-Cdk9) is almost exclusively located within (indicated by white in the merge) or at the edge (indicated by yellow in the merge) of nuclear speckles in activated primary CD4+ T cells (Fig. 7B). These data not only support our observations of active P-TEFb localization in HeLa cells, but also indicate that in cells undergoing high levels of transcription, most of the P-TEFb activity likely occurs at or very near to nuclear speckle regions.
Fig. 7.
Active P-TEFb localizes to nuclear speckles in primary activated CD4+ T lymphocytes. A: Resting CD4+ T lymphocytes were cytospun onto glass coverslips and fixed immediately (upper parts) or activated (1 h) with PMA + ionomycin, cytospun onto glass coverslips and fixed (lower parts). The fixed cells were triple immunolabeled for SC35 (blue), Cyclin T1 (red), and p-Cdk9 (green), and the DNA was counterstained with DAPI to visualize nuclei. As can be seen, both the level of Cdk9 T-loop phosphorylation and the expression of Cyclin T1 and SC35 are highly up-regulated after 1 h of cell activation (compare upper to lower parts). B: Merge parts of the activated cells in A, showing the areas of co-localization between SC35 with Cyclin T1 (indicated by purple), SC35 with p-Cdk9 (indicated by aqua), Cyclin T1 with p-Cdk9 (indicated by yellow) and the co-localization of all three proteins (indicated by white). Ascan be seen, almost all of the active P-TEFb (CyclinT1/p-Cdk9) islocated either within or at the periphery of nuclear speckles in activated Tlymphocytes (merge part). This is particularly evident in the enlargement of a speckle region from one of these cells (boxed area on merge part) shown at the right of the merge part. The boxed region has been rotated 90° clockwise in the speckle enlargement for aesthetics. Bar, 5 µm.
Discussion
Active P-TEFb localization in nuclear speckles
Although previous work has determined that P-TEFb localizes to nuclear speckles (Herrmann and Mancini, 2001), this is the first report to specifically examine the subnuclear distribution of the active form of P-TEFb containing a phosphorylated T-loop. The development of a phospho-Thr186-Cdk9 specific antiserum made this analysis possible. The immunofluorescent pattern seen with the p-Cdk9 antiserum is similar yet more defined when compared to the pattern seen with a pan-Cdk9 antiserum, indicating that the T-loop activated Cdk9 population can be evaluated by immunofluorescent microscopy separately from total Cdk9 (Fig. 1B).
A previous study found that although both Cyclin T1 and Cdk9 co-localize with nuclear speckles, the enrichment of Cyclin T1 within these domains was more pronounced than that of Cdk9, and that the Cyclin T1 subunit of P-TEFb is responsible for recruiting Cdk9 into nuclear speckles (Herrmann and Mancini, 2001). Our results are consistent with these data and indicate that the fraction of Cdk9 present in nuclear speckles is phosphorylated at the Thr186 position and is associated with Cyclin T1, and thus is an active form of P-TEFb capable of inducing transcriptional elongation. Our analysis of transiently expressed mutant Cdk9 proteins indicates that neither Thr186 phosphorylation nor Cdk9 catalytic function is required for speckle localization (Fig. 4) and reiterates that Cyclin T1 is the component that directs P-TEFb to nuclear speckles.
In contrast to the enrichment of active P-TEFb in nuclear speckles, we found that although numerous foci of p-Cdk9 are seen in punctae outside of the speckle domains, only a few foci per focal plane were associated with Cyclin T1, and most often these sites of active P-TEFb were located very near the boundary of a speckle. Importantly, these observations were seen in both HeLa cells and in primary activated CD4+ T lymphocytes. It is possible that the p-Cdk9 found outside of the nuclear speckles is associated in other complexes with proteins other than Cyclin T1. We have been unable to evaluate the co-localization of p-Cdk9 with the Cyclin T2 and Cyclin K regulatory subunits, as their low expression levels and the quality of available antisera against these proteins do not allow an analysis of their subnuclear localization by immunofluorescent microscopy.
Co-localization of p-Cdk9 and RNAPII in relation to nuclear speckles
It is generally agreed upon that nuclear speckles are not sites of active transcription. However, polyA+ RNA and all three phosphoforms of RNAPII have been found to localize to nuclear speckles, as well as a fraction of phospho-Ser2 RNAPII that remains stably associated with speckles after transcription inhibition (Bregman et al., 1994; Cmarko et al., 1999; Lamond and Spector, 2003; Xie et al., 2006).
We found that although p-Cdk9 co-localizes with all three forms of RNAPII, it appears to have the most extensive co-localization with the hyperphosphorylated forms. Furthermore, the majority of co-localization between p-Cdk9 and all three forms of RNAPII occurred at or near the boundaries of the speckle-like clusters of p-Cdk9 (Figs. 2 and 3). These observations raise the possibility that P-TEFb functions to activate transcriptional elongation at or very near the periphery of nuclear speckles. In fact, previous reports have demonstrated that while the interior of nuclear speckles are transcriptionally inactive, Br-UTP incorporation into RNA transcripts is detected along the periphery of the speckle domains (Cmarko et al., 1999; Xie et al., 2006). Moreover, speckles appear to assemble in close proximity to sites of active transcription and live-cell imaging experiments have shown that speckle components move from speckles via peripheral extrusions to nearby sites of active transcription (Misteli et al., 1997; Trinkle-Mulcahy and Lamond, 2008).
Nuclear speckles may be the sites of active P-TEFb exchange between the Brd4 and 7SK/HEXIM1 complexes
The proposed functional role for nuclear speckles is that they serve as storage and/or assembly sites for pre-mRNA processing and transcription factors (Lamond and Spector, 2003). However, it is unclear what possible regulatory roles these subnuclear regions may serve for the cell, particularly those pertaining to the process of transcriptional elongation. Our finding that active P-TEFb localizes to nuclear speckles prompted us to examine the extent of co-localization of the Cyclin T1/Brd4 and Cyclin T1/HEXIM1 regulatory complexes with these sites. We found that Brd4 associated Cyclin T1 co-localizes tightly with nuclear speckles in HeLa cells, with very few foci located outside these regions. Upon analyzing the subnuclear distribution of HEXIM1 associated Cyclin T1, we found that although there is co-localization of these complexes with nuclear speckles, the areas of overlap were predominantly seen running along the outer region of the speckles. Moreover, we consistently found HEXIM1 associated Cyclin T1 localized to an area encircling the outside of the speckle boundary in the majority of cells examined. Taken together these data imply that Brd4 predominantly binds to P-TEFb within nuclear speckles while the 7SK/HEXIM1 complex predominantly associates with P-TEFb outside of these domains. Indeed, one previous study employing HEXIM1-YFP fusion constructs determined that a mutant of HEXlM1 that does not bind to 7SK or Cyclin T1 is greatly excluded from speckles and diffuse throughout the nucleoplasm; however, another HEXIM1 mutant which can bind to Cyclin T1 but not 7SK became highly concentrated in nuclear speckles (Barboric et al., 2005).
In light of the data reported here, we propose a model in which active P-TEFb is associated with Brd4 in nuclear speckles and this form of P-TEFb is recruited to RNAPII transcription complexes to activate transcriptional elongation. The Brd4-associated P-TEFb may then be released from the RNAPII complex and bound by the 7SK/HEXIM1 snRNP, which can then recycle the active P-TEFb back to speckles. In this way, a pool of active P-TEFb can be readily recruited for an immediate stimulation of transcription or alternatively, active P-TEFb can be stored at speckles during periods where the transcriptional activity of the cell is low. It is not clear whether Brd4 remains associated with active P-TEFb and the transcribing RNAPII complex through the entire transcription cycle or if it is released once P-TEFb is recruited to the template. However, ChIP analyses have shown that the Brd4 distribution is similar to that of Cdk9 throughout entire transcription units (Jang et al., 2005; Yang et al., 2005). Nevertheless, our data support the notion that Brd4 may act as a global factor for delivering P-TEFb from nuclear speckles to transcription templates. Because nuclear speckles are highly dynamic structures in which the constituent components can cycle continuously between the speckles and other nuclear sites, it is conceivable that active P-TEFb associated with Brd4 is shuttled from speckles to transcription templates, and then may shuttle back to speckles via 7SK/HEXlM1 complexes.
In summary, we have shown that the active form of P-TEFb as defined by a phosphorylated T-loop localizes to nuclear speckles and associates with both negative (HEXIM1) and positive (Brd4) regulatory proteins, as well as a primary substrate (RNAPII), all within or very near to speckle domains. Collectively, our data suggest that nuclear speckles may be sites of exchange between the large and small P-TEFb complexes and support the idea that speckles may function as sites of higher-order assembly and/or storage for transcription factors.
Acknowledgments
We thank the personnel of the Baylor Integrated Microscopy Core for all their advice. We thank Cheng-Ming Chiang (UT Southwestern) and J. Wong (Baylor College of Medicine) for antibodies and Karen Chiang for help in preparing Figures. The work was supported by National Institutes of Health Grant A135381 to A.P.R.
Contract grant sponsor: National Institutes of Health;
Contract grant number: A135381.
Literature Cited
- Barboric M, Kohoutek J, Price JP, Blazek D, Price DH, Peterlin BM. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. EMBO J. 2005;24:4291–4303. doi: 10.1038/sj.emboj.7600883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumli S, Lolli G, Lowe ED, Troiani S, Rusconi L, Bullock AN, Debreczeni JE, Knapp S, Johnson LN. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 2008;27:1907–1918. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Biglione S, Byers SA, Price JP, Nguyen VT, Bensaude O, Price DH, Maury W. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology. 2007;4:47. doi: 10.1186/1742-4690-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman DB, Du L, Li Y, Ribisi S, Warren SL. Cytostellin distributes to nuclear regions enriched with splicing factors. J Cell Sci. 1994;107:387–396. doi: 10.1242/jcs.107.3.387. [DOI] [PubMed] [Google Scholar]
- Buratowski S. The CTD code. Nat Struct Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- Byers SA, Price JP, Cooper JJ, Li Q, Price DH. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J Biol Chem. 2005;280:16360–16367. doi: 10.1074/jbc.M500424200. [DOI] [PubMed] [Google Scholar]
- Chen R, Yang Z, Zhou Q. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J Biol Chem. 2004;279:4153–4160. doi: 10.1074/jbc.M310044200. [DOI] [PubMed] [Google Scholar]
- Chen R, Liu M, Li H, Xue Y, Ramey WN, He N, Ai N, Luo H, Zhu Y, Zhou N, Zhou Q. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 2008;22:1356–1368. doi: 10.1101/gad.1636008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmarko D, Verschure PJ, Martin TE, Dahmus ME, Krause S, FX XD, van Driel R, Fakan S. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol Biol Cell. 1999;10:211–223. doi: 10.1091/mbc.10.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden JL. Tails of RNA polymerase II. Trends Biochem Sci. 1990;15:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- Dahmus ME. The role of multisite phosphorylation in the regulation of RNA polymerase II activity. Prog Nucl Acid Res Mol Biol. 1994;48:143–179. doi: 10.1016/s0079-6603(08)60855-7. [DOI] [PubMed] [Google Scholar]
- Dahmus ME. Phosphorylation of the C-terminal domain of RNA polymerase II. Biochim Biophys Acta. 1995;1261:171–182. doi: 10.1016/0167-4781(94)00233-s. [DOI] [PubMed] [Google Scholar]
- Dames SA, Schonichen A, Schulte A, Barboric M, Peterlin BM, Grzesiek S, Geyer M. Structure of the Cyclin T binding domain of Hexim1 and molecular basis for its recognition of P-TEFb. Proc Natl Acad Sci USA. 2007;104:14312–14317. doi: 10.1073/pnas.0701848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu TJ, Peng J, Lee G, Price DH, Flores O. Cyclin K functions as a CDK9 regulatory subunit and participates in RNA polymerase II transcription. J Biol Chem. 1999;274:34527–34530. doi: 10.1074/jbc.274.49.34527. [DOI] [PubMed] [Google Scholar]
- Garriga J, Peng J, Parreno M, Price DH, Henderson EE, Grana X. Upregulation of cyclin T1/CDK9 complexes during T cell activation. Oncogene. 1998;17:3093–3102. doi: 10.1038/sj.onc.1202548. [DOI] [PubMed] [Google Scholar]
- Ghose R, Liou LY, Herrmann CH, Rice AP. Induction of TAK (cyclin T1/P-TEFb) in purified resting CD4(+) T lymphocytes by combination of cytokines. J Virol. 2001;75:11336–11343. doi: 10.1128/JVI.75.23.11336-11343.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann CH, Mancini MA. The Cdk9 and cyclin T subunits of TAK/P-TEFb localize to splicing factor-rich nuclear speckle regions. J Cell Sci. 2001;114:1491–1503. doi: 10.1242/jcs.114.8.1491. [DOI] [PubMed] [Google Scholar]
- Herrmann CH, Carroll RG, Wei P, Jones KA, Rice AP. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J Virol. 1998;72:9881–9888. doi: 10.1128/jvi.72.12.9881-9888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: A model for nuclear organelles. NatRev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Liou LY, Herrmann CH, Rice AP. Transient induction of cyclin T1 during human macrophage differentiation regulates human immunodeficiency virus type 1 Tat transactivation function. J Virol. 2002;76:10579–10587. doi: 10.1128/JVI.76.21.10579-10587.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou LY, Haaland RE, Herrmann CH, Rice AP. Cyclin T1 but not cyclin T2a is induced by a post-transcriptional mechanism in PAMP-activated monocyte-derived macrophages. J Leukoc Biol. 2006;79:388–396. doi: 10.1189/jlb.0805429. [DOI] [PubMed] [Google Scholar]
- Lochhead PA. Protein kinase activation loop autophosphorylation in cis: Overcoming a Catch-22 situation. Sci Signal. 2009;2:e4. doi: 10.1126/scisignal.254pe4. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- Michels AA, Bensaude O. RNA-driven cyclin-dependent kinase regulation: When CDK9/cyclin T subunits of P-TEFb meet their ribonucleoprotein partners. Biotechnol J. 2008;3:1022–1032. doi: 10.1002/biot.200800104. [DOI] [PubMed] [Google Scholar]
- Michels AA, Nguyen VT, Fraldi A, Labas V, Edwards M, Bonnet F, Lania L, Bensaude O. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol Cell Biol. 2003;23:4859–4869. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Clement JQ, Krainer AR, Wilkinson MF, Spector DL. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J Cell Biol. 1998;143:297–307. doi: 10.1083/jcb.143.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- Napolitano G, Licciardo P, Carbone R, Majello B, Lania L. CDK9 has the intrinsic property to shuttle between nucleus and cytoplasm, and enhanced expression of cyclin T1 promotes its nuclear localization. J Cell Physiol. 2002;192:209–215. doi: 10.1002/jcp.10130. [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- O’Keefe RT, Mayeda A, Sadowski CL, Krainer AR, Spector DL. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J Cell Biol. 1994;124:249–260. doi: 10.1083/jcb.124.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Peng J, Zhu Y, Milton JT, Price DH. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan R, Dow EC, Rice AP. Characterization of Cdk9 T-loop phosphorylation in resting and activated CD4(+) T lymphocytes. J Leukoc Biol. 2009;86:1345–1350. doi: 10.1189/jlb.0509309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice AP, Herrmann CH. Regulation of TAK/P-TEFb in CD4+ T lymphocytes and macrophages. Curr HIV Res. 2003;1:395–404. doi: 10.2174/1570162033485159. [DOI] [PubMed] [Google Scholar]
- Rother S, Strasser K. The RNA polymerase II CTD kinase Ctk1 functions in translation elongation. Genes Dev. 2007;21:1409–1421. doi: 10.1101/gad.428407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, III, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: The short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Sung TL, Rice AP. Effects of prostratin on Cyclin T1/P-TEFb function and the gene expression profile in primary resting CD4+ T cells. Retrovirology. 2006;3:66. doi: 10.1186/1742-4690-3-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson NE, Steinberg TH, Aronson DB, Burgess RR. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J Biol Chem. 1989;264:11511–11520. [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Lamond AI. Nuclear functions in space and time: Gene expression in a dynamic, constrained environment. FEBS Lett. 2008;582:1960–1970. doi: 10.1016/j.febslet.2008.04.029. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dow EC, Liang YY, Ramakrishnan R, Liu H, Sung TL, Lin X, Rice AP. Phosphatase PPM1A regulates phosphorylation of Thr-186 in the Cdk9 T-loop. J Biol Chem. 2008;283:33578–33584. doi: 10.1074/jbc.M807495200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren SL, Landolfi AS, Curtis C, Morrow JS. Cytostellin: A novel, highly conserved protein that undergoes continuous redistribution during the cell cycle. J Cell Sci. 1992;103:381–388. doi: 10.1242/jcs.103.2.381. [DOI] [PubMed] [Google Scholar]
- Xie SQ, Martin S, Guillot PV, Bentley DL, Pombo A. Splicing speckles are not reservoirs of RNA polymerase II, but contain an inactive form, phosphorylated on serine2 residues of the C-terminal domain. Mol Biol Cell. 2006;17:1723–1733. doi: 10.1091/mbc.E05-08-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Yik JH. The Yin and Yang of P-TEFb regulation: Implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol Mol Biol Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]