Abstract
A novel protective monoclonal antibody (mAb) that recognizes a lipopolysaccharide (LPS) epitope common between serotypes Ogawa and Inaba of the O1 serogroup of Vibrio cholerae was characterized and the potential to develop peptide mimics of this protective LPS epitope was investigated. mAb 72.1 recognizes both Ogawa and Inaba LPS and it is vibriocidal and protective in passive immunization against infection by strains of both serotypes. The cDNA-derived amino acid sequence of mAb 72.1 is closely related to the previously characterized mAb ZAC-3, which is thought to recognize an epitope in the lipid A core region of O1 LPS. In an attempt to develop a peptide mimic-based vaccine against V. cholerae, phage display libraries were screened with mAb 72.1 and 11 peptide mimics were identified. Remarkably, all of the peptide sequences identified from linear phage display libraries contained two cysteine residues, suggesting that mAb 72.1 preferentially binds to peptides constrained with a disulphide bond. One of the peptide mimics was immunologically characterized. Although immunization of mice with this peptide mimic conjugated to KLH elicited antibodies against the peptide itself, these antibodies did not cross-react with Ogawa or Inaba LPS. Effectiveness of a peptide mimic as a vaccine may depend on how well the peptide can mimic the carbohydrate interactions when binding to the anti-carbohydrate antibody. Thus, investigating how peptides and LPS bind to mAb 72.1 may be useful in improving current peptide mimics or designing more effective peptide mimics. Identification and characterization of novel protective anti-LPS antibodies may be useful in studying protective epitopes of LPS, which may help develop LPS-based therapeutics against V. cholerae.
INTRODUCTION
Cholera, caused by the Gram-negative bacterium Vibrio cholerae, remains a major health problem in many parts of the world. The disease is characterized by watery diarrhoea with a ‘rice water’ appearance. The long-term control of cholera depends on improving hygienic conditions, an uncontaminated water supply and appropriate sewage disposal. However, these improvements are a distant goal for many countries. Thus, the availability of an effective cholera vaccine is important for the prevention of cholera in affected areas (WHO, 2001). Currently available vaccines for cholera are based on killed, whole-cell, or live-attenuated formulations. The killed, whole-cell (WC and WCrBS) vaccines provide only partial short-term protection. The live-attenuated strains such as CVD 103-HgR have greatly improved efficacy in North American volunteers, but have not been proven efficacious in field trials (Hill et al., 2006; WHO, 2001). However, field trials of the live-attenuated strain Peru-15 have shown it to be safe, immunogenic and efficacious (Qadri et al., 2005).
Despite the success of live-attenuated vaccines, the search for a universally effective and safe vaccine is still ongoing. An alternative approach to vaccination is to develop defined, component formulations. LPS is one surface antigen to consider as a potential component of such a formulation. V. cholerae LPS induces protective humoral immune responses in humans and animal models during infection and vaccination. Thus, LPS is widely accepted as a protective immunogen for cholera vaccine development (Taylor et al., 2004). Typically, LPS is composed of three distinct domains: lipid A, core polysaccharide (inner and outer) and O-antigen, also known as O-specific polysaccharide (O-SP). Approximately 200 serogroups of V. cholerae have been identified based on the O-SP. However, only serogroup O1 and the newly emerged O139 have been associated with cholera epidemics. In contrast, intestinal and/or extraintestinal infections with non-O1/non-O139 serogroups or non-toxigenic O1 strains are rarely found and seem to be of less clinical significance (Reidl & Klose, 2002). After the O139 serogroup was first detected in 1992, it predominated as the cause of cholera in parts of Asia. However, currently the O139 serogroup accounts for only a minority of the cholera cases and the O1 serogroup is the predominant cause of cholera (Ryan et al., 2006). The O1 serogroup is divided into ‘classical’ and ‘El Tor’ biotypes. Each of these biotypes is further divided into Ogawa and Inaba serotypes. The O-SPs of the Ogawa and Inaba serotypes differ only by a single 2-O-methyl group, which is present in the terminal perosamine unit of the Ogawa O-SP and absent in the terminal perosamine residue of the Inaba O-SP (Chatterjee & Chaudhuri, 2003).
Although native LPS is immunogenic, it is highly toxic due to the lipid A component. Systemic LPS leads to inflammation, multiple organ failure, shock, and potentially death (Chatterjee & Chaudhuri, 2006). The O-SP by itself is poorly immunogenic (Gupta et al., 1992, 1998). In order to overcome the problems associated with LPS, synthetic O-SP sugar components can be conjugated to protein carriers (neoglycoconjugates) to recruit T cells. Although Ogawa neoglycoconjugates induced protective antibodies in mice (Chernyak et al., 2002), Inaba neoglycoconjugates induced only low-affinity antibodies that were not protective. This may be due to low intrinsic affinity of the epitope for Inaba-specific B-cell receptor. Another possibility is that Inaba-specific antibodies develop into protective antibodies only after extensive somatic hypermutations, which may be difficult to select (Meeks et al., 2004).
It is important to develop a V. cholerae LPS-based vaccine that can induce protective antibodies to both Inaba and Ogawa serotypes, as both serotypes cause disease. Individuals need to be immunized against both serotypes for optimal protection, as immune pressure in the infected population can drive serotype conversion during epidemics (Dalsgaard et al., 1997; Garg et al., 2000). Thus, it is desirable to include a protective LPS epitope that is common between the two serotypes in a subunit vaccine. One approach to include such a protective epitope in vaccine formulations is to identify peptide mimics of LPS by screening phage display libraries with an anti-LPS mAb that recognizes a protective epitope common to Ogawa and Inaba serotypes.
Several mAbs directed against V. cholerae O1 have been reported (Adams et al., 1988; Apter et al., 1993; Bougoudogo et al., 1995; Brayton et al., 1987; Colwell et al., 1992; Gustafsson et al., 1982; Gustafsson & Holme, 1983; Gustafsson, 1984; Lullau et al., 1996; Ramamurthy et al., 1995; Sugiyama et al., 1987; Winner et al., 1991). Most of these mAbs were studied for diagnostic purposes and only a few of them have been tested for their protective ability in mice. Three mAbs [S-20-4 (IgG1), A-20-6 (IgG1) and 2D6 (IgA mAb)] that are Ogawa-serotype specific, obtained by immunization with an Ogawa strain, have been associated with protection against the homologous serotype (Apter et al., 1993; Bougoudogo et al., 1995; Winner et al., 1991). Two mAbs (IgA ZAC-3 and IgG3 I-24-2) obtained by immunization with an Inaba strain are cross-reactive with both Ogawa and Inaba LPS. I-24-2 protects neonatal mice against V. cholerae O1 serotypes Ogawa and Inaba (Bougoudogo et al., 1995). The protective efficacy of ZAC-3 has not been reported. Inhibition studies revealed that the binding of I-24-2 to Ogawa and Inaba LPS may be mediated by a region located at the junction of the O-SP and core region of the LPS (Villeneuve et al., 1999; Wang et al., 1998). The ZAC-3 mAb binds to an epitope in the lipid A or core region but not to the O-SP (Lullau et al., 1996). In agreement with this reported specificity for the lipid A or core region, Wang et al. (1998) demonstrated that ZAC-3 does not bind to any synthetic ligand related to the O-SP of the Ogawa or Inaba serotypes.
Peptide mimics of surface carbohydrate structures of several pathogens and tumours have been identified by panning and screening phage display libraries with mAbs. Some of these peptide mimics have been used as immunogens to elicit cross-reactive carbohydrate-directed responses (De Bolle et al., 1999; Lesinski et al., 2001; Phalipon et al., 1997; Pincus et al., 1998; Shin et al., 2002)
In this study, we generated a protective mAb that recognizes an epitope common to both Ogawa and Inaba serotypes. The variable heavy and light chain sequences of this mAb are closely related to those of the previously characterized mAb ZAC-3, which recognizes an epitope located in either lipid A or the core region. In an attempt to utilize this protective epitope in vaccine formulations, phage display libraries were screened with this mAb to identify peptide mimics.
METHODS
Generation of mAbs.
The mAbs were generated with standard techniques following a protocol approved by the Institutional Animal Care and Use Committee. Four- to six-week-old female BALB/c mice were immunized intraperitoneally with a lysate containing LPS prepared from V. cholerae O395 (Ogawa) and Titermax adjuvant (Sigma). After three immunizations at monthly intervals, mice splenocytes were fused to NS1 cells (ATCC) using standard techniques and hybridoma supernatants were screened for reactivity by ELISA. Initial screening indicated that one of the hybridoma cell lines produced a mAb specific for LPS, designated mAb 72.1. This cell line was subcloned and further characterized.
Determination of mAb isotype.
Microtitre plates were coated with serial dilutions of mAb 72.1 in 0.1 M NaHCO3, pH 8.6, overnight at 4 °C. Plates were blocked with blocking buffer containing 5 mg BSA ml−1 and 0.1 M NaHCO3, pH 8.6. After washing the plate, isotype-specific antisera conjugated to horseradish peroxidase (HRP) (Southern Biotech) diluted 1 : 2000 in TBST (50 mM Tris/HCl, 150 mM NaCl, 0.5 % Tween-20) was added for 1 h. Following an additional washing step, the colorimetric substrate TMB (3,3′,5,5′-tetramethylbenzidine, Sigma) was added. After the colour developed, the reaction was terminated by addition of 3 M HCl, and absorbance was measured at 450 nm with a kinetic microplate reader (Molecular Devices).
Purification of mAbs.
Saturated (NH4)2SO4 was added to 2 l hybridoma supernatant until the final concentration reached 45 % (NH4)2SO4. The supernatant was then centrifuged at 4000 g for 1 h. The resulting pellet was resuspended in PBS, pH 7.4, and dialysed overnight against PBS at 4 °C. mAb 72.1 was further purified using a Montage Antibody Purification kit with PROSEP-A media (Millipore). Purified antibody was concentrated using Amicon Ultra-15 centrifugal filter devices with a 30 000 NMWL Ultracel regenerated cellulose membrane (Millipore) and dialysed against PBS. Purity of mAbs was determined by Coomassie-staining of SDS-PAGE gel and the antibody was quantified by Bradford assay (Bio-Rad).
Indirect ELISA.
Microtitre plates were coated with 5 μg ml−1 of V. cholerae Ogawa (a gift from Dr S. Kondo, Josai University, Japan) or Inaba (Sigma) LPS in 0.1 M Na2CO3/NaHCO3, pH 9.5, or 5 μg peptide-BSA ml−1 in 0.1 M NaHCO3, pH 8.6. Plates were blocked with blocking buffer containing 1 % fish gelatin (BioFX Laboratories) in PBS for 2 h at room temperature. Serial dilutions of serum or purified antibody were added to each plate and incubated at 4 °C overnight. After washing, bound antibodies were detected by HRP-conjugated goat anti-mouse IgG or IgM (Southern Biotech). The amount of bound antibodies was reported as absorbance at 450 nm (A450).
LPS competitive inhibition.
For the LPS/LPS competition assay, microtitre plates were coated with 5 μg ml−1 of Ogawa or Inaba LPS as described above. A constant amount of mAb 72.1 (0.4 μg ml−1) was added together with different concentrations of V. cholerae O1 Ogawa or Inaba LPS or Escherichia coli. The bound mAb 72.1 was detected with HRP-conjugated anti-IgG Ab.
Immuno-field emission scanning electron microscopy (FESEM).
For immunolabelling of the O395 Ogawa or 569B Inaba bacterial cells using mAb 72.1, 100 μl overnight culture was incubated on top of Formvar-coated coverslips for 1 h at 30 °C. The coverslips were fixed for 1 h in 4 % paraformaldehyde, 0.1 M sodium cacodylate at room temperature. Samples were washed twice in TBST before blocking in 3 % BSA/TBST for 1 h, and were then incubated with either mAb 72.1 or normal mouse serum (NMS) for 2 h. After several wash steps, the samples were incubated with goat anti-mouse secondary antibodies (Jackson ImmunoResearch Laboratories) conjugated to 10 nm colloidal gold particles at a 1 : 50 dilution for 30 min. Samples were washed again several times before a final fixation with 2.5 % glutaraldehyde, 0.1 M sodium cacodylate for 3 h. The samples were then washed with 0.1 M sodium cacodylate (pH 7.4), dehydrated through an ethanol series, and critical-point dried using a Samdri 795 (Tousimis). Dried samples were coated with 3 nm osmium, using an SPI plasma coater (SPI Supplies) before examination. Images were generated at 15 kV by an FEI XL-30 field emission gun scanning electron microscope (FEI Company).
Detection of vibriocidal antibody by the microtitre method.
A microtitre test was performed as described by Boutonnier et al. (2003). The vibriocidal assay is a well-accepted in vitro assay for assessing the capability of V. cholerae anti-LPS antibodies to facilitate complement-mediated killing. This assay measures the metabolic activity of the bacteria following treatment with antisera and complement. V. cholerae classical Ogawa strain O395 was inoculated into 2 ml APW (alkaline peptone water: 1.0 % peptone, 1.0 % NaCl, pH 8.6) and grown overnight at 37 °C. The culture was transferred to a pre-warmed nutrient agar plate and incubated for 90 min at 37 °C. Cold PBS was added on to the plate, gently swirled to resuspend the bacteria, and then transferred to a 15 ml conical tube. The OD600 of the bacterial suspension was adjusted to 0.9 with PBS to achieve a bacterial concentration of approximately 1×109 ml−1. Then 1 vol. bacterial suspension was mixed with 7 vols cold PBS and 2 vols guinea pig complement (Sigma), and incubated on ice for 20 min. Fifty microlitres of heat-inactivated, diluted, purified mAb 72.1 (1 mg ml−1) was placed in a round-bottom sterile microtitre plate and serially diluted in 1 : 2 increments in PBS. Fifty microlitres of complement-treated bacteria was added to each well and the plate was incubated uncovered in a humidified chamber for 2 h at 37 °C. Then 25 μl of an aqueous solution containing 1 vol. 1.0 % neotetrazoleum chloride (ICN Biochemicals) and 9 vols 2.7 % sodium succinate (ICN Biochemicals) was added to each well and the plate was incubated uncovered at room temperature for 15 min. Then the plate was placed in a humidified chamber at 4 °C overnight and the intensity of violet colour, after overnight incubation, indicating the presence of live metabolically active bacteria, was examined. The vibriocidal titre was reported as the reciprocal of the antibody dilution with the lowest concentration that caused bacterial killing, as indicated by a clear well. Data represent the mean end-point titres of three independent experiments.
Infant mouse challenge.
The infant mouse model for cholera was used to assess the protective quality of anti-LPS mAb 72.1 in vivo (Sun et al., 1990). V. cholerae O1 Ogawa strain O395 or Inaba strain 569B mixed with mAb 72.1 or NMS was orally administered into 4–5-day-old CD-1 infant mice, five in each group. Both strains were cultured overnight in LB medium at 37 °C and 10-fold serial dilutions were made in PBS. Each bacterial dilution was mixed 1 : 1 with purified antibody at 1 mg ml−1 or NMS and 50 μl was administered to each mouse by oral gavage. Challenged mice were kept at 30 °C and monitored after challenge, until the termination of the assay.
RNA isolation, cDNA synthesis and sequencing.
To determine the cDNA-derived amino acid sequence of variable heavy (VH) and variable light (VL) chains of mAb 72.1, total RNA was extracted from the hybridoma cells producing the mAb using TRIZOL (Invitrogen). The RNA was treated with DNase I (Ambion) and purified using RNeasy columns (Qiagen). cDNA was synthesized using TaqMan PCR Core Reagents kit (Roche). The reaction mixture (20 μl), containing maximally 1 μg total RNA, 2 μl 10× PCR buffer, 4 μl 25 mM MgCl2, 1 μl of each of the dNTPs (10 mM), 1 μl RNase inhibitor (20 units μl−1), 1 μl poly-(dT)15 primer (2 nM) and 1 μl Moloney murine leukaemia virus reverse transcriptase (50 units μl−1), with RNase-free water to bring the volume up to 20 μl, was incubated for 15 min at 42 °C. The reaction mixture was then directly used for PCR amplification. For VH amplification, primer for conserved framework one region, prMND85-72.1.VH.FR CTT CCG GAA TTC SAR GTN MAG CTG SAG SAG TCW GG, and IgG3 heavy chain primer, prMND84-72.1.VH.Ig3 GGA AGA TCT AGG GAC CAA GGG ATA GAC AGA TGG (mixed base code: R=A, G; Y=C, T; M=A, C; K=G, T; S=C, G; W=A, T; V=A, C, G; N=A, C, G, T) (Wang et al., 2000), were used. For VL amplification, primer specific for conserved framework one region, prMND82-72.1.VL.FR CCA GAT GTG AGC TCG TGA TGA CCC AGA CTC CA, and the κ chain-specific primer, prMND83-72.1.VL.κ GCC CGT CTA GAA TTA ACA CTC ATT CCT GTT GAA (Wang et al., 1998), were used. The reaction mixture contained 20 μl of the reaction mixture from the cDNA synthesis, 50 pmol forward and reverse primers, 8 μl 10× PCR buffer, 1 μl 25 mM MgCl2, 0.5 μl AmpliTaq DNA polymerase (5 units μl−1) and deionized water to bring the volume to 100 μl. The reaction mixture was subjected to 40 cycles of amplification. The PCR products were analysed on a 2.5 % Nusieve/Seakem gel. One hundred nanograms of the PCR product was subjected to dideoxyoligonucleotide termination reactions using a DNA sequencing kit (Perkin-Elmer) and the forward or reverse primers that were used for the amplification. The sequences of VL and VH were obtained by running the above reaction products through an automated DNA sequencer from Applied Biosystems.
Screening phage display libraries.
Phage display libraries in which 12 (X12) or seven (X7) amino acid random peptides or seven amino acid random peptides constrained by two cysteine residues (CX7C) displayed on the minor coat protein (pIII) of M13 phage were purchased from New England Biolabs (NEB). Phage were incubated with mAb 72.1 in solution, followed by affinity capture of the antibody–phage complexes onto protein A/G agarose beads as described by the manufacturer. Briefly, 50 μl protein A/G agarose (Pierce Biotechnology) was blocked for 1 h with blocking buffer (0.1 M NaHCO3, 5 mg BSA ml−1, pH 8.6) and washed with TBST. One microgram of mAb 72.1 and 2×1011 phage virions from a given phage display library were diluted with TBST to a final volume of 200 μl and incubated at room temperature for 20 min. The phage/antibody mixture was then transferred to the washed resin, incubated at room temperature for 15 min and washed with TBST to remove the unbound phage. The phage bound to resin were eluted with 1 ml glycine/HCl (pH 2.2) and rapidly neutralized with 150 μl 1 M Tris/HCl (pH 9.1). Eluted phage were titrated to determine the number of phage captured by the antibody-coated A/G agarose beads and were amplified by infecting E. coli strain ER2738 [F′ proA+B+ lacIq Δ(lacZ)M15 zzf : : Tn10(TetR)/fhuA2 glnV Δ(lac-proAB) thi-1 Δ(hsdS-mcrB)5, NEB]. Amplified phage were purified by precipitating the phage with PEG/NaCl (20 % polyethelene glycol, 2.5 M NaCl). Three or four rounds of panning were carried out until a large enrichment of phage was seen, indicated by a large increase in phage titre. After the enrichment, phage plaques were randomly picked and the gene III region was sequenced by automated dideoxyoligonucleotide sequencing as described in the NEB manual.
Phage-binding assays.
ELISA was used to assess the binding of phage to mAbs. First, 96-well microtitre plates were coated with serial dilutions of 72.1, S-20-4, A-20-6 or IgG3 isotype control antibody (Southern Biotech) in 0.1 M Na2CO3 (pH 8.6) overnight at 4 °C. Plates were blocked with blocking buffer containing 5 mg BSA ml−1. Approximately 1010 purified phage particles in TBST was placed in each of the wells and incubated for 1 h at room temperature. The bound phage were detected with HRP-conjugated anti-M13 (Pharmacia) antibodies diluted 1 : 3300 in TBST.
Phage competitive inhibition.
For the competition between LPS- and phage-displaying peptides, microtitre plates were coated with 0.05 μg mAb 72.1 ml−1, as described above. A constant amount of phage was added together with different concentrations of V. cholerae O1 Ogawa or Inaba LPS or E. coli LPS. The bound phage were detected with HRP-conjugated anti-M13 Ab.
Antibody binding to peptide-BSA.
Synthetic peptides representing peptide 3MB-1 (ACFFPNLSYC) and peptide 3ME-2 (SMCMHGGAYCFP) were chemically synthesized with GGGK at the C-terminus and the N-terminus acetylated. The disulphide bonds between the two cysteine groups were formed using Iodine deprotect. The cyclized peptide was conjugated to BSA or keyhole limpet haemocyanin (KLH) by the lysine at the C-terminus (Global Peptide). Binding of antibodies to peptide-BSA was determined by ELISA as described before.
Immunization of mice and serum collection.
Six-week-old female BALB/c mice (Charles River Laboratories), four in each group, were injected intraperitoneally with 50 μg 3MB-1-KLH in complete Freund's adjuvant (CFA) (Sigma), followed by two injections of 50 μg 3MB-1-KLH in incomplete Freund's adjuvant (IFA) (Sigma), 2 weeks apart. Mice injected with 50 μg KLH in CFA or IFA served as a control. Blood was collected before injection (pre-immune serum, P) and 10 days after every injection, representing primary (1′), secondary (2′) and tertiary (3′) sera.
RESULTS
Specificity of mAb 72.1
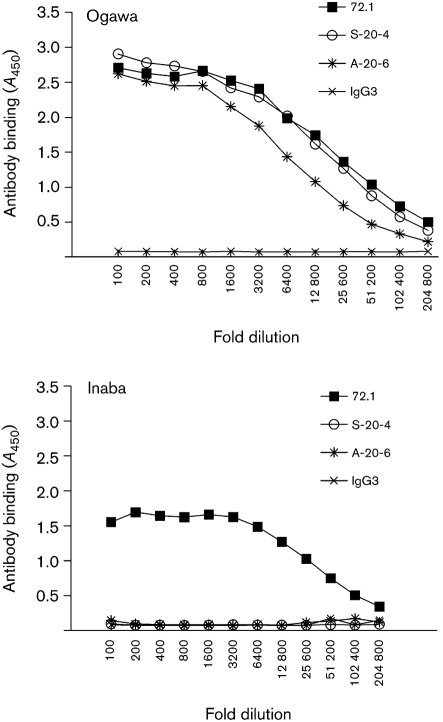
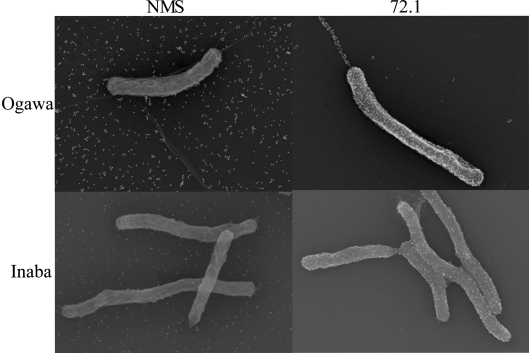
mAbs generated against Ogawa LPS were characterized for specificity using ELISA. One of the mAbs, 72.1, which has an isotype of IgG3 (see Methods), bound not only to Ogawa but also to Inaba LPS (Fig. 1). However, mAb 72.1 did not bind E. coli LPS (data not shown). Ogawa-specific mAbs, S-20-4 and A-20-6, which bound only to Ogawa LPS, served as controls. IgG3 isotype control antibody did not bind to Ogawa or Inaba LPS. In order to further confirm mAb 72.1 recognition of LPS, FESEM was performed. Consistent with mAb 72.1 specifically recognizing LPS in ELISA, mAb 72.1 bound to the bacterial cell surface and sheath flagella of both Ogawa and Inaba serotypes (Fig. 2). As a negative control, NMS did not bind to either serotype. These data demonstrate that mAb 72.1 recognizes an epitope of LPS common to both Ogawa and Inaba serotypes.
Fig. 1.
mAb 72.1 specifically recognizes Ogawa and Inaba serotype LPS as determined by ELISA. Plates were coated with Ogawa LPS or Inaba LPS. Serial dilutions of 1 mg ml−1 stocks of 72.1, S-20-4, A-20-6, IgG3 isotype control antibody were incubated in the coated wells. Bound antibodies were detected with anti-mouse IgG-HPR.
Fig. 2.
Detection of antibody binding to Ogawa and Inaba serotype bacteria by FESEM. mAb 72.1 specifically binds to Ogawa and Inaba serotype bacteria but NMS does not bind to bacteria of either serotype.
mAb 72.1 is vibriocidal and protective in the infant mouse
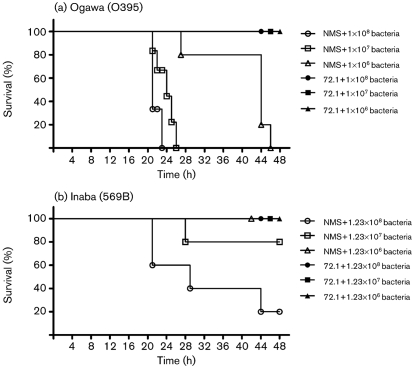
Purified mAb 72.1 was tested for its vibriocidal activity and for its ability to protect infant mice from V. cholerae O1 of either the Ogawa or Inaba serotypes. The purified mAb 72.1 (1 mg ml−1) killed both Ogawa and Inaba serotype V. cholerae by fixing complement. The mean vibriocidal titre for Ogawa and Inaba was 1280 and 1067, respectively. Consistent with this finding, anti-LPS mAb 72.1 (1 mg ml−1) was 100 % protective in infant mice when inoculated with various doses of V. cholerae of the Ogawa serotype (O395) and Inaba serotype (569B). All the doses of V. cholerae of Ogawa serotype killed 100 % of the control mice whereas the highest dose of V. cholerae of Inaba serotype killed 80 % of the control mice (Fig. 3). In previous studies, V. cholerae strain 569B has been shown to be attenuated in virulence due to the deletion of 1200 bp of DNA downstream from toxR, removing toxS (Miller et al., 1989). ToxR/S are inner-membrane regulatory proteins that activate the production of various virulence factors.
Fig. 3.
Protection of infant mice from experimental cholera infection by passive immunization with purified mAb 72.1. Percentage survival of infant mice following oral challenge with different doses of live V. cholerae Ogawa (O395) (a) or Inaba (569B) (b) and 1 mg ml−1 mAb 72.1 or NMS control. Five mice were used for each group. Survival curves were drawn using GraphPad Prism 5.0.
cDNA-derived variable heavy and light chain sequences of mAb 72.1
Since mAb 72.1 had several characteristics that were similar to previously characterized V. cholerae anti-LPS mAbs I-24-2 (IgG3) and ZAC-3 (IgA), cDNA-derived amino acid sequences of the heavy and light chain variable regions were determined and compared with mAbs I-24-2 and ZAC-3 (Fig. 4). Although the variable heavy chain sequence of mAb 72.1 (IgG3) was very similar to that of mAb ZAC-3 (86 % sequence identity), it greatly differed from mAb I-24-2. The light chain sequences of all three mAbs were more similar. The light chain of mAb 72.1 has 94 % and 77 % sequence identity with the ZAC-3 and I-24-3 light chain sequences, respectively.
Fig. 4.
Comparison of cDNA-derived variable heavy chain (VH) and variable light chain (VL) amino acid sequences of the anti-LPS mAb 72.1 (IgG3) with previously determined sequences of mAbs I-24-2 (IgG3) and ZAC-3 (IgA) (Wang et al., 1998). The heavy chain sequences of mAb 72.1 and mAb ZAC-3 are very similar but that of mAb I-24-2 is very different. The light chain sequences of all three mAbs are more similar. Dashes represent amino acids that are identical to mAb 72.1, and residues that differ from mAb 72.1 are indicated.
Selection of mAb 72.1-specific phage clones
Since LPS itself needs to be detoxified for any type of prophylactic or therapeutic use, we investigated the possibility of identifying peptide mimics of the protective epitope that is recognized by mAb 72.1. X12, X7 and CX7C phage display libraries were bio-panned against mAb 72.1. After enrichment, 12 plaques were randomly selected and sequenced. Eleven unique sequences that bound to mAb 72.1 were identified from three different phage display libraries (Table 1). Most of these sequences were found to occur multiple times amongst the enriched phage pools. Remarkably, all the sequences identified from linear X12 and X7 phage display libraries contained three to six amino acid residues conformationally constrained between two cysteine residues that form a disulphide bridge, as do the sequences from the cysteine-constrained CX7C phage display library. These results suggest that mAb 72.1 preferentially binds to peptides that are conformationally constrained. Although no single motif was present throughout all the peptides that were identified, tyrosine was found next to the second cysteine residue in almost all the sequences. Greater similarity was found among the peptides of the CX7C library than amongst those of the X12 library.
Table 1.
Binding of unique phage displayed peptides isolated by mAb 72.1 panning
| Library* | Unique sequences† | Antibody binding (A450)‡ | |||
|---|---|---|---|---|---|
| No antibody | IgG3 | S-20-4 | 72.1 | ||
| X12 | 3ME-1 ECLLLSKYCMPS | 0.263±0.024 | 0.486±0.166 | 0.472±0.193 | 3.216±0.094 |
| 3ME-2 SMCMHGGAYCFP | 0.213±0.046 | 0.505±0.195 | 0.444±0.100 | 3.113±0.022 | |
| 3ME-4 SSYCLRSVCLSD | 0.221±0.084 | 0.279±0.014 | 0.248±0.014 | 2.262±0.104 | |
| 3ME-6 WSSCVFSNSYCL | 0.152±0.038 | 0.232±0.055 | 0.231±0.047 | 2.824±0.034 | |
| 3ME-7 NQYCIYAGRYC | 0.258±0.131 | 0.283±0.041 | 0.307±0.006 | 2.561±0.014 | |
| 3ME-10 FCQPHLSCYILP | 0.195±0.084 | 0.219±0.065 | 0.173±0.038 | 2.580±0.014 | |
| 3ME-12 AAYCMTPYMCLH | 0.311±0.055 | 0.385±0.098 | 0.497±0.053 | 2.795±0.066 | |
| X7 | 4MC-1 CWNYCLP | 0.243±0.022 | 0.345±0.0037 | 0.325±0.065 | 2.697±0.015 |
| CX7C | 3MB-1 CFFPNLSYC | 0.266±0.011 | 0.439±0.149 | 0.467±0.122 | 2.702±0.071 |
| 3MB-4 CRQPGNNYC | 0.193±0.051 | 0.198±0.044 | 0.222±0.028 | 2.630±0.040 | |
| 3MB-9 CLFPNNSYC | 0.243±0.017 | 0.392±0.092 | 0.464±0.057 | 2.144±0.041 | |
| NA§ | 4P-8 NHNYPPLSLLTF | 0.248±0.064 | 0.432±0.031 | 2.978±0.013 | 0.349±0.055 |
*All the phage display libraries used have the random peptide expressed on pIII; X represents ‘randomized’ amino acids and C stands for fixed cysteines.
†Critical C residues are in bold type.
‡The indicated antibodies were coated onto wells of microtitre plates at 0.5 μg ml−1. The A450 values reflect the level of phage binding to the antibodies. Values are the means±sem of triplicate samples.
§Not applicable. The X12 library was panned with S-20-4 to identify phage clone 4P-8, which was used as a control (Dharmasena et al., 2007).
Binding properties of phage expressing pIII peptide mimics of the common epitope between Ogawa and Inaba LPS that is recognized by mAb 72.1
Each of the sequenced unique phage clones was analysed by ELISA to determine the degree of binding to antibodies 72.1, S-20-4, IgG3 isotype control and BSA (no antibodies). Of the unique sequences identified from the different phage display libraries that were used, 11 were capable of specifically binding to mAb 72.1, based on negligible binding to the IgG3 isotype control and Ogawa-specific mAb S-20-4 (Table 1). Phage clone 4P-8, which specifically binds to S-20-4, was used as a control phage (Dharmasena et al., 2007) and was not capable of binding to mAb 72.1.
Phage competitive inhibition
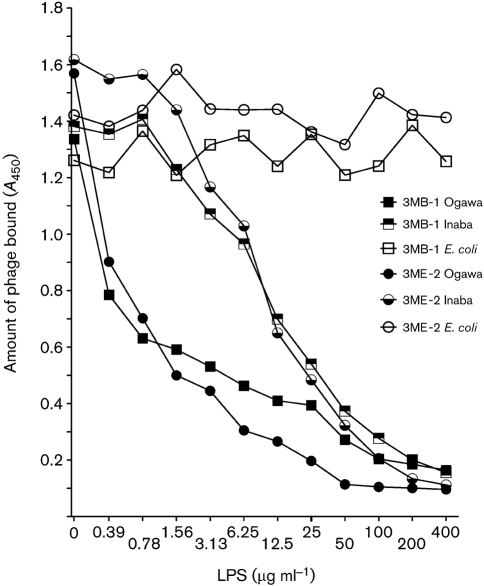
Out of the 11 unique sequences identified from different phage display libraries, two (3MB-1 and 3ME-2) that bound to mAb 72.1 with high affinity were selected for further studies. Another consideration was feasibility of chemical synthesis for subsequent immunization and binding studies. Both phage clones were capable of competing with Ogawa LPS and Inaba LPS for mAb 72.1 binding as tested by ELISA, suggesting that these phage clones encode peptide sequences that may bind to, or close to, the Ogawa and Inaba LPS binding site of mAb 72.1. Unlike V. cholerae Ogawa and Inaba LPS, E. coli LPS did not inhibit phage binding to mAb 72.1 (Fig. 5).
Fig. 5.
Competition between phage particles and LPS for binding to mAb 72.1. Phage binding to mAb 72.1 was decreased in the presence of increasing concentrations of Ogawa and Inaba LPS. Phage binding was not affected by the presence of E. coli LPS.
Antibody binding to peptide mimic-BSA conjugates
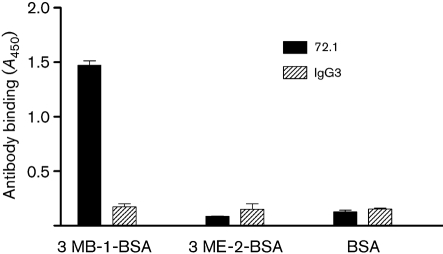
The 3MB-1 and 3ME-2 amino acid sequences were chemically synthesized and conjugated to BSA or KLH to further characterize their properties and to immunize mice. Synthetic peptides were used rather than the phage themselves because the peptides are a minor component in the context of pIII displayed on the phage (<5 molecules per phage). As shown in Fig. 6, mAb 72.1 bound to the synthetic 3MB-1-BSA, demonstrating that 3MB-1 does not have to be in the context of pIII to bind to mAb 72.1. Synthetic 3ME-2 conjugated to BSA did not bind to mAb 72.1, most likely due to a conformational difference of the peptide compared to when it is in the context of the phage.
Fig. 6.
ELISA of antibody binding to synthetic peptide conjugates. Plates were coated with 3MB-1-BSA, 3ME-2-BSA or BSA as a control. mAb 72.1 or IgG3 isotype control antibody was incubated in the coated wells. Bound antibodies were detected with anti-mouse IgG-HPR.
Immune responses elicited by peptide mimic-KLH conjugates
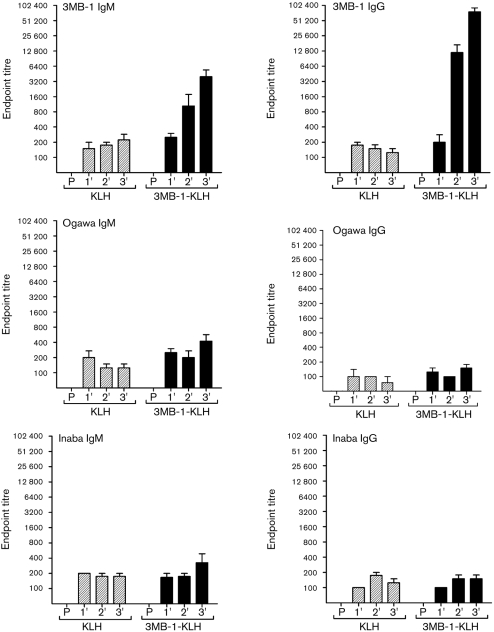
Since only 3MB-1 bound to mAb 72.1, 3MB-1-KLH was used to immunize mice to determine whether 3MB-1 could elicit cross-reactive anti-LPS antibodies. 3MB-1-KLH with CFA or IFA was injected into mice at 2 week intervals and serum was collected 10 days after each injection (see Methods). Each of the serum samples collected from 3MB-1-injected mice was tested by ELISA for the presence of anti-3MB-1 IgG and IgM. Serum collected from 3MB-1-KLH-injected mice showed very robust IgM and IgG antibody responses against 3MB-1, and antibody titres were significantly higher than those of pre-immune serum or serum collected from KLH-immunized mice. Anti-3MB-1 IgG and IgM antibody titres increased with the second and third 3MB-1-KLH injections. Each of the serum samples was also tested by ELISA to determine whether the anti-3MB-1 antibodies cross-reacted with Ogawa and Inaba LPS. Unexpectedly, anti-Ogawa and Inaba IgG and IgM antibody titres for serum collected from 3MB-1-KLH-immunized mice were not significantly higher than those for serum collected from KLH-immunized mice (Fig. 7).
Fig. 7.
IgM and IgG responses to 3MB-1, V. cholerae Ogawa LPS and V. cholerae Inaba LPS after intraperitoneal immunization of mice with KLH or 3MB-1-KLH. Plates were coated with 3MB-1-BSA, Ogawa LPS or Inaba LPS and serial dilutions of serum collected from 3MB-1-KLH- and KLH-vaccinated mice, or pre-immune serum (P), were incubated in the wells, followed by detection of 3MB-1-, Ogawa LPS- or Inaba LPS-bound antibodies by anti-mouse IgM-HPR or anti-mouse IgG-HPR. Data represent the means±sem of four values, representative of end-point titres for individual serum samples from each of the four mice in a group.
DISCUSSION
In this study we generated a mAb (72.1) that is vibriocidal and protects neonatal mice from V. cholerae O1 serotypes Ogawa and Inaba infection. The anti-LPS mAb 72.1 was obtained from a lymphocyte of mouse Peyer's patches following immunization of mice with a lysate that contained Ogawa LPS. cDNA-derived variable heavy and light chain amino acid sequences of mAb 72.1 were compared to previously determined sequences of mAbs ZAC-3 and I-24-2, which were generated following immunization of mice with a lysate containing Inaba LPS and which cross-reacted with both Ogawa and Inaba LPS. cDNA-derived variable heavy and light chain sequences of mAb ZAC-3 and variable light chain of mAb I-24-2 show similarity to mAb 72.1. However, the variable heavy chain of mAb I-24-2 differs extensively from those of 72.1 and ZAC-3. Previous studies have suggested that, while mAb ZAC-3 recognizes an epitope at the lipid A or core region, mAb I-24-2 recognizes an epitope at the junction of the core and O-SP region (Lullau et al., 1996; Villeneuve et al., 1999; Wang et al., 1998). Thus, mAbs ZAC-3 and I-24-2 may recognize different epitopes that are found on both O1 serotypes. Since the cDNA-derived variable heavy and light chain sequences of mAb 72.1, characterized in this study, show homology to mAb ZAC-3, it is possible that mAb 72.1 may also recognize an epitope in the lipid A or core region as does ZAC-3. However, further studies are necessary to determine whether mAb 72.1 recognizes the same epitope as mAb ZAC-3. Also ELISA and FESEM (Figs 1 and 2) suggest that mAb 72.1 binds more readily to Ogawa than Inaba LPS. The only known difference between Ogawa and Inaba LPS is at the terminal O-SP, where a single 2-O-methyl group is present in the terminal perosamine unit of the Ogawa O-SP and is absent in the terminal perosamine residue of the Inaba O-SP. Thus, we hypothesize that our new mAb 72.1 binds outside of the terminal perosamine, based on the fact that mAb 72.1 binds to both Ogawa and Inaba LPS.
In an attempt to determine if the protective epitope that is common to both Ogawa and Inaba serotype LPS can be utilized in a subunit vaccine against cholera, peptide mimics of this epitope were identified by screening phage display libraries with mAb 72.1. Eleven cyclic peptide mimics constrained by two cysteines were identified from three different phage display libraries. The frequency of cysteines in the 7-mer and 12-mer random phage display libraries from NEB that were used in this study is 0.5 %. Thus, having two cysteines in the random sequence is a rare occurrence. The cyclic peptides constrained with cystines may offer more conformational rigidity than linear peptides that are required to fit into the mAb 72.1 antigen-binding pocket. Although several other studies have identified cyclic peptide mimics from cystine-constrained phage display libraries (Brett et al., 2002; Moe et al., 1999; Tiwana et al., 2005), cyclic peptides are rarely identified from linear phage display libraries. There is one previous study that reported the identification of cyclic peptides with two cysteine residues from a linear phage display library (Coulon et al., 2004). In this study, a 10-mer random peptide library was screened with anti-colicin A mAb to determine its cognate epitope. The screen identified several groups of linear peptides and one group of peptides that contained two cysteines. Furthermore, the study demonstrated that the disulphide bond found in one group of the selected peptides was crucial for antibody recognition.
The conversion of peptides into efficient immunogens usually requires conjugation to an immunogenic carrier protein, presentation of the peptides as repeating units, or synthesis with T-cell epitopes. The two peptide mimics (3MB-1 and 3ME-2) that were selected for further studies here were chemically synthesized and conjugated to BSA for binding studies and to KLH for immunizations. 3MB-1 conjugated to BSA retained binding to mAb 72.1, suggesting the 3MB-1 conformation is not affected by its carrier protein. However, conjugation of 3ME-2 to BSA resulted in a loss of binding to mAb 72.1. This is likely to be due to conformational changes of the peptide when it is not in the context of the phage coat protein pIII.
The effectiveness of 3MB-1 in eliciting antibodies that cross-react with Ogawa and Inaba LPS was investigated. 3MB-1 was very effective as an immunogen in eliciting IgG and IgM antibodies against 3MB-1 itself. However, the anti-3MB-1 antibodies did not cross-react with either Ogawa or Inaba LPS at detectable levels. Thus, 3MB-1 is not a true mimotope. This lack of ability to induce a carbohydrate-reactive immune response is similar to what has been observed by others. Peptide mimics of Neisseria meningitidis group B capsular polysaccharide (CPS) that were identified from a mAb that does not cross-react with human tissue failed to elicit CPS cross-reactive antibodies (Lauvrak et al., 2004; Moe et al., 1999). Phalipon et al. (1997) identified 19 peptide mimics of Shigella flexneri serotype 5a LPS but out of 19 that were tested only two were able to induce cross-reactive anti-LPS antibody responses in mice. Scharff and co-workers identified a peptide mimetic (peptide PA1) of the capsular glucuronoxylomannan (GXM) of Cryptococcus neoformans by screening a phage display peptide library with the anti-cryptococcal polysaccharide mAb 2H1. However, PA1 failed to elicit significant titres of GXM cross-reactive antibodies (Valadon et al., 1996, 1998). In all these cases, selected peptides competed with the carbohydrate being mimicked, which suggests that the peptides bind to the same site or a site close to the carbohydrate-binding site on the anti-carbohydrate antibody. Still, carbohydrate-reactive immune responses by peptide immunizations are only rarely achieved. The inability of most peptide mimics to elicit cross-reactive immune responses against the native carbohydrate suggests that most widely used parameters for selecting peptide mimics, such as high-affinity binding to a relevant anti-carbohydrate antibody, competition with native carbohydrate antigen for antibody binding, or induction of a robust anti-peptide response, are insufficient and not predictive of whether a peptide is capable of eliciting an effective carbohydrate cross-reactive immune response. Although the features necessary for a peptide mimic to induce such an immune response are not clear, peptide binding to anti-carbohydrate antibodies by a similar mechanism as the carbohydrate, at least to some extent, are likely to be required for a peptide to effectively function as a mimotope. If the peptide and carbohydrate ligands are bound by similar groups on the antibody, structural mimicry is achieved. However, if the peptide and carbohydrate ligands are bound by different groups on the antibody, then the nature of the mimicry is functional and less likely to work appropriately from a vaccine standpoint. For example, previous studies have suggested the importance of structural mimicry in vaccine design. A peptide mimic of tumour-associated carbohydrate antigen ganglioside GD2, identified from a phage display library, was redesigned by structure-assisted computer modelling to increase the structural mimicry. The redesigned peptide was more efficient than the original peptide at eliciting cross-reactive antibodies to carbohydrate antigen (Monzavi-Karbassi et al., 2007). In another example, a peptide mimic of tumour-associated carbohydrate antigen Lewis Y was determined to be a structural mimic based on structure-assisted computer modelling studies. This peptide mimic was also efficient at eliciting cross-reactive antibodies to Lewis Y carbohydrate antigen (Luo et al., 2000).
In our previous studies, a peptide mimic of V. cholerae Ogawa LPS was found to be a partial structural mimic. Although this peptide mimic elicited cross-reactive antibodies against Ogawa LPS, those antibodies were not vibriocidal and could not passively protect neonatal mice from V. cholerae O1 serotype Ogawa infection. Since this peptide mimic is a partial structural mimic, it is only capable of eliciting low-affinity anti-LPS antibodies that are not vibriocidal or protective. However, priming mice with LPS prior to boosting with the peptide mimic resulted in prolonged LPS-reactive IgG and IgM antibody responses, as well as vibriocidal titres, and provided a much greater degree of protection than priming with LPS alone. Thus, if pre-existing immunity to LPS, elicited by either natural exposure or vaccination, exists, it could be extended by vaccination with such a peptide mimic (Dharmasena et al., 2007).
In the current study we characterized a novel anti-LPS mAb that recognizes an epitope common to both Ogawa and Inaba serotype LPS. The anti-LPS antibody was used to identify peptide mimics and one of the peptides was conjugated to KLH yet failed to elicit antibodies that cross-react with Ogawa or Inaba LPS. Investigation of how LPS and peptides bind mAb 72.1 may facilitate the design of an effective peptide mimic-based vaccine that protects against all O1 serogroup V. cholerae infections and will likely require crystallographic-structure and protein-modelling approaches. Apart from identifying peptide mimics, the characterization of novel anti-LPS antibodies is useful in studying protective epitopes of LPS. The protective epitope recognized by mAb 72.1 that we characterized in this study is of great importance, as a vaccine based on this protective epitope would protect against the majority of the V. cholerae infections (Ryan et al., 2006). mAb 72.1 may facilitate defining of this important protective epitope and help develop LPS-based therapeutics against V. cholerae.
Acknowledgments
We gratefully acknowledge Dr Peter M. Morganelli for generating hybridomas. This work was supported by NIH grant AI 025096 to R. K. T.
Abbreviations
CFA, complete Freund's adjuvant
FESEM, immuno-field emission scanning electron microscopy
HRP, horseradish peroxidase
IFA, incomplete Freund's adjuvant
KLH, keyhole limpet haemocyanin
NMS, normal mouse serum
O-SP, O-specific polysaccharide
References
- Adams, L. B., Henk, M. C. & Siebeling, R. J. (1988). Detection of Vibrio cholerae with monoclonal antibodies specific for serovar O1 lipopolysaccharide. J Clin Microbiol 26, 1801–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apter, F. M., Michetti, P., Winner, L. S., III, Mack, J. A., Mekalanos, J. J. & Neutra, M. R. (1993). Analysis of the roles of antilipopolysaccharide and anti-cholera toxin immunoglobulin A (IgA) antibodies in protection against Vibrio cholerae and cholera toxin by use of monoclonal IgA antibodies in vivo. Infect Immun 61, 5279–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougoudogo, F., Vely, F., Nato, F., Boutonnier, A., Gounon, P., Mazie, J.-C. & Fournier, J.-M. (1995). Protective activities of serum immunoglobulin G on the mucosal surface to Vibrio cholerae O1. Bull Inst Pasteur 93, 273–283. [Google Scholar]
- Boutonnier, A., Dassy, B., Dumenil, R., Guenole, A., Ratsitorahina, M., Migliani, R. & Fournier, J. M. (2003). A simple and convenient microtiter plate assay for the detection of bactericidal antibodies to Vibrio cholerae O1 and Vibrio cholerae O139. J Microbiol Methods 55, 745–753. [DOI] [PubMed] [Google Scholar]
- Brayton, P. R., Tamplin, M. L., Huq, A. & Colwell, R. R. (1987). Enumeration of Vibrio cholerae O1 in Bangladesh waters by fluorescent-antibody direct viable count. Appl Environ Microbiol 53, 2862–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett, P. J., Tiwana, H., Feavers, I. M. & Charalambous, B. M. (2002). Characterization of oligopeptides that cross-react with carbohydrate-specific antibodies by real time kinetics, in-solution competition enzyme-linked immunosorbent assay, and immunological analyses. J Biol Chem 277, 20468–20476. [DOI] [PubMed] [Google Scholar]
- Chatterjee, S. N. & Chaudhuri, K. (2003). Lipopolysaccharides of Vibrio cholerae. I. Physical and chemical characterization. Biochim Biophys Acta 1639, 65–79. [DOI] [PubMed] [Google Scholar]
- Chatterjee, S. N. & Chaudhuri, K. (2006). Lipopolysaccharides of Vibrio cholerae: III. Biological functions. Biochim Biophys Acta 1762, 1–16. [DOI] [PubMed] [Google Scholar]
- Chernyak, A., Kondo, S., Wade, T. K., Meeks, M. D., Alzari, P. M., Fournier, J. M., Taylor, R. K., Kovac, P. & Wade, W. F. (2002). Induction of protective immunity by synthetic Vibrio cholerae hexasaccharide derived from V. cholerae O1 Ogawa lipopolysaccharide bound to a protein carrier. J Infect Dis 185, 950–962. [DOI] [PubMed] [Google Scholar]
- Colwell, R. R., Hasan, J. A., Huq, A., Loomis, L., Siebeling, R. J., Torres, M., Galvez, S., Islam, S., Tamplin, M. T. & Bernstein, D. (1992). Development and evaluation of a rapid, simple, sensitive, monoclonal antibody-based co-agglutination test for direct detection of Vibrio cholerae O1. FEMS Microbiol Lett 76, 215–219. [DOI] [PubMed] [Google Scholar]
- Coulon, S., Metais, J. Y., Chartier, M., Briand, J. P. & Baty, D. (2004). Cyclic peptides selected by phage display mimic the natural epitope recognized by a monoclonal anti-colicin A antibody. J Pept Sci 10, 648–658. [DOI] [PubMed] [Google Scholar]
- Dalsgaard, A., Skov, M. N., Serichantalergs, O., Echeverria, P., Meza, R. & Taylor, D. N. (1997). Molecular evolution of Vibrio cholerae O1 strains isolated in Lima, Peru, from 1991 to 1995. J Clin Microbiol 35, 1151–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bolle, X., Laurent, T., Tibor, A., Godfroid, F., Weynants, V., Letesson, J. J. & Mertens, P. (1999). Antigenic properties of peptidic mimics for epitopes of the lipopolysaccharide from Brucella. J Mol Biol 294, 181–191. [DOI] [PubMed] [Google Scholar]
- Dharmasena, M. N., Jewell, D. A. & Taylor, R. K. (2007). Development of peptide mimics of a protective epitope of Vibrio cholerae Ogawa O-antigen and investigation of the structural basis of peptide mimicry. J Biol Chem 282, 33805–33816. [DOI] [PubMed] [Google Scholar]
- Garg, P., Nandy, R. K., Chaudhury, P., Chowdhury, N. R., De, K., Ramamurthy, T., Yamasaki, S., Bhattacharya, S. K., Takeda, Y. & Nair, G. B. (2000). Emergence of Vibrio cholerae O1 biotype El Tor serotype Inaba from the prevailing O1 Ogawa serotype strains in India. J Clin Microbiol 38, 4249–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, R. K., Szu, S. C., Finkelstein, R. A. & Robbins, J. B. (1992). Synthesis, characterization, and some immunological properties of conjugates composed of the detoxified lipopolysaccharide of Vibrio cholerae O1 serotype Inaba bound to cholera toxin. Infect Immun 60, 3201–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, R. K., Taylor, D. N., Bryla, D. A., Robbins, J. B. & Szu, S. C. (1998). Phase 1 evaluation of Vibrio cholerae O1, serotype Inaba, polysaccharide-cholera toxin conjugates in adult volunteers. Infect Immun 66, 3095–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson, B. (1984). Monoclonal antibody-based enzyme-linked immunosorbent assays for identification and serotyping of Vibrio cholerae O1. J Clin Microbiol 20, 1180–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson, B. & Holme, T. (1983). Monoclonal antibodies against group- and type-specific lipopolysaccharide antigens of Vibrio cholerae O : 1. J Clin Microbiol 18, 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson, B., Rosen, A. & Holme, T. (1982). Monoclonal antibodies against Vibrio cholerae lipopolysaccharide. Infect Immun 38, 449–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, D. R., Ford, L. & Lalloo, D. G. (2006). Oral cholera vaccines: use in clinical practice. Lancet Infect Dis 6, 361–373. [DOI] [PubMed] [Google Scholar]
- Lauvrak, V., Berntzen, G., Heggelund, U., Herstad, T. K., Sandin, R. H., Dalseg, R., Rosenqvist, E., Sandlie, I. & Michaelsen, T. E. (2004). Selection and characterization of cyclic peptides that bind to a monoclonal antibody against meningococcal L3,7,9 lipopolysaccharides. Scand J Immunol 59, 373–384. [DOI] [PubMed] [Google Scholar]
- Lesinski, G. B., Smithson, S. L., Srivastava, N., Chen, D., Widera, G. & Westerink, M. A. (2001). A DNA vaccine encoding a peptide mimic of Streptococcus pneumoniae serotype 4 capsular polysaccharide induces specific anti-carbohydrate antibodies in Balb/c mice. Vaccine 19, 1717–1726. [DOI] [PubMed] [Google Scholar]
- Lullau, E., Heyse, S., Vogel, H., Marison, I., von Stockar, U., Kraehenbuhl, J. P. & Corthesy, B. (1996). Antigen binding properties of purified immunoglobulin A and reconstituted secretory immunoglobulin A antibodies. J Biol Chem 271, 16300–16309. [DOI] [PubMed] [Google Scholar]
- Luo, P., Canziani, G., Cunto-Amesty, G. & Kieber-Emmons, T. (2000). A molecular basis for functional peptide mimicry of a carbohydrate antigen. J Biol Chem 275, 16146–16154. [DOI] [PubMed] [Google Scholar]
- Meeks, M. D., Saksena, R., Ma, X., Wade, T. K., Taylor, R. K., Kovac, P. & Wade, W. F. (2004). Synthetic fragments of Vibrio cholerae O1 Inaba O-specific polysaccharide bound to a protein carrier are immunogenic in mice but do not induce protective antibodies. Infect Immun 72, 4090–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, V. L., DiRita, V. J. & Mekalanos, J. J. (1989). Identification of toxS, a regulatory gene whose product enhances toxR-mediated activation of the cholera toxin promoter. J Bacteriol 171, 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moe, G. R., Tan, S. & Granoff, D. M. (1999). Molecular mimetics of polysaccharide epitopes as vaccine candidates for prevention of Neisseria meningitidis serogroup B disease. FEMS Immunol Med Microbiol 26, 209–226. [DOI] [PubMed] [Google Scholar]
- Monzavi-Karbassi, B., Hennings, L. J., Artaud, C., Liu, T., Jousheghany, F., Pashov, A., Murali, R., Hutchins, L. F. & Kieber-Emmons, T. (2007). Preclinical studies of carbohydrate mimetic peptide vaccines for breast cancer and melanoma. Vaccine 25, 3022–3031. [DOI] [PubMed] [Google Scholar]
- Phalipon, A., Folgori, A., Arondel, J., Sgaramella, G., Fortugno, P., Cortese, R., Sansonetti, P. J. & Felici, F. (1997). Induction of anti-carbohydrate antibodies by phage library-selected peptide mimics. Eur J Immunol 27, 2620–2625. [DOI] [PubMed] [Google Scholar]
- Pincus, S. H., Smith, M. J., Jennings, H. J., Burritt, J. B. & Glee, P. M. (1998). Peptides that mimic the group B streptococcal type III capsular polysaccharide antigen. J Immunol 160, 293–298. [PubMed] [Google Scholar]
- Qadri, F., Chowdhury, M. I., Faruque, S. M., Salam, M. A., Ahmed, T., Begum, Y. A., Saha, A., Alam, M. S. & Zaman, K. (2005). Randomized, controlled study of the safety and immunogenicity of Peru-15, a live attenuated oral vaccine candidate for cholera, in adult volunteers in Bangladesh. J Infect Dis 192, 573–579. [DOI] [PubMed] [Google Scholar]
- Ramamurthy, T., Garg, S. & Nair, G. B. (1995). Monoclonal antibodies against Ogawa specific and Ogawa-Inaba common antigenic determinants of Vibrio cholerae O1 and their diagnostic utility. Indian J Med Res 101, 10–12. [PubMed] [Google Scholar]
- Reidl, J. & Klose, K. E. (2002). Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol Rev 26, 125–139. [DOI] [PubMed] [Google Scholar]
- Ryan, E. T., Calderwood, S. B. & Qadri, F. (2006). Live attenuated oral cholera vaccines. Expert Rev Vaccines 5, 483–494. [DOI] [PubMed] [Google Scholar]
- Shin, J. S., Yu, J., Lin, J., Zhong, L., Bren, K. L. & Nahm, M. H. (2002). Peptide mimotopes of pneumococcal capsular polysaccharide of 6B serotype: a peptide mimotope can bind to two unrelated antibodies. J Immunol 168, 6273–6278. [DOI] [PubMed] [Google Scholar]
- Sugiyama, J., Gondaira, F., Matsuda, J., Soga, M. & Terada, Y. (1987). New method for serological typing of Vibrio cholerae 0:1 [corrected] using a monoclonal antibody-sensitized latex agglutination test. Microbiol Immunol 31, 387–391. [DOI] [PubMed] [Google Scholar]
- Sun, D. X., Mekalanos, J. J. & Taylor, R. K. (1990). Antibodies directed against the toxin-coregulated pilus isolated from Vibrio cholerae provide protection in the infant mouse experimental cholera model. J Infect Dis 161, 1231–1236. [DOI] [PubMed] [Google Scholar]
- Taylor, R. K., Kirn, T. J., Bose, N., Stonehouse, E., Tripathi, S. A., Kovac, P. & Wade, W. F. (2004). Progress towards development of a cholera subunit vaccine. Chem Biodivers 1, 1036–1057. [DOI] [PubMed] [Google Scholar]
- Tiwana, H., Clow, K. J., Hall, C., Feavers, I. M. & Charalambous, B. M. (2005). The immunogenicity of a conformationally restricted peptide mimetic of meningococcal lipooligosaccharide. Scand J Immunol 62, 385–392. [DOI] [PubMed] [Google Scholar]
- Valadon, P., Nussbaum, G., Boyd, L. F., Margulies, D. H. & Scharff, M. D. (1996). Peptide libraries define the fine specificity of anti-polysaccharide antibodies to Cryptococcus neoformans. J Mol Biol 261, 11–22. [DOI] [PubMed] [Google Scholar]
- Valadon, P., Nussbaum, G., Oh, J. & Scharff, M. D. (1998). Aspects of antigen mimicry revealed by immunization with a peptide mimetic of Cryptococcus neoformans polysaccharide. J Immunol 161, 1829–1836. [PubMed] [Google Scholar]
- Villeneuve, S., Boutonnier, A., Mulard, L. A. & Fournier, J. M. (1999). Immunochemical characterization of an Ogawa-Inaba common antigenic determinant of Vibrio cholerae O1. Microbiology 145, 2477–2484. [DOI] [PubMed] [Google Scholar]
- Wang, J., Villeneuve, S., Zhang, J., Lei, P., Miller, C. E., Lafaye, P., Nato, F., Szu, S. C., Karpas, A. & other authors (1998). On the antigenic determinants of the lipopolysaccharides of Vibrio cholerae O : 1, serotypes Ogawa and Inaba. J Biol Chem 273, 2777–2783. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Raifu, M., Howard, M., Smith, L., Hansen, D., Goldsby, R. & Ratner, D. (2000). Universal PCR amplification of mouse immunoglobulin gene variable regions: the design of degenerate primers and an assessment of the effect of DNA polymerase 3′ to 5′ exonuclease activity. J Immunol Methods 233, 167–177. [DOI] [PubMed] [Google Scholar]
- WHO (2001). Weekly epidemiological record. Bulletin 76, 117–124. [Google Scholar]
- Winner, L., III, Mack, J., Weltzin, R., Mekalanos, J. J., Kraehenbuhl, J. P. & Neutra, M. R. (1991). New model for analysis of mucosal immunity: intestinal secretion of specific monoclonal immunoglobulin A from hybridoma tumors protects against Vibrio cholerae infection. Infect Immun 59, 977–982. [DOI] [PMC free article] [PubMed] [Google Scholar]